Cell-Free Expression of a Therapeutic Protein Serratiopeptidase
Abstract
1. Introduction
2. Results and Discussion
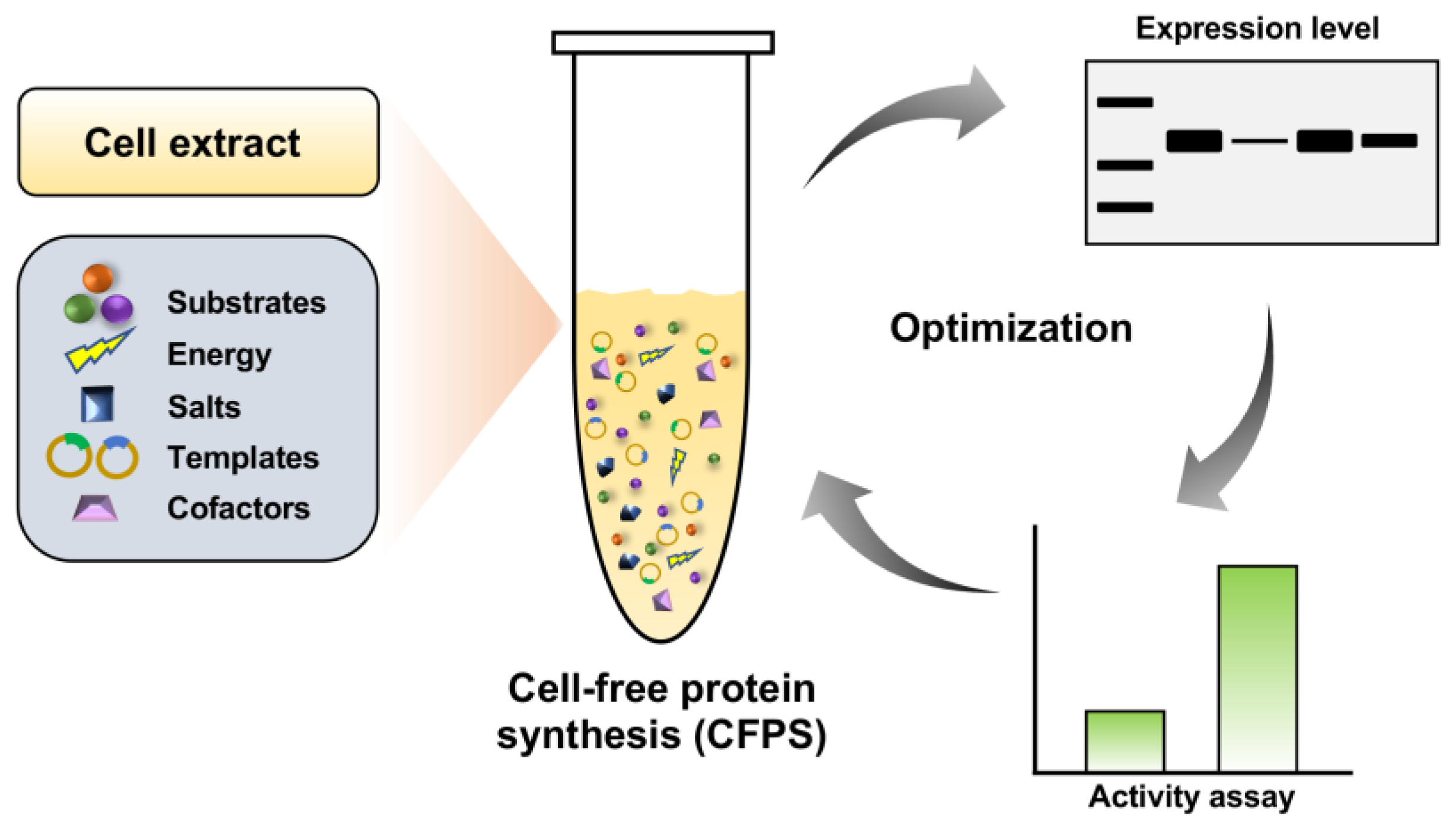
2.1. Cell-Free Expression of Serratiopeptidase
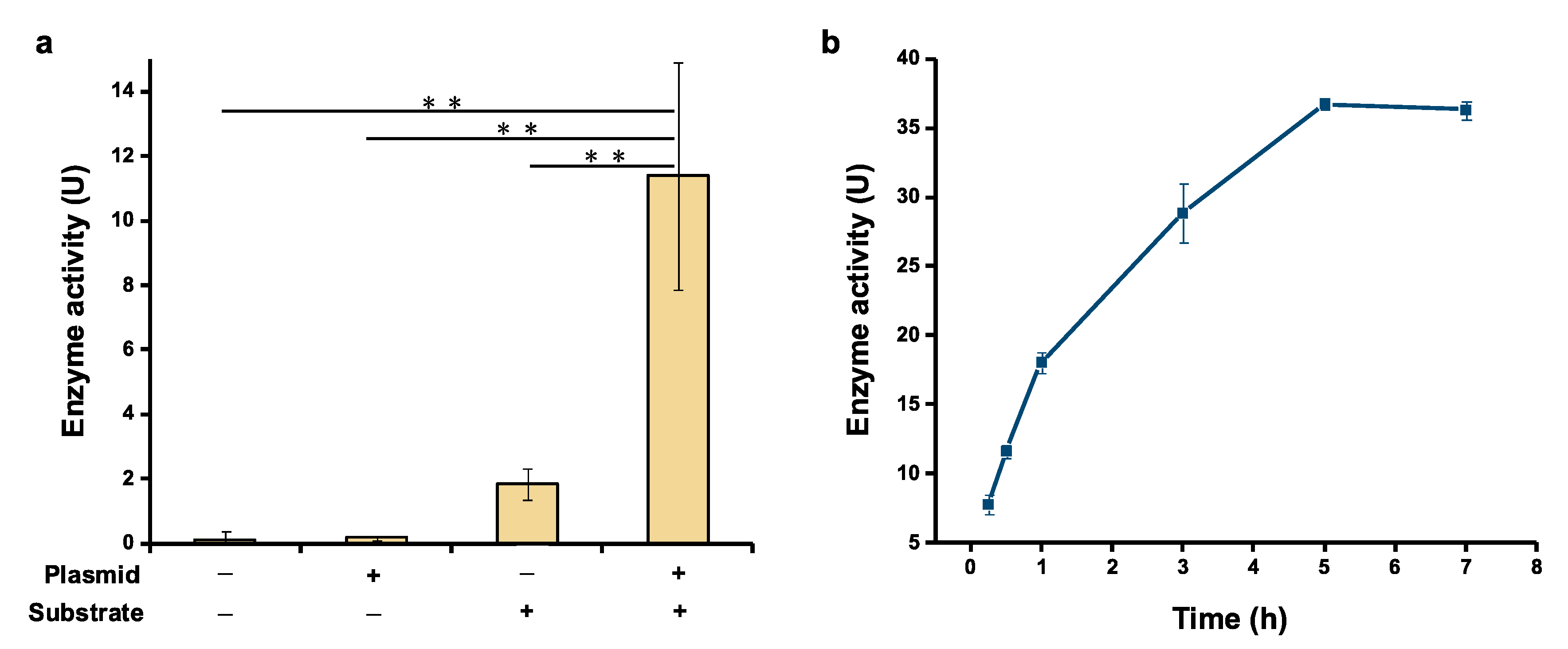
2.2. Activity Assay of Cell-Free Expressed Serratiopeptidase
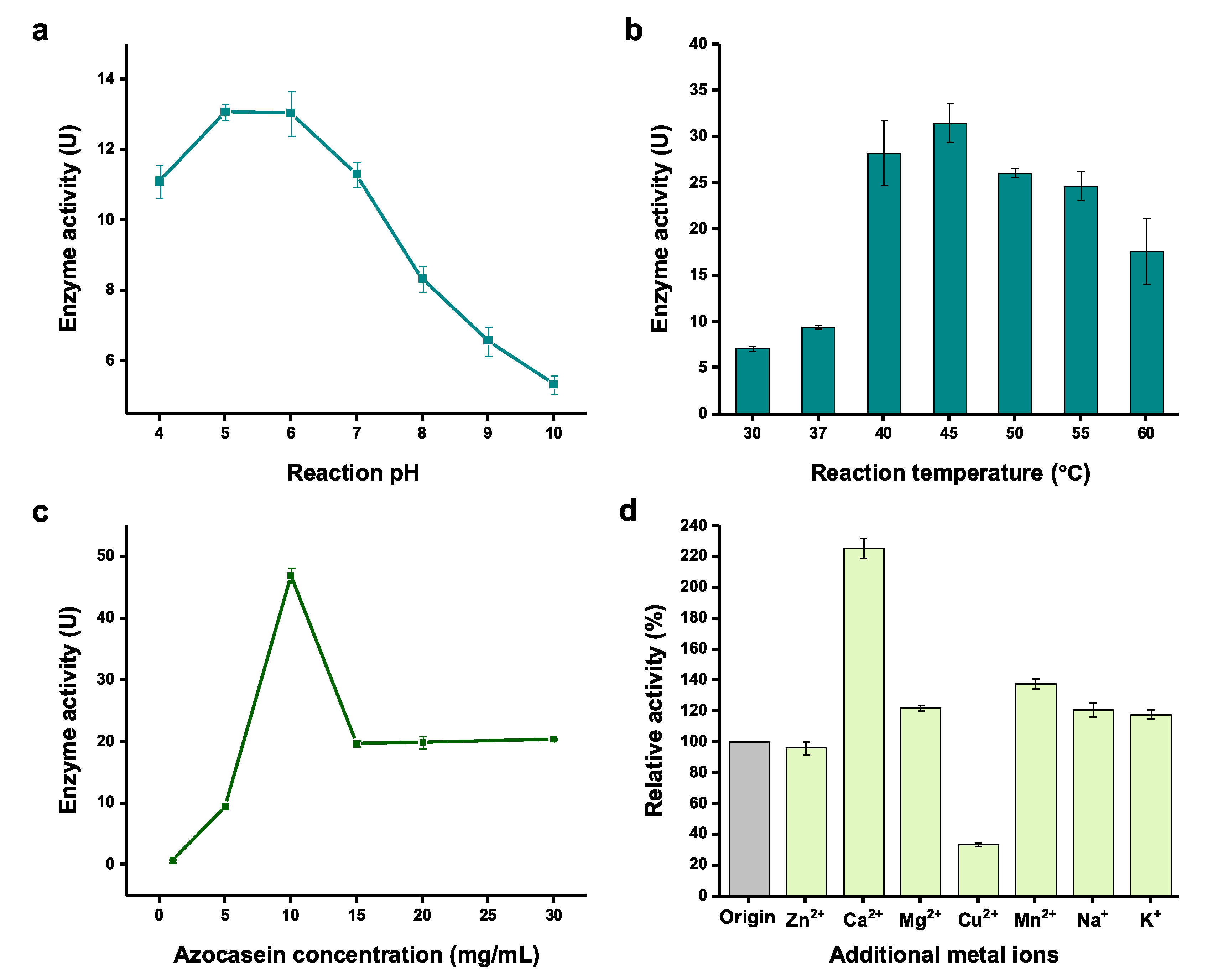
2.3. Effect of Physicochemical Factors on Serratiopeptidase Activity
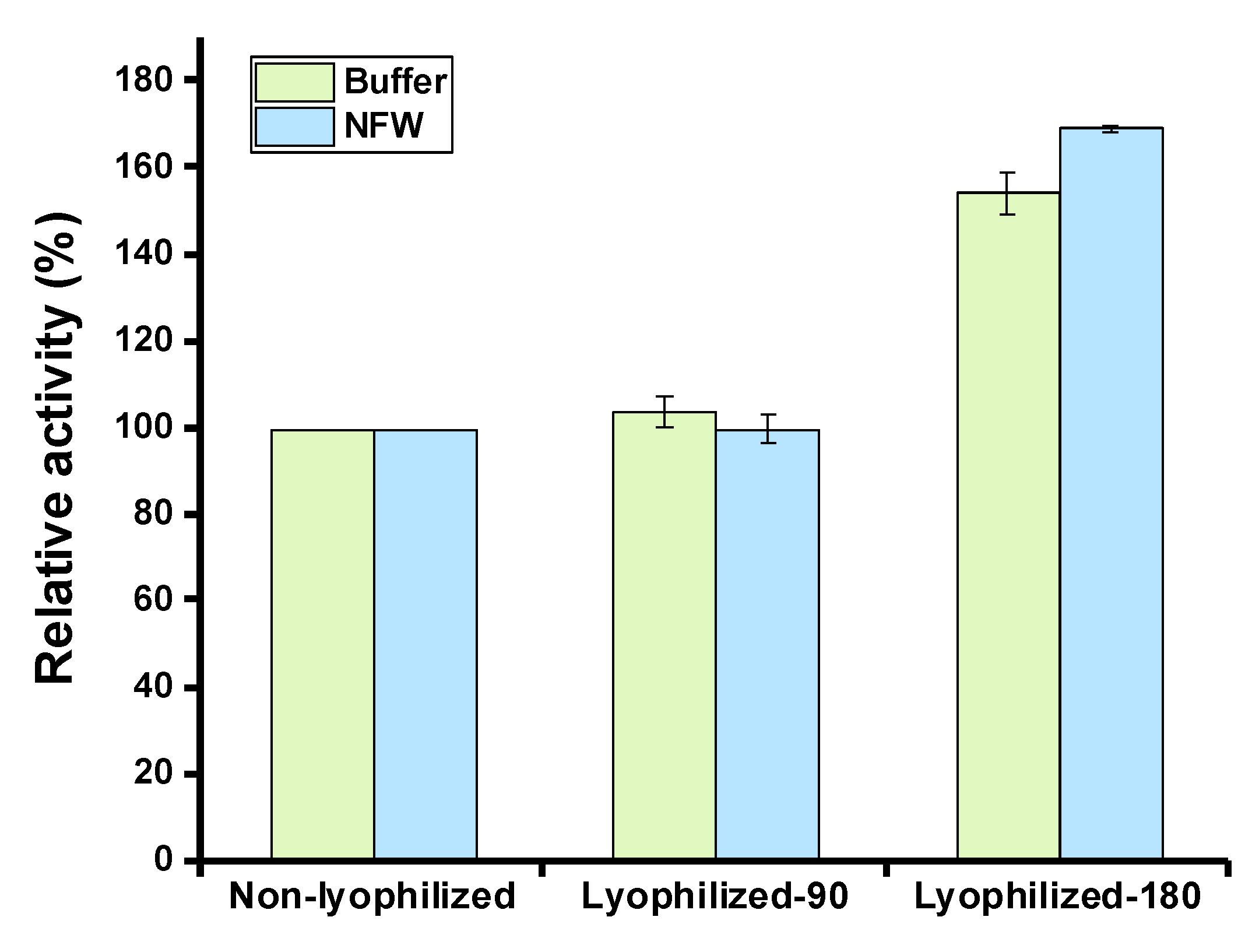
2.4. Lyophilization of Cell-Free Expressed Serratiopeptidase
3. Materials and Methods
3.1. Strains, Media, and Plasmids
3.2. Cell Extract Preparation
3.3. Cell-Free Protein Synthesis (CFPS) and Western-Blot Analysis
3.4. Activity Assay of Serratiopeptidase
3.5. Effect of Reaction Time on Serratiopeptidase Activity
3.6. Effect of Physicochemical Factors on Serratiopeptidase Activity
3.7. Detection of Serratiopeptidase Activity after Lyophilization
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Reshma, C.V. Microbial enzymes: Therapeutic applications. Microbiol. Res. J. Int. 2019, 27, 1–8. [Google Scholar] [CrossRef]
- Benedini, L. Advanced protein drugs and formulations. Curr. Protein Pept. Sci. 2022, 23, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Hashimoto, Y.; Mikami, M.; Yamanaka, E.; Soma, T.; Hino, M.; Azuma, A.; Kudoh, S. Effect of the proteolytic enzyme serrapeptase in patients with chronic airway disease. Respirology 2003, 8, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Mecikoglu, M.; Saygi, B.; Yildirim, Y.; Karadag-Saygi, E.; Ramadan, S.S.; Esemenli, T. The effect of proteolytic enzyme serratiopeptidase in the treatment of experimental implant-related infection. J. Bone Jt. Surg. Am. 2006, 88, 1208–1214. [Google Scholar] [CrossRef]
- Hines, D.A.; Saurugger, P.N.; Ihler, G.M.; Benedik, M.J. Genetic analysis of extracellular proteins of Serratia marcescens. J. Bacteriol. 1988, 170, 4141–4146. [Google Scholar] [CrossRef]
