Abstract
Klebsiella is a common dangerous pathogen for humans and animals and is widely present in the digestive system. The genus Klebsiella is ubiquitous, as it is endemic to surface water, soil, and sewage. In this study, 70 samples were obtained from soil-dwelling invertebrates from September 2021 to March 2022 from Taif and Shafa in different altitudinal regions of Saudi Arabia. Fifteen of these samples were identified as Klebsiella spp. The Klebsiella isolates were genetically identified as Klebsiella pneumoniae using rDNA sequencing. The antimicrobial susceptibility of the Klebsiella isolates was determined. Amplification of virulence genes was performed using PCR. In this study, 16S rDNA sequencing showed a similarity from 98% to 100% with related K. pneumonia from the NCBI database, and the sequences were deposited in the NCBI GenBank under accession numbers ON077036 to ON077050. The growth inhibition properties of ethanolic and methanolic extracts of the medicinal plant Rhazya stricta’s leaves against K. pneumoniae strains using the minimum inhibitory concentration (MIC) method and disc diffusion were evaluated. In addition, the biofilm inhibitory potential of these extracts was investigated using crystal violet. HPLC analysis identified 19 components divided into 6 flavonoids, 11 phenolic acids, stilbene (resveratrol), and quinol, and revealed variations in the number of components and their quantities between extracts. Both extracts demonstrated interesting antibacterial properties against K. pneumoniae isolates. The 2 extracts also showed strong biofilm inhibitory activities, with percentages of inhibition extending from 81.5% to 98.7% and from 35.1% to 85.8% for the ethanolic and methanolic extracts, respectively. Rhazya stricta leaf extract revealed powerful antibacterial and antibiofilm activities against K. pneumoniae isolates and could be a good candidate for the treatment or prevention of K. pneumonia-related infections.
1. Introduction
The gastrointestinal tract of invertebrates is an ideal location for microflora; the stomach is rich in bacteria, while the proctodaeal region is rich in fungi [1]. From the hindgut of Glomeris species, six bacteria, six actinomycetes, and two fungal strains were isolated [2]. Pseudomonas stutzeri and Pseudomonas putida survive passage through the gut of a millipede, Pachyiulus flavipes, and increase fresh excrement [3]. Bacteria recovered from the gut of millipedes (Ommatoiulus sabulosus) include Klebsiella, Bacillus, and Corynebacterium species, while actinomycetes (such as Micromonospora sp.) are known to accumulate in the hindgut and are likely to participate in the breakdown of chitin [4]. Fecal pellets consist of dense populations of micro-organisms [2]. The genus Klebsiella, a severe opportunistic pathogen belonging to the family Enterobacteriaceae, is a major pathogen associated with urinary, respiratory, gastrointestinal, and skin infections in humans [5]. Klebsiella pneumoniae control is impaired by the frequent multidrug-resistant phenotype and genotype, representing a major threat to neonates, the elderly, and immuno-compromised patients [6,7]. Klebsiella is ubiquitous in terms of habitat association. It is also a resident or transient flora, particularly in the gastrointestinal tract of some invertebrates [2]. In addition, Klebsiella species frequently acquire antibiotic resistance genes for all classes of antibiotics and are considered the first microorganism to help in spreading resistance and virulence genes [8,9,10]. Similar to other opportunistic pathogens, K. pneumoniae is a ubiquitous bacterium that thrives in environmental compartments (e.g., soil, plants, and waterways) [11,12]. While sometimes K. pneumoniae bacteria are present in human and animal waste sources and, therefore, can be considered environmental pollutants, at other times, these K. pneumoniae strains are environmental strains that appear in their natural habitat [12,13]. Water, vegetation, and soil have been described as the native environments for K. pneumoniae [2]. Phenotypic and genotypic traits were compared between isolates of K. pneumoniae obtained from hospitals and those obtained from the natural environment [14]. K. pneumoniae is a prominent hospital-acquired pathogen, as well as a significant foodborne pathogen that may cause liver abscesses, pneumonia, septicemia, and diarrhea [15,16,17]. K. pneumoniae has been recognized as a major food-borne pathogen due to its prevalence outside of the medical environment, where it is often detected in cooked meals, raw vegetables, powdered infant formula, fish, meat, and street foods [18,19,20,21,22,23]. It is important to investigate the common and distinct genomic traits of clinical and environmental strains of K. pneumoniae [24]. Although the distinction between clinical strains and environmental strains is difficult, the characterization of strains from both origins is crucial to assess the harmful effect of both types and to study the evolution and acquisition of new genetic traits from one source to the other, as well as to infer pathways of transmission from the environment to humans [14].
K. pneumoniae isolates were subjected to a medicinal plant extract, Rhazya stricta, which is an economically important medicinal plant. In Saudi Arabia and many Asian countries, R. stricta and its metabolites are traditionally used for the treatment of cancer, skin diseases, hypertension, rheumatism, sore throat, syphilis, parasitic infections, inflammatory conditions, and fever [25,26]. Various parts of R. stricta contain many phytochemical constituents, such as alkaloids, flavonoids, triterpenes, and volatile bases [25,27], which display potential antimicrobial and biological activities [25]. Furthermore, leaf and fruit extracts of R. stricta have shown antimicrobial properties against many multidrug-resistant human pathogens [26,28]. However, the antibiofilm activity of R. stricta has not been explored.
In the present study, K. pneumoniae strains were isolated from invertebrate animals collected from different regions of Taif in Saudi Arabia. Therefore, the main aim of this study was to classify and characterize K. pneumoniae isolates obtained from invertebrate animals, and to evaluate the antibacterial and antibiofilm properties of the phenolic components in R. stricta leaf ethanolic and methanolic extracts against K. pneumoniae isolates and their virulence gene profiles.
