Coordination of the N-Terminal Heme in the Non-Classical Peroxidase from Escherichia coli
Abstract
1. Introduction
2. Results and Discussion
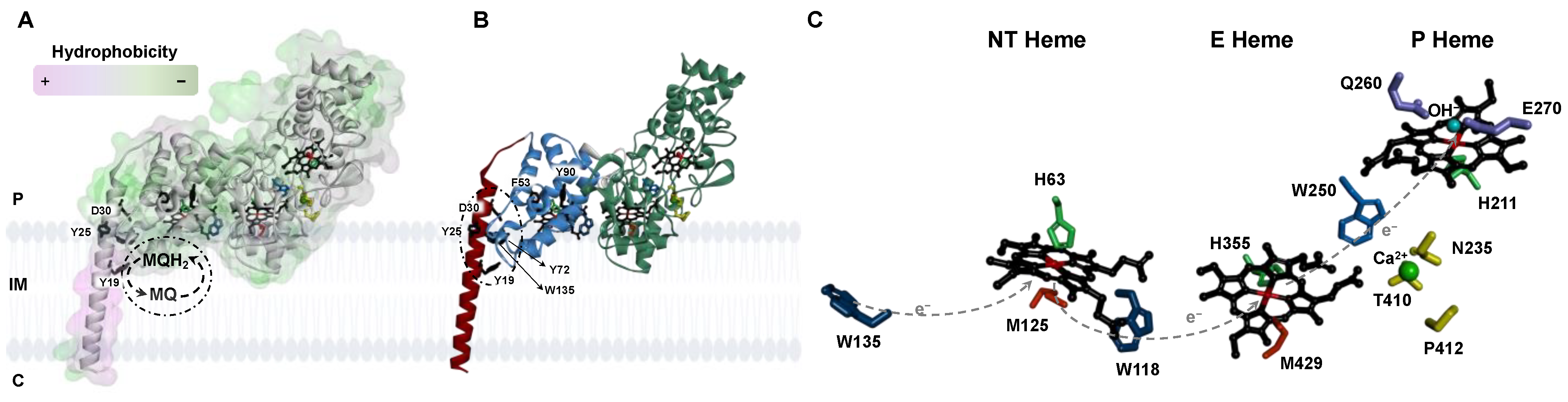
2.1. Analysis of the Primary Sequence and Structural Model
2.2. Isolation and Characterization of YhjA Variants
2.2.1. Protein Isolation
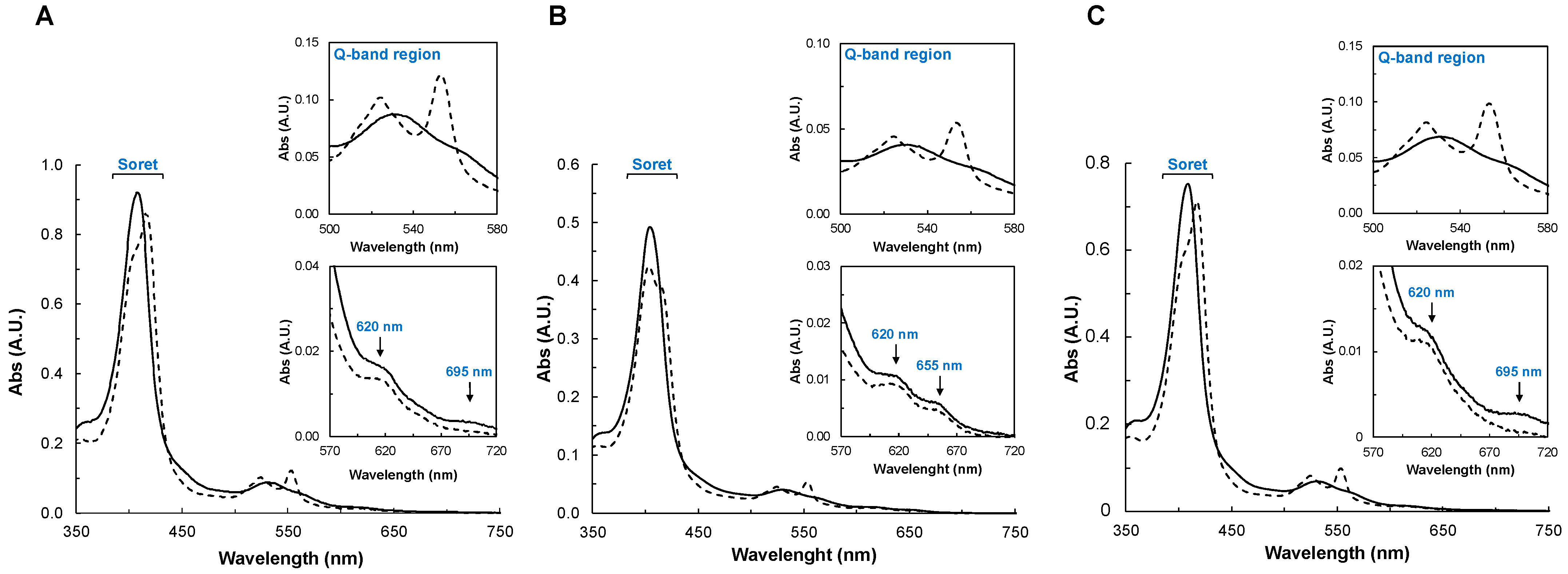
2.2.2. UV-Visible Spectroscopy
2.2.3. Spectral Changes in the Presence of Peroxides
2.2.4. Enzymatic Activity of the YhjA WT and Variants
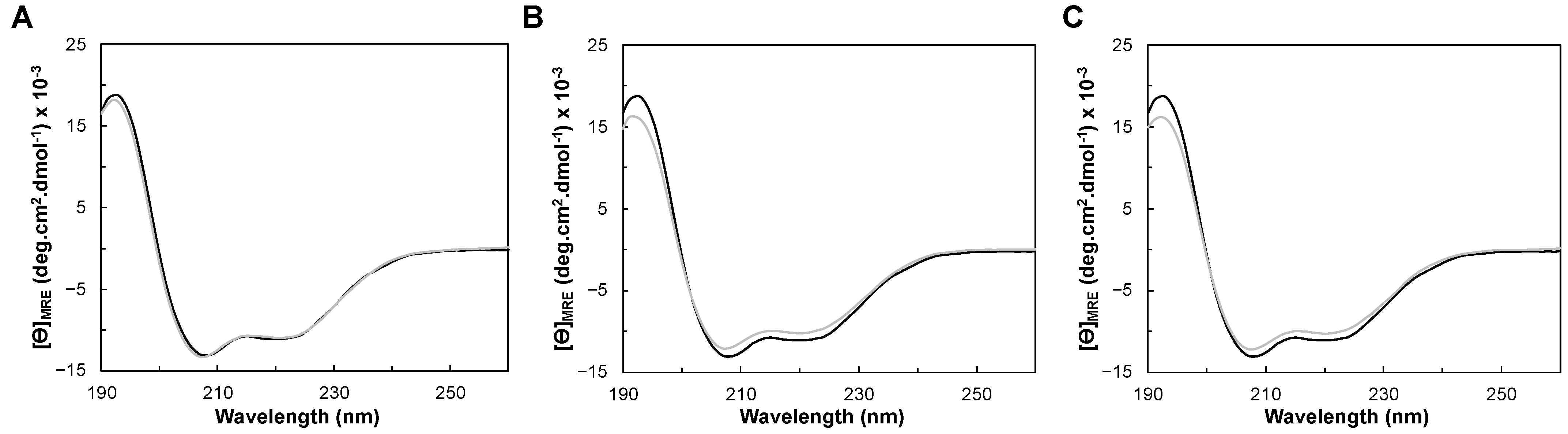
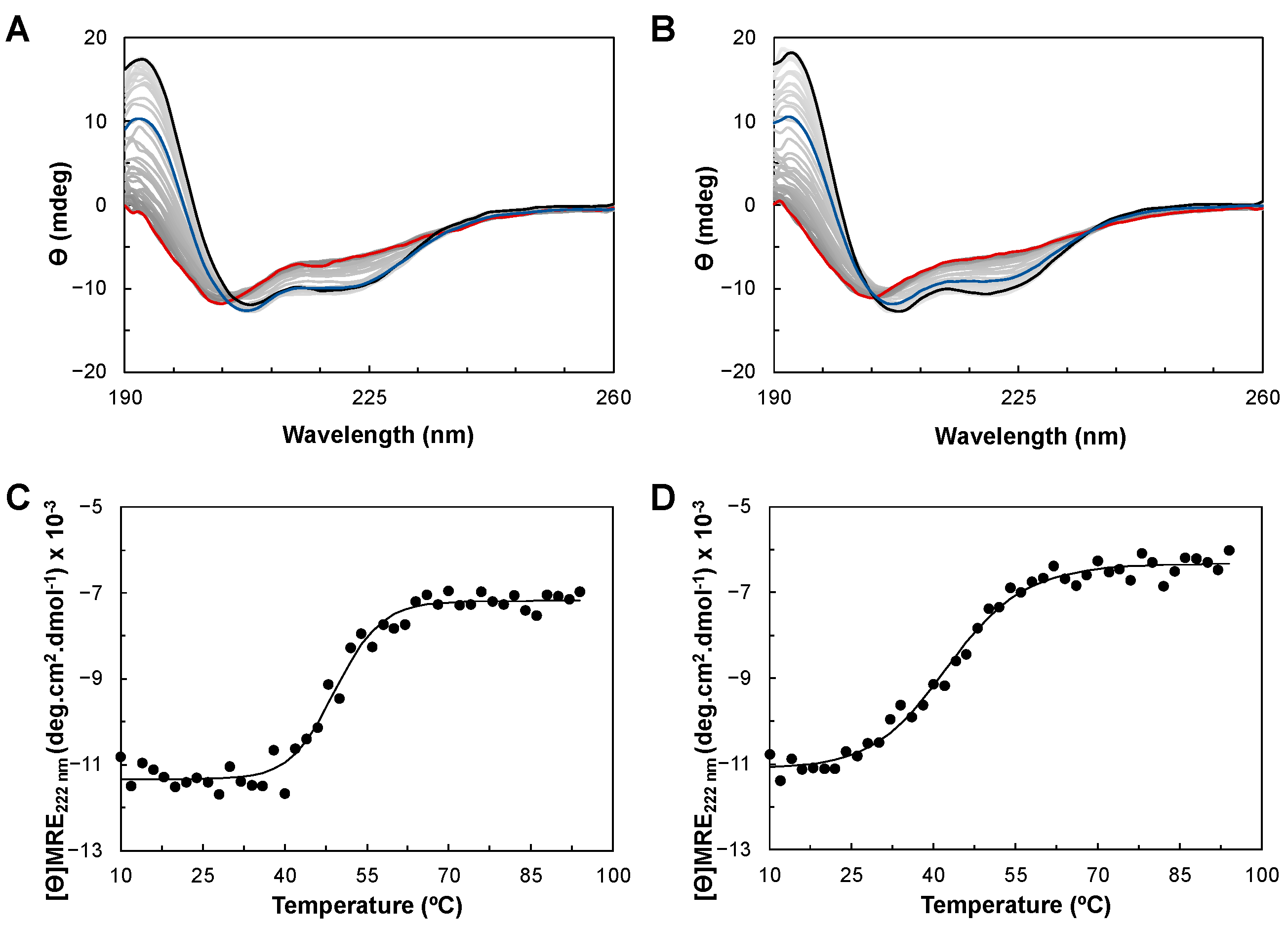
2.3. Circular Dichroism of YhjA WT and Variants
2.3.1. Folding Status and Secondary Structure Content
2.3.2. Thermostability
3. Materials and Methods
3.1. Bioinformatic Analysis
3.2. Generation of the Variants
3.3. Protein Production and Isolation
3.4. Spectroscopic Characterization
3.5. Kinetic Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Nisbet, E.G.; Sleep, N.H. The habitat and nature of early life. Nature 2001, 409, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar] [PubMed]
- Seaver, L.C.; Imlay, J.A. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 2004, 279, 48742–48750. [Google Scholar] [CrossRef]
- Parsonage, D.; Youngblood, D.S.; Sarma, G.N.; Wood, Z.A.; Karplus, P.A.; Poole, L.B. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 2005, 44, 10583–10592. [Google Scholar] [CrossRef] [PubMed]
- Christman, M.F.; Storz, G.; Ames, B.N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 1989, 86, 3484–3488. [Google Scholar] [CrossRef]
- Seaver, L.C.; Imlay, J.A. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 2001, 183, 7182–7189. [Google Scholar] [CrossRef]
- Aslund, F.; Zheng, M.; Beckwith, J.; Storz, G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 1999, 96, 6161–6165. [Google Scholar] [CrossRef]