- Nakahama, K.; Yoshimura, K.; Marumoto, R.; Kikuchi, M.; Lee, I.S.; Hase, T.; Matsubara, H. Cloning and sequencing of Serratia protease gene. Nucleic Acids Res. 1986, 14, 5843–5855. [Google Scholar] [CrossRef]
- Ethiraj, S.; Gopinath, S. Production, purification, characterization, immobilization, and application of serrapeptase: A review. Front. Biol. 2017, 12, 333–348. [Google Scholar] [CrossRef]
- Chappi, D.M.; Suresh, K.V.; Patil, M.R.; Desai, R.; Tauro, D.P.; Bharani, K.N.S.S.; Parkar, M.I.; Babaji, H.V. Comparison of clinical efficacy of methylprednisolone and serratiopeptidase for reduction of postoperative sequelae after lower third molar surgery. J. Clin. Exp. Dent. 2015, 7, e197–e202. [Google Scholar] [CrossRef]
- Tiwari, M. The role of serratiopeptidase in the resolution of inflammation. Asian J. Pharm. Sci. 2017, 12, 209–215. [Google Scholar] [CrossRef]
- Kumar, M.P.S. The emerging role of serratiopeptidase in oral surgery: Literature update. Asian J. Pharm. Clin. Res. 2018, 11, 19. [Google Scholar] [CrossRef]
- Bracale, G.; Selvetella, L. Clinical study of the efficacy of and tolerance to seaprose S in inflammatory venous disease. Controlled study versus serratiopeptidase. Minerva Cardioangiol. 1996, 44, 515–524. [Google Scholar] [PubMed]
- Sharma, C.; Jha, N.K.; Meeran, M.F.N.; Patil, C.R.; Goyal, S.N.; Ojha, S. Serratiopeptidase, a serine protease anti-inflammatory, fibrinolytic, and mucolytic drug, can be a useful adjuvant for management in COVID-19. Front. Pharmacol. 2021, 12, 603997. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.R.; Devi, C.S. Serratiopeptidase: An integrated view of multifaceted therapeutic enzyme. Biomolecules 2022, 12, 1468. [Google Scholar] [CrossRef] [PubMed]