2. Results
2.1. Isolation and Identification of K. pneumoniae Isolates
2.1.1. Isolation of K. pneumoniae Isolates
Fifteen isolates were obtained from different invertebrate animals (millipedes and isopods) collected from the Taif governorate and identified as K. pneumoniae. The location and invertebrates are presented in Table 1. Nine K. pneumoniae isolates were obtained from millipede guts, three of which (KTU-10, KTU-11, and KTU-12) were collected from Wady Ghazal, Taif, and six (KTU-1, KTU-2, KTU-3, KTU-13, KTU-14, and KTU-15) from Al-Shafa, Taif. Six K. pneumoniae isolates (KTU-4, KTU-5, KTU-6, KTU-7, KTU-8, and KTU-9) were isolated from the gut of an isopod, Porcellio laevis, collected from the Taif University Garden in Hawia, Taif, Saudi Arabia.

Table 1.
Source and locations of Klebsiella pneumoniae that were isolated from some invertebrates in Taif, Saudi Arabia.
2.1.2. Molecular Genotyping of K. pneumoniae
The 16S rRNA gene of all K. pneumoniae isolates was amplified and sequenced, and specific fragments were aligned and compared with the available 16S rRNA sequences for other K. pneumoniae isolates in the NCBI database. The sequences of the K. pneumoniae isolates were deposited in the NCBI GenBank under accession numbers ON077036 to ON077050. The BLAST results showed that the partial 16S rRNA sequences were more similar to other sequences from the NCBI database. The similarity matrix among the K. pneumoniae isolates and related strains from the NCBI database ranged from 98 to 100%. For example, the K. pneumoniae KTU-11 isolate with accession number ON077046 has low similarity to K. pneumoniae strains. The K. pneumoniae KTU-1 isolate with accession number ON077036 is moderately similar to the K. pneumoniae strain MT-379622 and the K. pneumoniae strain MN-314311. The K. pneumoniae KTU-15 isolate with accession number ON077050 has high similarity to the K. pneumoniae strain MN749610, with approximately 100% similarity (Table 2, Figure 1).

Table 2.
NCBI BLAST query for Klebsiella pneumoniae isolated from invertebrates in Taif, Saudi Arabia.
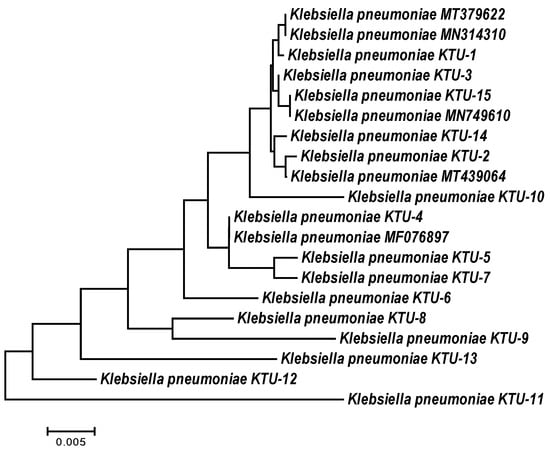
Figure 1.
Neighbor-joining phylogeny tree based on 16S rDNA gene sequences of Klebsiella pneumoniae isolates collected from some invertebrates in Taif, Saudi Arabia, with 1000 bootstraps.
2.1.3. Antimicrobial Susceptibility
Klebsiella pneumoniae was tested for antimicrobial susceptibility to 12 types of antibiotics. The overall susceptibility, intermediate susceptibility, and resistance values were determined (Table 3). Most K. pneumoniae strains showed a high percentage of resistance to carbecillin (100%), oxacillin (100%), cefoxitin (100%), amoxicillin (100%), and penicillin (93.3%). Erythromycin (80%), amkacillin (53.3%), ampicillin (40%), and cefrizine (40%) indicated moderate susceptibility. On the other hand, intermediate resistance was found to sulfamethoxazole/Trimethoprim (26.7%). Moreover, all the K. pneumoniae isolates were sensitive to ciprofloxacin and gentamicin.

Table 3.
Antibiotic resistance profile of Klebsiella pneumoniae isolates.
2.1.4. Detection of Virulence Genes in K. pneumoniae
The existence of antibiotic-resistant genes is shown in Figure 2 and Table 4. The virulence genes AcrAB, mdtk, OmpK35, FimH, and RmpA were recorded in all K. pneumoniae isolates (Table 4). The K1 gene, which is responsible for the formation of capsule and K genotypes, was found in only 3 isolates of K. pneumoniae, KTU-7, KTU-8, and KTU-11, representing 15% of the isolates. The K. pneumoniae KTU-8 and KTU-10 isolates have the most investigated virulence genes. OmpK35 plays a role in K. pneumoniae infection and virulence. The Aea gene was found in all K. pneumoniae isolates, except KTU-5, KTU-8, KTU-9, and KTU-11. TolC was also found in all K. pneumoniae isolates, except K. pneumoniae KTU-9 and KTU-11. Moreover, the SHV and TEM genes were found in all K. pneumoniae isolates, whereas the CTX gene was found in two isolates, K. pneumoniae KTU-8 and KTU-10.
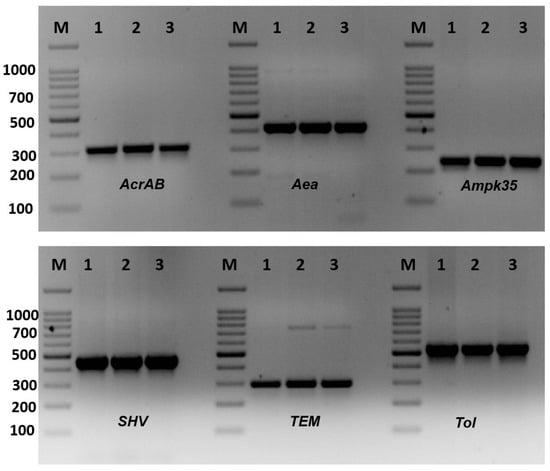
Figure 2.
