Modulatory Effects of Atractylodin and β-Eudesmol on Human Cytochrome P450 Enzymes: Potential Drug-Drug Interactions
Abstract
1. Introduction
2. Results
2.1. In Vitro Inhibitory Effects of Atractylodin and β-Eudesmol on Five Major CYP450 Enzymes
2.2. Ex Vixo Modulatory Effects of Atractylodin and β-Eudesmol on the Expression Levels of mCYP1A2 and m3A11 mRNA, Proteins and Enzyme Activities
2.2.1. mRNA Expression
2.2.2. Protein Expression
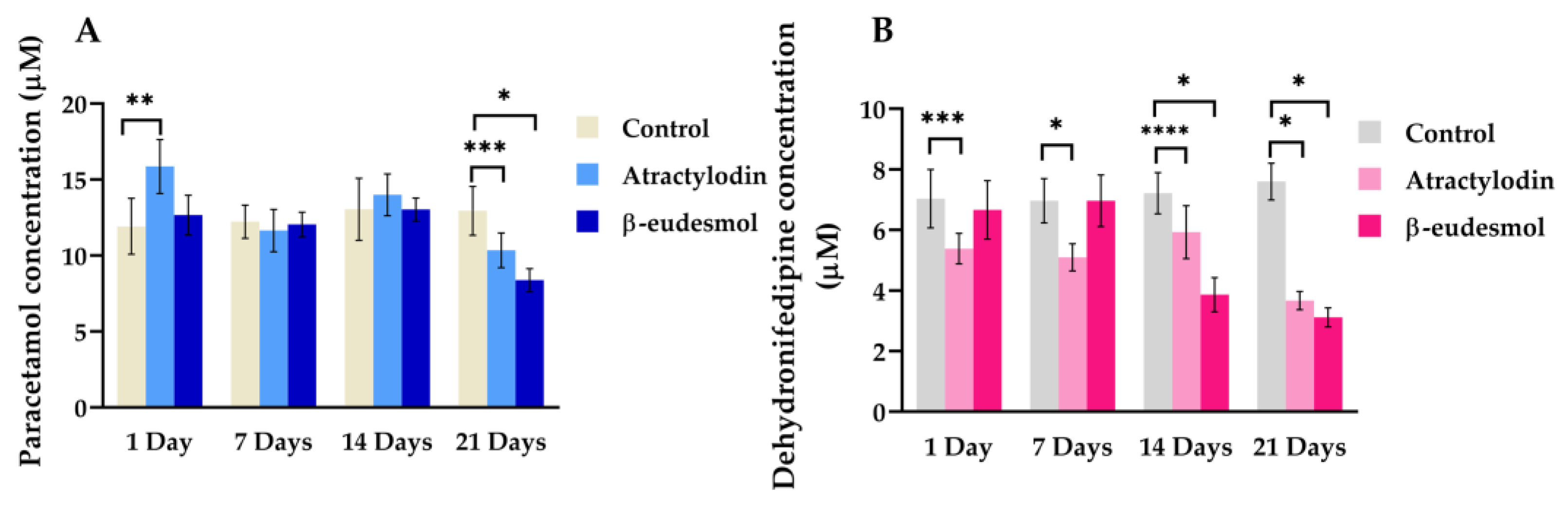
2.2.3. Enzyme Activities
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Treatments and Animals
4.3. In Vitro Inhibitory Effects on rCYP450s
4.4. Ex Vixo Modulatory Effects of Atractylodin and β-Eudesmol on the Expression Levels of mCYP1A2 and m3A11 mRNA, Proteins and Enzyme Activities
4.4.1. Quantitation of mRNA by Real-Time PCR
4.4.2. Preparation of Liver Microsomes and Determination of CYP450 Activities
4.4.3. Western Blot Analysis for Protein Quantification
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Houghton, P.J. The Role of Plants in Traditional Medicine and Current Therapy. J. Altern. Complement. Med. 1995, 1, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Welz, A.N.; Emberger-Klein, A.; Menrad, K. Why people use herbal medicine: Insights from a focus-group study in Germany. BMC Complement. Altern. Med. 2018, 18, 92. [Google Scholar] [CrossRef] [PubMed]
- Genovese, S.; Epifano, F.; Curini, M.; Menger, D.; Zembruski, N.C.; Weiss, J. In vitro effects of natural prenyloxycinnamic acids on human cytochrome P450 isozyme activity and expression. Phytomed. Int. J. Phytother. Phytopharm. 2011, 18, 586–591. [Google Scholar] [CrossRef]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [CrossRef]
- Ouyang, Z.; Yang, L.; Su, S.-L.; Feng, X.; Wang, M. Fingerprint of volatile oil of Atractylodes lancea by GC-MS. Yao Xue Xue Bao Acta Pharm. Sin. 2007, 42, 968–972. [Google Scholar]
- Koonrungsesomboon, N.; Na-Bangchang, K.; Karbwang, J. Therapeutic potential and pharmacological activities of Atractylodes lancea (Thunb.) DC. Asian Pac. J. Trop. Med. 2014, 7, 421–428. [Google Scholar] [CrossRef]
- Plengsuriyakarn, T.; Viyanant, V.; Eursitthichai, V.; Picha, P.; Kupradinun, P.; Itharat, A.; Na-Bangchang, K. Anticancer activities against cholangiocarcinoma, toxicity and pharmacological activities of Thai medicinal plants in animal models. BMC Complement. Altern. Med. 2012, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Plengsuriyakarn, T.; Karbwang, J.; Na-Bangchang, K. Anticancer activity using positron emission tomography-computed tomography and pharmacokinetics of β-eudesmol in human cholangiocarcinoma xenografted nude mouse model. Clin. Exp. Pharmacol. Physiol. 2015, 42, 293–304. [Google Scholar] [CrossRef]
- Na-Bangchang, K.; Plengsuriyakarn, T.; Karbwang, J. Research and Development of Atractylodes lancea (Thunb) DC. as a Promising Candidate for Cholangiocarcinoma Chemotherapeutics. Evid. Based Complement. Altern. Med. eCAM 2017, 2017, 5929234. [Google Scholar] [CrossRef]