- Rouhani, M.; Valizadeh, V.; Molasalehi, S.; Norouzian, D. Production and expression optimization of heterologous serratiopeptidase. Iran. J. Public Health 2020, 49, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Doshi, P.; Bhargava, P.; Singh, V.; Pathak, C.; Joshi, C.; Joshi, M. Escherichia coli strain engineering for enhanced production of serratiopeptidase for therapeutic applications. Int. J. Biol. Macromol. 2020, 160, 1050–1060. [Google Scholar] [CrossRef]
- Srivastava, V.; Mishra, S.; Chaudhuri, T.K. Enhanced production of recombinant serratiopeptidase in Escherichia coli and its characterization as a potential biosimilar to native biotherapeutic counterpart. Microb. Cell Fact. 2019, 18, 215. [Google Scholar] [CrossRef]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef]
- Focke, P.J.; Hein, C.; Hoffmann, B.; Matulef, K.; Bernhard, F.; Dötsch, V.; Valiyaveetil, F.I. Combining in vitro folding with cell free protein synthesis for membrane protein expression. Biochemistry 2016, 55, 4212–4219. [Google Scholar] [CrossRef]
- Rosenblum, G.; Cooperman, B.S. Engine out of the chassis: Cell-free protein synthesis and its uses. FEBS Lett. 2014, 588, 261–268. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Jewett, M.C. Cell-Free Protein Synthesis: An Emerging Technology for Understanding, Harnessing, and Expanding the Capabilities of Biological Systems. In Synthetic Biology: Parts, Devices and Applications; John Wiley & Sons: Willie, CO, USA, 2018; Chapter 15. [Google Scholar]
- Li, J.; Lawton, T.J.; Kostecki, J.S.; Nisthal, A.; Fang, J.; Mayo, S.L.; Rosenzweig, A.C.; Jewett, M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 2016, 11, 212–218. [Google Scholar] [CrossRef]
- Goering, A.W.; Li, J.; McClure, R.A.; Thomson, R.J.; Jewett, M.C.; Kelleher, N.L. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol. 2017, 6, 39–44. [Google Scholar] [CrossRef]
- Zhuang, L.; Huang, S.; Liu, W.Q.; Karim, A.S.; Jewett, M.C.; Li, J. Total in vitro biosynthesis of the nonribosomal macrolactone peptide valinomycin. Metab. Eng. 2020, 60, 37–44. [Google Scholar] [CrossRef]