Amplification of virulence genes of Klebsiella pneumoniae isolates by single PCR. Amplification of AcrAB gene (312 bp), amplification of Aea gene (410 bp), amplification of Ompk35 gene (241 bp), amplification of SHV gene (436 bp), amplification of TEM gene (295), and amplification of Tol genes (527). M: 100 bp DNA ladder, 1–3: some positive isolates.

Table 4.
Virulence genes (AcrAB, TolC, mdtk, Ompk35, FimH, RmpA, Aea, SHVM, TEM, K1, and K2) in Klebsiella pneumoniae isolates.
2.2. The Potential of R. stricta Extract against K. pneumoniae
2.2.1. Chemical Composition of R. stricta Leaf Extracts
The chemical composition of the ethanolic and methanolic extracts of R. stricta are listed in Table 5. Nineteen components were obtained from the HPLC analysis of these extracts; they were divided into six flavonoids, eleven phenolic acids, stilbene (resveratrol), and quinol. In total, 17 compounds were detected in each extract, with a quantity of 15,292.89 mg/kg and 33,050.65 mg/kg for the ethanolic and methanolic extracts, respectively, indicating that the methanolic extract is richer in phenolic compounds than the ethanolic extract.

Table 5.
Chemical compositions of R. stricta extracts (mg/kg).
The results revealed variations between the two extracts in terms of the number and quantity of components. The major compounds of R. stricta ethanolic extract are Quinol, Resveratrol, p-Coumaric acid, Benzoic acid, Rutin, Quercitin, Myricetin, and Kaempferol. However, the major compounds of methanolic extract are Resveratrol, Benzoic acid, Ferulic acid, Rutin, Quercitin, Neringein, and Kaempferol. Both R. stricta extracts are rich in flavonoids (11,104.27 mg/kg and 21,357.93 mg/kg for the ethanolic and methanolic extracts, respectively) compared to other phenolic compounds (4188.62 mg/kg and 11,692.72 mg/kg for the ethanolic and methanolic extracts, respectively).
2.2.2. Antibacterial Activity of R. stricta Extracts against K. pneumoniae
Disc Diffusion
The ethanolic and methanolic extracts of R. stricta leaves were examined for their antimicrobial activity against K. pneumoniae isolates (Table 6). First, the disc diffusion method showed that the two extracts were active against all isolates, despite the variation in the type of inhibitory action. R. stricta ethanolic extract demonstrated strong inhibitory activity against 40% of the strains, compared to the methanolic extract, which showed a strong inhibitory action on 33.3% of the isolates. As shown in Table 6, the ethanolic extract was slightly more effective than the methanolic extract against K. pneumoniae isolates.

Table 6.
Antimicrobial activity of Rhazya stricta leaves extract against Klebsiella isolates.
Determination of (MIC) and (MBC)
The antimicrobial activities of the ethanolic and methanolic extracts of R. stricta leaves were also investigated using MIC and MBC for the 15 K. pneumoniae isolates. For the ethanolic extract, the MIC ranged from 0.122 to 0.970 mg/mL, whereas the MBC ranged from 0.224 mg/mL to 1.9 mg/mL. For the methanolic extract of R. stricta leaves, the MIC values varied from 0.224 mg/mL to 1.9 mg/mL, while the MBC values ranged from 0.448 mg/mL to 3.9 mg/mL. Accordingly, the ethanolic extract had the greatest antibacterial activity against K. pneumoniae isolates compared with the methanolic extract.
2.3. Biofilm Formation and Inhibition
2.3.1. Biofilm Formation on Polystyrene Surface
The bacterial isolates were inspected for their ability to produce biofilms on polystyrene surfaces (Table 7). The results showed that all K. pneumoniae strains were capable of producing biofilms and were allocated as follows: 26.7% were highly positive biofilm producers, with OD570 values varying from 1.015 to 1.060, and 73.3% were low-grade positive, with OD570 values ranging from 0.442 to 0.808.

Table 7.
Antibiofilm potentialities of ethanolic and methanolic Rhazya stricta leaf extracts against Klebsiella isolates.
2.3.2. Biofilm Inhibition
The ability of R. stricta ethanolic and methanolic extracts to inhibit biofilm formation by K. pneumoniae isolates is shown in Table 7. Isolates showing potential for biofilm formation were selected for this experiment. Fifteen strains were classified as low-grade and highly positive biofilms, and both extracts demonstrated strong biofilm inhibition activity.
2.3.3. Antibiofilm Activity
The present investigation revealed that the ethanolic extract of R. stricta leaves has strong biofilm inhibition activity on all the isolates (15 strains), with the percentage of inhibition extending from 81.5% to 98.7%. Overall, 4 out of 5 highly positive isolates (80%) were biofilm-negative. In addition, 10 low-grade positive isolates (75%) changed to biofilm-negative after treatment.
Biofilm inhibitory activities were also observed for the methanolic extract, with most isolates ranging from 35.1% to 85.8%. Furthermore, the same results observed for the four highly positive biofilm isolates treated with the ethanolic extract were obtained after treatment with the methanolic extract. However, 4 low-grade positive isolates (26.7%) were biofilm-negative. Table 7 shows that isolate No. 10 conserved its initial biofilm phenotype after treatment with the 2 extracts, despite the large decrease in the amount of biofilm; however, the methanolic extract did not affect the ability of isolate No. 1 to form a biofilm. No significant correlation was detected between the MIC and antibiofilm activity for either the methanolic or the ethanolic extract of R. stricta leaves.
3. Discussion
Recently, 16S rRNA gene sequencing has been used as an alternative method for the molecular detection of various microbes, including K. pneumoniae [4]. This gene is found in all bacteria and, hence, ensures the accurate identification of bacteria at the genus and species levels [29]. Thus, sequencing can be reasonably applied to the preparation of many microbes, especially those isolated from the external environment or from other animals. In the present study, 16S rRNA gene sequencing displayed similarities between K. pneumoniae isolated from invertebrates and those obtained from GenBank, indicating that sequencing has the potential to be more sensitive than culture-dependent morphological and microscopic identification [30].