- Na-Bangchang, K.; Kulma, I.; Plengsuriyakarn, T.; Tharavanij, T.; Kotawng, K.; Chemung, A.; Muhamad, N.; Karbwang, J. Phase I clinical trial to evaluate the safety and pharmacokinetics of capsule formulation of the standardized extract of Atractylodes lancea. J. Tradit. Complement. Med. 2021, 11, 343–355. [Google Scholar] [CrossRef]
- Mathema, V.B.; Chaijaroenkul, W.; Na-Bangchang, K. Cytotoxic activity and molecular targets of atractylodin in cholangiocarcinoma cells. J. Pharm. Pharmacol. 2019, 71, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Martviset, P.; Panrit, L.; Chantree, P.; Muhamad, P.; Na-Bangchang, K. Suppression of Cholangiocarcinoma Cell Growth and Proliferation by Atractylodes lancea (Thunb) DC. through ERK-Signaling Cascade. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 3633–3640. [Google Scholar] [CrossRef] [PubMed]
- Mahavorasirikul, W.; Viyanant, V.; Chaijaroenkul, W.; Itharat, A.; Na-Bangchang, K. Cytotoxic activity of Thai medicinal plants against human cholangiocarcinoma, laryngeal and hepatocarcinoma cells in vitro. BMC Complement. Altern. Med. 2010, 10, 55. [Google Scholar] [CrossRef]
- Acharya, B.; Chajaroenkul, W.; Na-Bangchang, K. β-Eudesmol Inhibits the Migration of Cholangiocarcinoma Cells by Suppressing Epithelial-Mesenchymal Transition via PI3K/AKT and p38MAPK Modulation. Asian Pac. J. Cancer Prev. APJCP 2022, 23, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.; Chaijaroenkul, W.; Na-Bangchang, K. Atractylodin inhibited the migration and induced autophagy in cholangiocarcinoma cells via PI3K/AKT/mTOR and p38MAPK signaling pathways. J. Pharm. Pharmacol. 2021, 73, 1191–1200. [Google Scholar] [CrossRef]
- Sumsakul, W.; Mahavorasirikul, W.; Na-Bangchang, K. Inhibitory Activities of Thai Medicinal Plants with Promising Activities Against Malaria and Cholangiocarcinoma on Human Cytochrome P450. Phytother. Res. PTR 2015, 29, 1926–1933. [Google Scholar] [CrossRef]
- Pao, L.H.; Hu, O.Y.; Fan, H.Y.; Lin, C.C.; Liu, L.C.; Huang, P.W. Herb-drug interaction of 50 Chinese herbal medicines on CYP3A4 activity in vitro and in vivo. Am. J. Chin. Med. 2012, 40, 57–73. [Google Scholar] [CrossRef]
- Ramasamy, S.; Kiew, L.V.; Chung, L.Y. Inhibition of human cytochrome P450 enzymes by Bacopa monnieri standardized extract and constituents. Molecules 2014, 19, 2588–2601. [Google Scholar] [CrossRef]
- Samer, C.F.; Lorenzini, K.I.; Rollason, V.; Daali, Y.; Desmeules, J.A. Applications of CYP450 testing in the clinical setting. Mol. Diagn. Ther. 2013, 17, 165–184. [Google Scholar] [CrossRef]
- Zhou, S.F.; Liu, J.P.; Chowbay, B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab. Rev. 2009, 41, 89–295. [Google Scholar] [CrossRef]