- Partridge, J.D.; Poole, R.K.; Green, J. The Escherichia coli yhjA gene, encoding a predicted cytochrome c peroxidase, is regulated by FNR and OxyR. Microbiology 2007, 153, 1499–1509. [Google Scholar] [CrossRef]
- Nóbrega, C.S. Biochemical and Physiological Insights into Bacterial Cytochrome c Peroxidases from Escherichia coli and Neisseria gonorrhoeae; NOVA University Lisbon: Lisbon, Portugal, 2017. [Google Scholar]
- Nobrega, C.S.; Devreese, B.; Pauleta, S.R. YhjA—An Escherichia coli trihemic enzyme with quinol peroxidase activity. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 411–422. [Google Scholar] [CrossRef]
- Khademian, M.; Imlay, J.A. Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc. Natl. Acad. Sci. USA 2017, 114, E6922–E6931. [Google Scholar] [CrossRef]
- Bayramoglu, B.; Toubiana, D.; Gillor, O. Genome-wide transcription profiling of aerobic and anaerobic Escherichia coli biofilm and planktonic cultures. FEMS Microbiol. Lett. 2017, 364, fnx006. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, D.S.; Oliveira, R.N.S.; Pauleta, S.R. Bacterial peroxidases—Multivalent enzymes that enable the use of hydrogen peroxide for microaerobic and anaerobic proliferation. Coord. Chem. Rev. 2023, 485, 215114. [Google Scholar] [CrossRef]
- Nóbrega, C.S.; Pauleta, S.R. Reduction of hydrogen peroxide in gram-negative bacteria-bacterial peroxidases. Adv. Microb. Physiol. 2019, 74, 415–464. [Google Scholar] [CrossRef]
- Pauleta, S.R.; Guerlesquin, F.; Goodhew, C.F.; Devreese, B.; Van Beeumen, J.; Pereira, A.S.; Moura, I.; Pettigrew, G.W. Paracoccus pantotrophus pseudoazurin is an electron donor to cytochrome c peroxidase. Biochemistry 2004, 43, 11214–11225. [Google Scholar] [CrossRef] [PubMed]