Klebsiella pneumoniae is a public health problem worldwide. This bacterium is the most prominent antibiotic-resistant acquired pathogen. Infections can spread from person to person through the respiratory system, the environment, or by using contaminated medical equipment [4]. Therefore, the discovery of new therapeutic agents, especially natural products, against K. pneumoniae is highly important.
Currently, plant compounds have emerged as potential candidates, given the interest of scientists to search for antimicrobial and antibiofilm drugs. Among these, R. stricta has gained attention because of its medicinal uses [25]. In this study, the potential antibacterial properties of ethanolic and methanolic extracts of R. stricta against K. pneumoniae isolated from invertebrates were investigated. The isolates were investigated by growth inhibition assays. Experiments showed that the extracts of R. stricta leaves have strong antibacterial activity [27,31].
The high ability of R. stricta leaf extracts to prevent the growth and multiplication of this bacterium, observed in this study, may be attributed to the phenols and flavonoid compounds found in these extracts [32,33]. It has been shown that flavonoids, such as quercetin [32], kaempferol, and catechin [26], exhibit great growth inhibition activity against K. pneumoniae isolates. Flavonoids, which are the major components of these extracts, are responsible for the inhibition of nucleic acid synthesis [26], damage to the cytoplasmic membrane through the alteration of its function [32,33], inhibition of energy metabolism by the alteration of the cytoplasmic membrane, and inhibition of the energy supply for bacteria [26]. In addition, the inhibition of cell membrane synthesis and the aggregatory effect on whole bacterial cells have also been reported [31]. Several studies have demonstrated the antibacterial properties of phenolic acids, especially caffeic acid, ferulic acid, coumaric acid, and chlorogenic acid, which have antibacterial activities [34,35]. Phenolic acids damage the K. pneumoniae cell wall, leading to cytoplasmic leakage and changes in bacterial cell morphology [26,34,35]. Moreover, the high K. pneumoniae growth inhibition activity seems to be due to the synergetic effect of flavonoids and other phenolic compounds present in the R. stricta leaf extracts.
In the present study, the ethanolic extract of R. stricta leaves was more effective against K. pneumoniae isolates than the methanolic extract, despite its lower abundance of flavonoids and phenols. This can be attributed to quinol and chlorogenic acid, which do not exist in the methanolic extract, and/or to myricetin and p-Coumaric acid, which are more abundant in the ethanolic extract. Accordingly, Xie et al. [36] reported that myricetin displayed the most significant antimicrobial activity of all the flavonoids and exhibited extensive activity against K. pneumoniae and many other pathogenic bacteria [26,37]. Furthermore, myricetin inhibits Escherichia coli DnaB helicase, which plays a major role in DNA replication and elongation [38]. In addition, p-Coumaric acid effectively inhibited the growth of K. pneumoniae and other pathogenic bacteria. p-Coumaric acid is responsible for the disruption of bacterial cell membranes and the inhibition of cellular functions by binding to bacterial genomic DNA [38]. Ma et al. [39] reported that quinol exhibited relatively strong antibacterial activity against K. pneumoniae by destroying the bacterial cell membrane and cell wall, increasing permeability, and influencing the expression of genes. However, chlorogenic acid does not show significant antibacterial activity [26].
Klebsiella pneumoniae isolates were examined for their ability to develop biofilms on polystyrene surfaces, and the experiment demonstrated that 23.33% of the isolates were strong biofilm producers, while 50% were low-grade positive producers. This finding demonstrates the high potential of K. pneumoniae strains to produce biofilms, confirming that K. pneumoniae is the most prevalent bacterium in biofilm-associated infections [40]. Biofilm, as an important virulence factor, is responsible for more than 65% of nosocomial infections and 80% of microbial infections [41]. Biofilms are associated with nasal colonization of the respiratory system, endocarditis, soft tissue infections, urinary tract infections, and other diseases [4]. Biofilms are also a severe issue in the field of urology because they are responsible for the persistence of bacteria in the genitourinary tract over the long term [37]. K. pneumoniae biofilms have been associated with medical equipment and chronic infections, and the presence of biofilms makes bacteria more resistant to antibiotics and phagocytosis, making their treatment more difficult [37]. Therefore, the discovery of novel therapeutic strategies for biofilm inhibition is important. Extracts of R. stricta leaves were tested for their ability to inhibit biofilm formation by K. pneumoniae. Antibiofilm examination showed that both plant extracts displayed strong biofilm inhibitory activity, with a 98.7% reduction in the amount of biofilm produced. This activity is largely due to flavonoids as a major component, in addition to other phenolic compounds found in the extracts. This result emphasizes the findings of Nielsen et al. [37], who reported that flavonoids are responsible for the reduction of bacterial adhesion, biofilm formation, and the inhibition of quorum sensing (cell-to-cell communication system in the biofilm formation signal receptors TraR and RhlR). Furthermore, a decrease in the amount of biofilm could be considered a reduction in K. pneumoniae virulence, which is in agreement with Saadatian et al. [42], who mentioned that flavonoids inhibit bacterial virulence factors. Moreover, Xie et al. [33] showed that flavonoids, such as quercetin, kaempferol, naringenin, and apigenin, suppress the activity of autoinducer-2, which is responsible for cell-to-cell communication and, consequently, reduces biofilm synthesis. In this study, the ethanolic extract also displayed biofilm inhibitory properties more than the methanolic extract, in addition to its growth inhibition activity, indicating that the components involved in growth inhibition are the same as those associated with biofilm inhibition, and that myricetin inhibits biofilm formation by K. pneumoniae [36]. Additionally, Saadatian et al. [42] revealed that flavonoids efficiently inhibited the bacterial biofilm matrix by targeting Bap-like amyloids. Myricetin also inhibits curli-dependent biofilm formation in E. coli [37].