- Cali, J.J.; Ma, D.; Sobol, M.; Simpson, D.J.; Frackman, S.; Good, T.D.; Daily, W.J.; Liu, D. Luminogenic cytochrome P450 assays. Expert Opin. Drug Metab. Toxicol. 2006, 2, 629–645. [Google Scholar] [CrossRef]
- Krenc, D.; Na-Bangchang, K. Spectroscopic observations of β-eudesmol binding to human cytochrome P450 isoforms 3A4 and 1A2, but not to isoforms 2C9, 2C19, and 2D6. Xenobiotica 2022, 52, 199–208. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Guidance for Industry Drug Interaction Studies—Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2012.
- Nelson, D.R.; Zeldin, D.C.; Hoffman, S.M.; Maltais, L.J.; Wain, H.M.; Nebert, D.W. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alterna-tive-splice variants. Pharmacogenetics 2004, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.; Jansson, I.; Schenkman, J.B.; Sarfarazi, M.; Stoilov, I. Comparative expression profiling of 40 mouse cyto-chrome P450 genes in embryonic and adult tissues. Arch. Biochem. Biophys. 2003, 414, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Yanagimoto, T.; Itoh, S.; Muller-Enoch, D.; Kamataki, T. Mouse liver cytochrome P-450 (P-450IIIAM1): Its cDNA cloning and inducibility by dexamethasone. Biochim. Biophys. Acta 1992, 1130, 329–332. [Google Scholar] [CrossRef]
- Yanagimoto, T.; Itoh, S.; Sawada, M.; Kamataki, T. Mouse cytochrome P450 (Cyp3a11): Predominant expression in liver and capacity to activate aflatoxin B1. Arch. Biochem. Biophys. 1997, 340, 215–218. [Google Scholar] [CrossRef]
- Turpeinen, M.; Ghiciuc, C.; Opritoui, M.; Tursas, L.; Pelkonen, O.; Pasanen, M. Predictive value of animal models for human cytochrome P450 (CYP)-mediated metabolism: A comparative study in vitro. Xenobiotica 2007, 37, 1367–1377. [Google Scholar] [CrossRef]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef]
- Eaton, D.L.; Gallagher, E.P.; Bammler, T.K.; Kunze, K.L. Role of cytochrome P4501A2 in chemical carcinogenesis: Implications for human variability in expression and enzyme activity. Pharmacogenetics 1995, 5, 259–274. [Google Scholar] [CrossRef]
- Zhou, S.-F.; Yang, L.-P.; Zhou, Z.-W.; Liu, Y.-H.; Chan, E. Insights into the Substrate Specificity, Inhibitors, Regulation, and Polymorphisms and the Clinical Impact of Human Cytochrome P450 1A2. AAPS J. 2009, 11, 481–494. [Google Scholar] [CrossRef]
- Patocka, J.; Jun, D.; Kuca, K. Possible role of hydroxylated metabolites of tacrine in drug toxicity and therapy of Alzheimer’s disease. Curr. Drug Metab. 2008, 9, 332–335. [Google Scholar] [CrossRef]
- Kato, H. Computational prediction of cytochrome P450 inhibition and induction. Drug Metab. Pharmacokinet. 2020, 35, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Juvonen, R.O.; Jokinen, E.M.; Javaid, A.; Lehtonen, M.; Raunio, H.; Pentikäinen, O.T. Inhibition of human CYP1 enzymes by a classical inhibitor α-naphthoflavone and a novel inhibitor N-(3, 5-dichlorophenyl)cyclopropanecarboxamide: An in vitro and in silico study. Chem. Biol. Drug Des. 2020, 95, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Unger, M. Pharmacokinetic drug interactions by herbal drugs: Critical evaluation and clinical relevance. Wien. Med. Woch-Enschr. 