- Sachse, R.; Dondapati, S.K.; Fenz, S.F.; Schmidt, T.; Kubick, S. Membrane protein synthesis in cell-free systems: From bio-mimetic systems to bio-membranes. FEBS Lett. 2014, 588, 2774–2781. [Google Scholar] [CrossRef]
- Bundy, B.C.; Swartz, J.R. Efficient disulfide bond formation in virus-like particles. J. Biotechnol. 2011, 154, 230–239. [Google Scholar] [CrossRef]
- Garamella, J.; Marshall, R.; Rustad, M.; Noireaux, V. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol. 2016, 5, 344–355. [Google Scholar] [CrossRef]
- Westbrook, A.; Tang, X.; Marshall, R.; Maxwell, C.S.; Chappell, J.; Agrawal, D.K.; Dunlop, M.J.; Noireaux, V.; Beisel, C.L.; Lucks, J.; et al. Distinct timescales of RNA regulators enable the construction of a genetic pulse generator. Biotechnol. Bioeng. 2019, 116, 1139–1151. [Google Scholar] [CrossRef]
- Kelwick, R.; Ricci, L.; Chee, S.M.; Bell, D.; Webb, A.J.; Freemont, P.S. Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth. Biol. 2018, 3, ysy016. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Anderson, K.C.; Jewett, M.C. Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol. 2016, 5, 1578–1588. [Google Scholar] [CrossRef]
- Karim, A.S.; Dudley, Q.M.; Juminaga, A.; Yuan, Y.; Crowe, S.A.; Heggestad, J.T.; Garg, S.; Abdalla, T.; Grubbe, W.S.; Rasor, B.J.; et al. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat. Chem. Biol. 2020, 16, 912–919. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Liu, W. Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products. Synth. Syst. Biotechnol. 2018, 3, 83–89. [Google Scholar] [CrossRef]
- Feng, J.; Yang, C.; Zhao, Z.; Xu, J.; Li, J.; Li, P. Application of cell-free protein synthesis system for the biosynthesis of L-theanine. ACS Synth. Biol. 2021, 10, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, W.Q.; Xu, H.; Ji, X.; Liu, Y.; Li, J. Cell-free expression of NO synthase and P450 enzyme for the biosynthesis of an unnatural amino acid L-4-nitrotryptophan. Synth. Syst. Biotechnol. 