Deletion of OmpK36 or OmpK35/OmpK36 can reduce the virulence of highly contagious K. pneumoniae strains and increase their susceptibility to neutrophil phagocytosis [43]. In our study, OmpK35 porin-coding genes were simultaneously detected in all K. pneumoniae isolates. A direct correlation between efflux pumps and the virulence of pathogenic bacteria was reported by Padilla et al. [44]. Most strains of intestinal bacteria contain genes that encode iron absorption systems, such as enterochelin or aerobactin. Iron plays an important role, as it can inhibit T-cell proliferation, in addition to promoting iron absorption. The rmpA, wabG, uge, Ycfm, fimh1, EntB, Ybt-irp2, and kfu genes have been reported in most antibiotic-resistant K. pneumoniae isolates [43]. The most pathogenic genes lead to high-pathogenicity strains that contain virulence genes prevalent in Klebsiella species [5]. ESBLs are now found in all Enterobacteriaceae species worldwide [45]. The ESBL genes TEM and SHV were found in all K. pneumoniae isolates in this investigation, and only three of them harbored the CTX gene. The number of CTX-M-producing K. pneumoniae strains has also increased [5,45].
4. Materials and Methods
4.1. Isolation and Identification of K. pneumoniae Strains
4.1.1. Isolation of K. pneumoniae Strains
Seventy samples were isolated from soil-dwelling invertebrates between September 2021 and March 2022. Digestive tracts were obtained from Archispirostreptus syriacus (millipede), Porcellio laevis (swift woodlouse), and Armadillidium sp. (isopods). The bacterial isolates were obtained using the dilution method, where gut contents were diluted and spread on nutrient agar media and incubated for 24 h at 37 °C. Morphologically identified Klebsiella isolates were also genetically identified using 16S rDNA sequencing.
4.1.2. Application of 16S rDNA Gene Sequencing
Genomic DNA was isolated from all K. pneumoniae isolates using a DNA extraction kit (Gena Bioscience, Jena, Germany), according to the manufacturer’s instructions. One fragment of the DNA (approximately 1465 bp) was amplified from the 16S rDNA gene [30]. The pieces were punctuated using a QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced using a DNA Analyzer 3146 Applied Bioscience (Applied Biosystems, Waltham, MA, USA). The sequencing texts were edited and compiled using the DNASTAR software (Laser gene 17.3, Madison, WI, USA). BLAST searches were performed using the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi accessed on 7 March 2023).
4.1.3. Antimicrobial Susceptibility Test
The antibiotic sensitivity of K. pneumoniae strains was studied using the disc diffusion method, according to Clinical and Laboratory Standards Institute (CLSI) guidelines [46]. This study was carried out using 12 commercially available antibiotics: sulfamethoxazole/trimethoprim (25 µg), ampicillin (10 µg), carbecillin (100 µg), amkacillin (30 µg), cefatrizine (10 µg), oxacillin (5 µg), penicillin (10 µg), ciprofloxacin (5 µg), gentamicin (10 µg), cefoxitin (30 µg), erythromycin (15 µg), and amoxicillin/clavulanic acid (30 µg).
4.1.4. Detection of Virulence and Antibiotic Resistance Genes of K. pneumoniae
Twelve PCR reactions were performed to detect the presence of virulence genes (acrAB, tolC, mdtk, Ompk35, fimH, rmpA, aea, k1, and k2) in K. pneumoniae isolates [5], and antibiotic resistance genes, primer sequences, amplification conditions, and amplicon sizes were used as explained [20]. PCR was performed using the GoTaq® Green Master Mix (Promega, Maddison, WI, USA). The expected sizes of the amplicons were ascertained by electrophoresis on 1.5% agarose gel with an appropriate molecular size marker (100 bp DNA ladder, MBI, Fermentas, LT, USA).
4.2. Leaf Extraction of R. stricta, HPLC Analysis, and Antibacterial Activity
4.2.1. R. stricta Leaves Collection and Extraction Procedure
Fresh leaves of R. stricta were collected in September 2021 from their natural habitat at Taif-Makkah Road. The plant’s fresh leaves were air dried and ground into fine powder, then extracted using 100 mL of 95% ethanol and methanol separately at room temperature for 3 days. Each extract was centrifuged at 7000 rpm for 15 min and filtered 3 times with Whatman filter paper No. 1 to obtain a pure filtrate. The filtrate was passed through a Buchner funnel using a rotary vacuum evaporator (Dai-Han Inc., Seoul, Republic of Korea) at 30 °C, then the extracts (pellets) were dissolved in an aqueous solution of dimethylsulfoxide 1% (DMSO) [47]. The extracts were subjected to HPLC analysis to separate their components.
4.2.2. HPLC (High Performance Liquid Chromatography) Analysis
Phenolic compounds were detected in the tested extracts as previously described [47], with fine modifications, using an Agilent 1260 infinity HPLC Series (Agilent, Santa Clara, CA, USA) equipped with a quaternary pump. Kinetex® 5 µm EVO C18 100 mm × 4.6 mm (Phenomenex, Torrance, CA, USA) was used as the column and operated at 30 °C. The separation was carried out using a ternary linear elution gradient with (A) HPLC grade water with 0.2% and H3PO4 (v/v), (B) methanol, and (C) acetonitrile. Subsequently, 20 µL of the extract was injected. The AVWD detector (Agilent, Santa Clara, CA, USA) was set at 284 nm to detect phenols and flavonoids.
4.2.3. Antibacterial Activity of R. stricta Extracts
Disc Diffusion
The antibacterial properties of the R. stricta leaf extracts were assessed in triplicate using the agar disc diffusion method [23]. K. pneumoniae cells were allowed to grow for 24 h at 37 °C in a Mueller–Hinton liquid medium. The K. pneumoniae suspension was prepared in saline water, adjusted to 0.5 turbidity standards, and distributed in Mueller–Hinton agar (MHA, Oxoid, Basingstoke, UK). A sterile filter disc was impregnated with R. stricta leaf extract (10 μL/disc) placed on the agar surface. The MHA plates were kept for 2 h at 4 °C before their incubation at 37 °C for 24 h. The antimicrobial properties were categorized by measuring the zone of cell growth inhibition around the discs. The inhibitory activity was evaluated as previously described [26].