2010, 160, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Westerink, W.M.; Schoonen, W.G. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2007, 21, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 1–17. [Google Scholar] [CrossRef]
- Zuber, R.; Anzenbacherová, E.; Anzenbacher, P. Cytochromes P450 and experimental models of drug metabolism. J. Cell. Mol. Med. 2002, 6, 189–198. [Google Scholar] [CrossRef]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. The National Academies Collection: Reports funded by National Institutes of Health. In Guide for the Care and Use of Laboratory Animals; National Academy of Sciences: Washington, DC, USA, 2011. [Google Scholar]
- Sumsakul, W.; Plengsuriyakarn, T.; Na-Bangchang, K. Pharmacokinetics, toxicity, and cytochrome P450 modulatory activity of plumbagin. BMC Pharmacol. Toxicol. 2016, 17, 50. [Google Scholar] [CrossRef]


| Compounds | Median IC50 (Range) | ||||
|---|---|---|---|---|---|
| rCYP1A2 | rCYP2C9 | rCYP2C19 | rCYP2D6 | rCYP3A4 | |
| Atractylodin | 497.2 (460–561.4) µM | >685.9 µM | >685.9 µM | >685.9 µM | >685.9 µM |
| 90.6 (83.8–102.3) µg/ml | >125 µg/ml | >125 µg/ml | >125 µg/ml | >125 µg/ml | |
| β-Eudesmol | >562.1 µM | 533.3 (443.4–539.2) µM | 172.7 (167.3–180.3) µM | >562.1 µM | 218.6 (204.6–223.5) µM |
| >125 µg/ml | 118.6 (98.6–119.9) µg/ml | 38.4 (37.2–40.1) µg/ml | >125 µg/ml | 48.6 (45.5–49.7) µg/ml | |
| Inhibitors | 0.18 (0.13–0.19) µM | 0.24 (0.21–0.25) µM | 2.07 (1.96–2.33) µM | 0.015 (0.013–0.016) µM | 0.10 (0.10–0.11) µM |
| ∝-Naphthoflavone | Sulfaphenazole | Troglitazone | Quinidine | Ketoconazole | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiengsusuk, A.; Plengsuriyakarn, T.; Na-Bangchang, K. Modulatory Effects of Atractylodin and β-Eudesmol on Human Cytochrome P450 Enzymes: Potential Drug-Drug Interactions. Molecules 2023, 28, 3140. https://doi.org/10.3390/molecules28073140
Thiengsusuk A, Plengsuriyakarn T, Na-Bangchang K. Modulatory Effects of Atractylodin and β-Eudesmol on Human Cytochrome P450 Enzymes: Potential Drug-Drug Interactions. Molecules. 2023; 28(7):3140. https://doi.org/10.3390/molecules28073140
Chicago/Turabian StyleThiengsusuk, Artitaya, Tullayakorn Plengsuriyakarn, and Kesara Na-Bangchang. 2023. "Modulatory Effects of Atractylodin and β-Eudesmol on Human Cytochrome P450 Enzymes: Potential Drug-Drug Interactions" Molecules 28, no. 7: 3140. https://doi.org/10.3390/molecules28073140
APA StyleThiengsusuk, A., Plengsuriyakarn, T., & Na-Bangchang, K. (2023). Modulatory Effects of Atractylodin and β-Eudesmol on Human Cytochrome P450 Enzymes: Potential Drug-Drug Interactions. Molecules, 28(7), 3140. https://doi.org/10.3390/molecules28073140