2022, 7, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Paper-based synthetic gene networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Lu, Y.; Welsh, J.P.; Swartz, J.R. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl. Acad. Sci. USA 2014, 111, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Min, S.E.; Lee, K.H.; Park, S.W.; Yoo, T.H.; Oh, C.H.; Park, J.H.; Yang, S.Y.; Kim, Y.S.; Kim, D.M. Cell-free production and streamlined assay of cytosol-penetrating antibodies. Biotechnol. Bioeng. 2016, 113, 2107–2112. [Google Scholar] [CrossRef]
- Hong, S.H.; Ntai, I.; Haimovich, A.D.; Kelleher, N.L.; Isaacs, F.J.; Jewett, M.C. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth. Biol. 2014, 3, 398–409. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Hershewe, J.M.; Stark, J.C.; Kightlinger, W.; Kath, J.E.; Jaroentomeechai, T.; Natarajan, A.; DeLisa, M.P.; Jewett, M.C. A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol. Bioeng. 2018, 115, 739–750. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.A.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004, 5, 75–86. [Google Scholar] [CrossRef]
- Marblestone, J.G.; Edavettal, S.C.; Lim, Y.; Lim, P.; Zuo, X.; Butt, T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006, 15, 182–189. [Google Scholar] [CrossRef]
- Panavas, T.; Sanders, C.; Butt, T.R. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 2009, 497, 303–317. [Google Scholar] [PubMed]
- Hamada, K.; Hata, Y.; Katsuya, Y.; Hiramatsu, H.; Fujiwara, T.; Katsube, Y. Crystal structure of Serratia protease, a zinc-dependent proteinase from Serratia sp. E-15, containing a beta-sheet coil motif at 2.0 A resolution. J. Biochem. 1996, 119, 844. [Google Scholar] [CrossRef]