- Pauleta, S.R.; Cooper, A.; Nutley, M.; Errington, N.; Harding, S.; Guerlesquin, F.; Goodhew, C.F.; Moura, I.; Moura, J.J.; Pettigrew, G.W. A copper protein and a cytochrome bind at the same site on bacterial cytochrome c peroxidase. Biochemistry 2004, 43, 14566–14576. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, C.S.; Pauleta, S.R. Interaction between Neisseria gonorrhoeae bacterial peroxidase and its electron donor, the lipid-modified azurin. FEBS Lett. 2018, 592, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, P.M.; Pauleta, S.R.; Goncalves, M.L.; Pettigrew, G.W.; Moura, I.; Dos Santos, M.M.; Moura, J.J. Mediated catalysis of Paracoccus pantotrophus cytochrome c peroxidase by P. pantotrophus pseudoazurin: Kinetics of intermolecular electron transfer. J. Biol. Inorg. Chem. 2007, 12, 691–698. [Google Scholar] [CrossRef]
- Pettigrew, G.W.; Pauleta, S.R.; Goodhew, C.F.; Cooper, A.; Nutley, M.; Jumel, K.; Harding, S.E.; Costa, C.; Krippahl, L.; Moura, I.; et al. Electron transfer complexes of cytochrome c peroxidase from Paracoccus denitrificans containing more than one cytochrome. Biochemistry 2003, 42, 11968–11981. [Google Scholar] [CrossRef]
- Abe, T.; Kawarai, T.; Takahashi, Y.; Konishi, K. Enzymatic kinetics of the quinol peroxidase of an aggressive periodontopathic bacterium. J. Biochem. 2017, 161, 513–520. [Google Scholar] [CrossRef]
- Atack, J.M.; Kelly, D.J. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv. Microb. Physiol. 2007, 52, 73–106. [Google Scholar] [CrossRef]
- Balodite, E.; Strazdina, I.; Galinina, N.; McLean, S.; Rutkis, R.; Poole, R.K.; Kalnenieks, U. Structure of the Zymomonas mobilis respiratory chain: Oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiology 2014, 160, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Takashima, E.; Konishi, K. Molecular characterization of the membrane-bound quinol peroxidase functionally connected to the respiratory chain. FEBS J. 2007, 274, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Takashima, E.; Konishi, K. Characterization