Determination of Minimal Inhibitory Concentrations (MICs) and Minimal Bactericidal Concentrations (MBCs)
MIC is the lowest concentration of the extract at which the growth of K. pneumoniae cells is inhibited. However, MBCs have the lowest concentrations of the extract that killed ≥ 99.9% of the initial K. pneumoniae cells. MIC and MBC were carried out 3 times using a 96-well microtiter plate (Nunc, Roskilde, Denmark) [29]. The K. pneumoniae suspension was prepared from an overnight culture diluted to 0.5 McFarland. Then, serial dilutions of both methanolic and ethanolic R. stricta leaf extracts were prepared in 5 mL of nutrient broth with concentrations extended from 0.012 to 50 mg/mL. Microtiter plates were prepared by placing 95 μL of nutrient broth and 5 μL of the K. pneumoniae inoculum in them, in addition to 100 μL of the respective dilutions of the extracts. The negative control contained 5 μL of bacterial inoculum and 195 μL of nutrient broth without the R. stricta extract. After incubation of the plates at 37 °C for 18–24 h, the MIC and MBC were determined [28]. MBC was determined by subculturing 20 μL of the clear wells of the MIC test on MHA.
4.3. Biofilm Formation and Inhibition
4.3.1. Biofilm Formation
The potential of K. pneumoniae strains to develop biofilms on U-bottomed, 96-well, microtiter polystyrene plates was tested using a crystal violet assay [28]. Briefly, K. pneumoniae cells were grown in a Trypticase Soy broth (TSB) media overnight at 37 °C. Then, 200 μL of the diluted culture (1:100) in TSB, supplemented with 2% (w/v) glucose, was transferred to a microtiter plate with wells containing sterile TSB as controls. After 24h of incubation at 37 °C, the cultures were removed, and the plates were washed 2 times with phosphate buffer saline and dried in an inverted position. Adherent cells were fixed with 95% ethanol and stained with 100 μL of 1% crystal violet (Merck, Lyon, France) for 5 min. The wells were then washed with 300 μL of sterile distilled water and left to dry in air. The biofilm produced was determined.
4.3.2. Biofilm Inhibition
R. stricta leaf methanolic and ethanolic extracts were tested for their ability to reduce the development of biofilms by K. pneumoniae isolates at MIC. In total, 100 µL of the extracts in TSB (2% glucose) were added to microtiter plate wells containing 100 μL of bacterial suspension (108 CFU/mL) in each well. The negative control wells contained tryptic TSB and sterile water. Biofilm formation was determined using the crystal violet assay [28]. The percentage biofilm inhibition was calculated [47].
% Inhibition = 100 − ((OD570 sample)/(OD570 control) × 100)
This analysis was repeated three times.
4.4. Statistical Analysis
Three replicates were used for each of the treatments, and in each replicate, at least four plants were used, and the significance of the difference between the mean values was calculated. One-way analysis of variance (ANOVA) was used to perform the analysis of all data, and the significance of the difference among the treatments was determined according to the least significant difference (LSD) [47].
5. Conclusions
Soil invertebrates are important organisms harboring a lot of internal microflora in their digestive tract that need to be intensively studied. They already have useful microflora for the soil, but they may harbor pathogenic bacteria through their feeding habits, which may be harmful for humans. Therefore, we used leaf extracts of the wild plant R. stricta against the pathogen K. pneumoniae. Strong biofilm inhibitory activity and interesting antibacterial characteristics were shown by the extracts against K. pneumoniae isolates. R. stricta leaf extracts may be useful for treating or preventing K. pneumonia infections.
Author Contributions
Conceptualization, M.M.H. (Mohamed M. Hassan), B.A., T.M., M.F.A., M.M.H. (Montaser M. Hassan) and A.A.M.; methodology, M.M.H. (Mohamed M. Hassan), B.A., T.M., M.F.A., M.M.H. (Mohamed M. Hassan), J.A.A.-O. and A.A.M.; software, M.M.H. (Mohamed M. Hassan) and M.M.E.; validation, M.M.H. (Mohamed M. Hassan), B.A., T.M., M.F.A., R.H.K., M.M.H. (Montaser M. Hassan), A.A.M. and M.M.E.; formal analysis, M.M.H. (Mohamed M. Hassan), J.A.A.-O. and M.M.E.; investigation, M.M.H. (Mohamed M. Hassan), B.A., T.M., M.F.A., M.M.H. (Mohamed M. Hassan) and A.A.M.; resources, M.M.H. (Mohamed M. Hassan) and M.M.E.; data curation, M.M.H. (Mohamed M. Hassan) and M.M.E.; writing—original draft preparation, M.M.H. (Mohamed M. Hassan), J.A.A.-O. and M.M.E.; writing—review and editing, M.M.H. (Mohamed M. Hassan) and M.M.E.; visualization, M.M.H. (Mohamed M. Hassan) and M.M.E.; supervision, M.M.H. (Mohamed M. Hassan); project administration, M.M.H. (Mohamed M. Hassan); funding acquisition, M.M.H. (Mohamed M. Hassan), B.A. and M.F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Dean of Scientific Research through the High-Altitude Research Centre at Taif University, Taif, Saudi Arabia, under project number 1-442-43.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors extend their appreciation to Taif University for funding the current work through the High-Altitude Research Centre, under project number 1-442-43.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sridhar, K.R.; Ashwini, K.M. Diversity, restoration and conservation of millipedes. In Biodiversity in India; Pullaiah, T., Ed.; Regency Publications: New Delhi, India, 2016; Volume 5, Chapter 1; pp. 1–38. [Google Scholar]