- Wang, W. Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 2000, 203, 1–60. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Y.; Liu, W.Q.; Wu, C.; Li, J. Designing modular cell-free systems for tunable biotransformation of L-phenylalanine to aromatic compounds. Front. Bioeng. Biotechnol. 2021, 9, 730663. [Google Scholar] [CrossRef]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M.; Gold, D.S.; Heinsohn, H.G.; Murray, C.J. Microscale to manufacturing scale-up of cell-free cytokine production − a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Kwon, Y.C.; Jewett, M.C. Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng. 2017, 114, 1343–1353. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Needham, H.; Polizzi, K.M.; Freemont, P.S. Streptomyces venezuelae TX-TL—A next generation cell-free synthetic biology tool. Biotechnol. J. 2017, 12, 1600678. [Google Scholar] [CrossRef]
- Xu, H.; Liu, W.Q.; Li, J. Translation related factors improve the productivity of a Streptomyces-based cell-free protein synthesis system. ACS Synth. Biol. 2020, 9, 1221–1224. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Chee, S.M.; Toh, M.; Coode, S.; Chengan, K.; Capel, P.; Corre, C.; de Los Santos, E.L.; Freemont, P.S. A Streptomyces venezuelae cell-free toolkit for synthetic biology. ACS Synth. Biol. 2021, 10, 402–411. [Google Scholar] [CrossRef]
- Xu, H.; Yang, C.; Tian, X.; Chen, Y.; Liu, W.Q.; Li, J. Regulatory part engineering for high-yield protein synthesis in an all-Streptomyces-based cell-free expression system. ACS Synth. Biol. 2022, 11, 570–578. [Google Scholar] [CrossRef]
- Kelwick, R.; Webb, A.J.; MacDonald, J.T.; Freemont, P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 2016, 38, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, J.; Jewett, M.C. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol. 2018, 3, ysy003. [Google Scholar] [CrossRef] [PubMed]
- Des Soye, B.J.; Davidson, S.R.; Weinstock, M.T.; Gibson, D.G.; Jewett, M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 2018, 7, 2245–2255. [Google Scholar] [CrossRef]