of a quinol peroxidase mutant in Aggregatibacter actinomycetemcomitans. FEMS Microbiol. Lett. 2008, 286, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Takashima, E.; Yamada, H.; Yamashita, T.; Matsushita, K.; Konishi, K. Recombinant expression and redox properties of triheme c membrane-bound quinol peroxidase. FEMS Microbiol. Lett. 2010, 302, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, C.S.; Raposo, M.; Van Driessche, G.; Devreese, B.; Pauleta, S.R. Biochemical characterization of the bacterial peroxidase from the human pathogen Neisseria gonorrhoeae. J. Inorg. Biochem. 2017, 171, 108–119. [Google Scholar] [CrossRef]
- Kaushik, S.; He, H.; Dalbey, R.E. Bacterial signal peptides-navigating the journey of proteins. Front. Physiol 2022, 13, 933153. [Google Scholar] [CrossRef]
- Nóbrega, C.S.; Carvalho, A.L.; Romão, M.J.; Pauleta, S.R. Structural characterization of Neisseria gonorrhoeae bacterial peroxidase—Insights into the catalytic cycle of bacterial peroxidases. Int. J. Mol. Sci. 2023, 24, 6246. [Google Scholar] [CrossRef]
- Iverson, T.M.; Luna-Chavez, C.; Croal, L.R.; Cecchini, G.; Rees, D.C. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J. Biol. Chem. 2002, 277, 16124–16130. [Google Scholar] [CrossRef]
- Hellwig, P.; Yano, T.; Ohnishi, T.; Gennis, R.B. Identification of the residues involved in stabilization of the semiquinone radical in the high-affinity ubiquinone binding site in cytochrome bo(3) from Escherichia coli by site-directed mutagenesis and EPR spectroscopy. Biochemistry 2002, 41, 10675–10679. [Google Scholar] [CrossRef]
- Tran, Q.M.; Rothery, R.A.; Maklashina, E.; Cecchini, G.; Weiner, J.H. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J. Biol. Chem. 2006, 281, 32310–32317. [Google Scholar] [CrossRef]