- Byzov, B.A.; Claus, H.; Tretyakova, E.B.; Zvyagintsev, D.G.; Filip, Z. Effects of soil invertebrates on the survival of some 28 genetically engineered bacteria in leaf litter and soil. Biol. Fertlity Soils 1996, 23, 221–228. [Google Scholar] [CrossRef]
- Zenova, G.M.; Babkina, N.I.; Polyanskaya, L.M.; Zvyagintsev, D.G. Actinomycetes in the intestinal tract of soil invertebrates fed with vermicompost or litter. Microbiology 1996, 65, 360–365. [Google Scholar]
- Alsanie, W.F. Molecular diversity and profile analysis of virulence-associated genes in some Klebsiella pneumoniae isolates. Pract. Lab. Med. 2020, 19, e00152. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, A.K.; Farag, M.M.; Abbadi, S.H.; Hassan, M.M.; Gaber, A.; Abdel-Moneima, A.S. Antibiotic resistance profile and random amplification typing of β-lactamase-producing Enterobacteriaceae from the local area of Al-Taif and nearby cities in Saudi Arabia. Asian Biomed. 2016, 10, 219–228. [Google Scholar]
- Ranjbar, R.; Fatahian Kelishadrokhi, A.; Chehelgerdi, M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect. Drug. Resist. 2019, 12, 603–611. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Li, Y.; Shen, Q.; Jiang, W.; Zhao, K.; He, Y.; Dai, P.; Nie, Z.; Xu, X.; et al. Diversity and frequency of resistance and virulence genes in blaKPC and blaNDM co-producing Klebsiella pneumoniae strains from China. Infect. Drug. Resist. 2019, 12, 2819–2826. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, K.; Chen, W.; Chen, J.; Zheng, J.; Liu, C.; Cheng, L.; Zhou, W.; Shen, H.; Cao, X. Epidemiological characteristics of carbapenem-resistant Enterobacteriaceae collected from 17 hospitals in Nanjing district of China. Antimicrob. Resist. Infect. Control 2020, 9, 15. [Google Scholar] [CrossRef]
- Bagley, S.T. Habitat association of Klebsiella species. Infect. Control 1985, 6, 52–58. [Google Scholar] [CrossRef]
- Yang, F.; Deng, B.; Liao, W.; Wang, P.; Chen, P.; Wei, J. High rate of multi resistant Klebsiella pneumoniae from human and animal origin. Infect. Drug. Resist. 2019, 12, 2729–2737. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Berglund, B.; Zou, H.; Zheng, B.; Börjesson, S.; Ji, X.; Ottoson, J.; Lundborg, C.S.; Li, X.; Nilsson, L.E. Characterization of clinically relevant strains of extended-spectrum β-Lactamase-Producing Klebsiella pneumoniae occurring in environmental sources in a rural area of China by using whole-genome sequencing. Front. Microbiol. 2019, 12, 211. [Google Scholar] [CrossRef]
- Rocha, J.; Ferreira, C.; Mil-Homens, D.; Busquets, A.; Fialho, A.M.; Henriques, I.; Gomila, M.; Manaia, C.M. Third generation cephalosporin-resistant Klebsiella pneumoniae thriving in patients and in wastewater: What do they have in common? BMC Genom. 2022, 23, 72. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Xu, W.Y. An investigation of food poisoning caused by Klebsiella pneumoniae. Chin. Pract. Med. 2013, 8, 275–276. [Google Scholar]
- Cao, X.; Xu, X.; Zhang, Z.; Han, S.; Chen, J.; Zhang, K. Molecular characterization of clinical multidrug-resistant Klebsiella pneumoniae isolates. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 16. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, S.; Zhan, L.; Jin, Y.; Duan, J.; Hao, Z.; Lv, J.; Qi, X.; Chen, L.; Kreiswirth, B.N.; et al. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front. Cell. Infect. Microbiol. 2017, 7, 24. [Google Scholar] [CrossRef]
- Haryani, Y.; Noorzaleha, A.S.; Fatimah, A.B.; Noorjahan, B.A.; Patrick, G.B.; Shamsinar, A.T.; Laila, R.A.S.; Son, R. Incidence of Klebsiella pneumoniae in street foods sold in Malaysia and their characterization by antibiotic resistance, plasmid profiling, and RAPD–PCR analysis. Food Control 2007, 18, 847–853. [Google Scholar] [CrossRef]
- Sun, F.; Wu, D.; Qiu, Z.; Jin, M.; Wang, X.; Li, J. Development of real time PCR systems based on SYBR Green fro specific detection and quantification of Klebsiella pneumoniae in infant formula. Food Control 2010, 21, 487–491. [Google Scholar] [CrossRef]
- Puspanadan, S.; Afsahhejri, L.; Loo, Y.Y.; Nillian, E.; Kuan, C.H.; Goh, S.G.; Chang, W.S.; Lye, Y.L.; John, Y.H.T.; Rukayadi, Y.; et al. Detection of Klebsiella pneumoniae in raw vegetables using most probable number-polymerase chain reaction (MPN-PCR). Int. Food Res. J. 2012, 19, 1757–1762. [Google Scholar]
- Overdevest, I.T.; Heck, M.; van der Zwaluw, K.; Huijsdens, X.; van Santen, M.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Savelkoul, P.; et al. Extended-spectrum β-lactamase producing Klebsiella spp. in chicken meat and humans: A comparison of typing methods. Clin. Microbiol. Infect. 2014, 20, 251–255. [Google Scholar] [CrossRef]
- Kim, H.S.; Chon, J.W.; Kim, Y.J.; Kim, D.H.; Kim, M.S.; Seo, K.H. Prevalence and characterization of extended-spectrum-β-lactamase-producing Escherichia coli, and Klebsiella pneumoniae, in ready-to-eat vegetables. Int. J. Food Microbiol. 2015, 207, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Dvis, G.S.; Price, L.B. Recent research examining links among Klebsiella pneumoniae from food, food animals, and human extraintestinal infections. Curr. Environ. Health Rep. 2016, 3, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Albeshri, A.; Baeshen, N.A.; Bouback, T.A.; Aljaddawi, A.A.A. Review of Rhazya stricta decne phytochemistry, bioactivities, pharmacological activities, toxicity, and folkloric medicinal uses. Plants 2021, 10, 2508. [Google Scholar] [CrossRef] [PubMed]