- Aw, R.; Polizzi, K.M. Biosensor-assisted engineering of a high-yield Pichia pastoris cell-free protein synthesis platform. Biotechnol. Bioeng. 2019, 116, 656–666. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, X.; Wang, T.; Guo, W.; Lu, Y. Development and comparison of cell-free protein synthesis systems derived from typical bacterial chassis. Bioresour. Bioprocess. 2021, 8, 58. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, W.Q.; Li, J. Establishing a eukaryotic Pichia pastoris cell-free protein synthesis system. Front. Bioeng. Biotechnol. 2020, 8, 536. [Google Scholar] [CrossRef]
- Yang, C.; Yang, M.; Zhao, W.; Ding, Y.; Wang, Y.; Li, J. Establishing a Klebsiella pneumoniae-based cell-free protein synthesis system. Molecules 2022, 27, 4684. [Google Scholar] [CrossRef]

| Construction of Plasmid | Primer (5′→3′) | Restriction Site a |
|---|---|---|
| pJL1-serratio | ||
| Forward | CATCATCATCATCACCATATGGCGGCGACCACCGGCTATGATG | NdeI |
| Reverse | TTTGTTAGCAGCCGGTCGACTTACACAATAAAATCGGTCGCCAC | SalI |
| pET28a-SUMO-serratio | ||
| Forward | GATTGGTGGTACCGAGCTCATGGCGGCGACCACCGGCTATG | SacI |
| Reverse | CGAGTGCGGCCGCAAGCTTTTACACAATAAAATCGGTCG | HindIII |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.; Yang, M.; Liu, W.; Li, J. Cell-Free Expression of a Therapeutic Protein Serratiopeptidase. Molecules 2023, 28, 3132. https://doi.org/10.3390/molecules28073132
Meng Y, Yang M, Liu W, Li J. Cell-Free Expression of a Therapeutic Protein Serratiopeptidase. Molecules. 2023; 28(7):3132. https://doi.org/10.3390/molecules28073132
Chicago/Turabian StyleMeng, Yaru, Miaomiao Yang, Wanqiu Liu, and Jian Li. 2023. "Cell-Free Expression of a Therapeutic Protein Serratiopeptidase" Molecules 28, no. 7: 3132. https://doi.org/10.3390/molecules28073132
APA StyleMeng, Y., Yang, M., Liu, W., & Li, J. (2023). Cell-Free Expression of a Therapeutic Protein Serratiopeptidase. Molecules, 28(7), 3132. https://doi.org/10.3390/molecules28073132