- De Smet, L.; Savvides, S.N.; Van Horen, E.; Pettigrew, G.; Van Beeumen, J.J. Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases. J. Biol. Chem. 2006, 281, 4371–4379. [Google Scholar] [CrossRef]
- Hsiao, H.C.; Boycheva, S.; Watmough, N.J.; Brittain, T. Activation of the cytochrome c peroxidase of Pseudomonas aeruginosa. The role of a heme-linked protein loop: A mutagenesis study. J. Inorg. Biochem. 2007, 101, 1133–1139. [Google Scholar] [CrossRef]
- Shimizu, H.; Schuller, D.J.; Lanzilotta, W.N.; Sundaramoorthy, M.; Arciero, D.M.; Hooper, A.B.; Poulos, T.L. Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases. Biochemistry 2001, 40, 13483–13490. [Google Scholar] [CrossRef]
- Echalier, A.; Goodhew, C.F.; Pettigrew, G.W.; Fulop, V. Activation and catalysis of the di-heme cytochrome c peroxidase from Paracoccus pantotrophus. Structure 2006, 14, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Echalier, A.; Brittain, T.; Wright, J.; Boycheva, S.; Mortuza, G.B.; Fulop, V.; Watmough, N.J. Redox-linked structural changes associated with the formation of a catalytically competent form of the diheme cytochrome c peroxidase from Pseudomonas aeruginosa. Biochemistry 2008, 47, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Fulop, V.; Ridout, C.J.; Greenwood, C.; Hajdu, J. Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa. Structure 1995, 3, 1225–1233. [Google Scholar] [CrossRef]
- Fülöp, V.; Watmough, N.J.; Ferguson, S.J. Structure and enzymology of two bacterial diheme enzymes: Cytochrome cd1 nitrite reductase and cytochrome c peroxidase. Adv. Inorg. Chem. 2001, 51, 163–204. [Google Scholar]
- Foote, N.; Peterson, J.; Gadsby, P.M.; Greenwood, C.; Thomson, A.J. Redox-linked spin-state changes in the di-haem cytochrome c-551 peroxidase from Pseudomonas aeruginosa. Biochem. J. 1985, 230, 227–237. [Google Scholar] [CrossRef]