- El-Tarras, A.A.; El-Awady, A.M.; Hassan, M.M. Evaluation of the genetic effects of the in vitro antimicrobial activities of Rhazya stricta leaf extract using molecular techniques and scanning electron microscope. Afr. J. Biotech. 2013, 12, 3171–3180. [Google Scholar]
- Marwat, S.K.; Usman, K.; Shah, S.S.; Anwar, N.; Ullah, I. A review of phytochemistry, bioactivities and ethnomedicinal uses of Rhazya stricta Decsne (Apocynaceae). Afr. J. Microbiol. Res. 2012, 6, 1629–1641. [Google Scholar]
- Raziuddin, K.; Baeshen, M.N.; Kulvinder, S.S.; Roop, S.B.; Al-Hejin, A.; Nabih, A.B. Antibacterial activities of Rhazya stricta leaf extracts against multidrug-resistant human pathogens. Biotechnol. Biotechnol. Equip. 2018, 30, 1016–1025. [Google Scholar]
- Hassan, M.M.; Soliman, M.M.; Alotaibi, S.S.; Sayed, S.; El-Shehawi, A.M.; Ben-Abdallah, F. Ameliorative impacts of rough cocklebur leaf extracts against methicillin-resistant Staphylococcus aureus. Fres Env. Bull. 2022, 31, 6553–6560. [Google Scholar]
- Alsanie, W.F.; Felemban, E.M.; Farid, M.A.; Hassan, M.M.; Sabry, A.; Gaber, A. Molecular identification and phylogenetic analysis of multidrug-resistant bacteria using 16S rDNA sequencing. J. Pure Appl. Microbiol. 2018, 12, 489–496. [Google Scholar] [CrossRef]
- Beigomi, M.; Shahraki-Mojahed, L.; Heydari-Sadegh, B.; Dahmardeh, N.; Rouhani, R.; Javadian, F. Evaluation of antimicrobial activity of Rhazya stricta (Apocynaceae) extract prepared with different solvents on Staphylococcus aureus (Staphylococcaceae) isolated from humans. Int. J. Adv. Biol. Biomed. Res. 2021, 9, 241–253. [Google Scholar]
- Macé, S.; Hansen, L.; Rupasinghe, H. Anti-bacterial activity of phenolic compounds against Streptococcus pyogenes. Medicines 2017, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yang, W.; Tang, F.; Chen, X.; Ren, L. Antibacterial activities of flavonoids: Structure-activity relationship and mechanism. Cur. Med. Chem. 2021, 22, 132–149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, M.; Zhao, Z.; Yu, S. The antibiotic activity and mechanisms of sugarcane (Saccharum officinarum L.) bagasse extract against food-borne pathogens. Food Chem. 2015, 185, 112–118. [Google Scholar] [CrossRef]
- Lima, V.N.; Oliveira-Tintino, C.D.; Santos, E.S.; Morais, L.P.; Tintino, S.R.; Freitas, T.S.; Geraldo, Y.S.; Pereira, R.L.; Cruz, R.P.; Menezes, I.R. Antimicrobial and enhancement of the antibiotic activity by phenolic compounds: Gallic acid, caffeic acid and pyrogallol. Microb. Pathog. 2016, 99, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, J.; Xiao, A.; Liu, L. Antibacterial activity of polyphenols: Structure-activity relationship and influence of hyperglycemic condition. Molecules 2017, 22, 1913. [Google Scholar] [CrossRef]
- Nielsen, D.W.; Klimavicz, J.; Cavender, T.; Wannemuehler, Y.; Barbieri, N.L.; Nolan, L.K.; Logue, C.M. The impact of media, phylogenetic classification, and E. coli pathotypes on biofilm formation in extraintestinal and commensal E. coli from humans and animals. Front. Microbiol. 2018, 9, 902. [Google Scholar] [CrossRef]
- Zaixiang, L.; Hongxin, W.; Shengqi, R.; Juntao, S.; Chaoyang, M.; Jing, L. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Cont. 2012, 25, 550–554. [Google Scholar]
- Ma, C.; He, N.; Zhao, Y.; Xia, D.; Wei, J.; Kang, W. Antimicrobial mechanism of hydroquinone. Appl. Biochem. Biotechnol. 2019, 189, 1291–1303. [Google Scholar] [CrossRef]
- Ben Abdallah, F.; Lagha, R.; Gaber, A. Biofilm inhibition and eradication properties of medicinal plant essential oils against methicillin-resistant Staphylococcus aureus clinical isolates. Pharmaceuticals 2020, 13, 369. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Int. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Saadatian, F.A.; Nowroozi, J.; Eslami, G.; Sabokbar, A. RAPD PCR profile, antibiotic resistance, prevalence of armA gene, and detection of KPC enzyme in Klebsiella pneumoniae isolates. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 6183162. [Google Scholar] [CrossRef] [PubMed]
- Aljanaby, A. Role of rmpA, wabG, uge, Ycfm, fimh1, EntB, Ybt-irp2 and kfu genes in pathogenicity of Klebsiella pneumoniae: An overview. Int. J. Chemtech. Res. 2017, 10, 391–398. [Google Scholar]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Gharrah, M.M.; El-Mahdy, A.M.; Barwa, R.F. Association between Virulence Factors and Extended Spectrum Beta-Lactamase Producing Klebsiella pneumoniae Compared to Nonproducing Isolates. Interdiscip. Perspect. Infect. Dis. 2017, 2017, 7279830. [Google Scholar] [CrossRef] [PubMed]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement M100–S25; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2018; p. 240. [Google Scholar]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agriculture Research, 2nd ed.; John Willey: New York, NY, USA, 1984; p. 680. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).