- Reedy, C.J.; Elvekrog, M.M.; Gibney, B.R. Development of a heme protein structure-electrochemical function database. Nucleic Acids Res. 2008, 36, D307–D313. [Google Scholar] [CrossRef] [PubMed]
- Borsook, H.; Keighley, G. Oxidation-reduction potential of ascorbic acid (Vitamin C). Proc. Natl. Acad. Sci. USA 1933, 19, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Maiocco, S.J. Biophysical Characterization of Electron Transfer Proteins Containing Multiple Metallocofactors: Investigation of the AdoMet Radical and Cytochrome c Peroxidase Enzyme Superfamilies; Boston University: Boston, MA, USA, 2016. [Google Scholar]
- Reeder, B.J. The redox activity of hemoglobins: From physiologic functions to pathologic mechanisms. Antioxid. Redox Signal. 2010, 13, 1087–1123. [Google Scholar] [CrossRef] [PubMed]
- Belikova, N.A.; Vladimirov, Y.A.; Osipov, A.N.; Kapralov, A.A.; Tyurin, V.A.; Potapovich, M.V.; Basova, L.V.; Peterson, J.; Kurnikov, I.V.; Kagan, V.E. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry 2006, 45, 4998–5009. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Ronnberg, M.; Ellfolk, N.; Soininen, R. Circular dichroism studies on cytochrome c peroxidase and cytochrome c-551 of Pseudomonas aeruginosa. Biochim. Biophys. Acta 1979, 578, 392–400. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyaki, E.; Kun, J.; Moussong, E.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Klose, D.P.; Wallace, B.A.; Janes, R.W. 2Struc: The secondary structure server. Bioinformatics 2010, 26, 2624–2625. [Google Scholar] [CrossRef]
- Pettigrew, G.W.; Goodhew, C.F.; Cooper, A.; Nutley, M.; Jumel, K.; Harding, S.E. The electron transfer complexes of cytochrome c peroxidase from Paracoccus denitrificans. Biochemistry 2003, 42, 2046–2055. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Bernhofer, M.; Dallago, C.; Karl, T.; Satagopam, V.; Heinzinger, M.; Littmann, M.; Olenyi, T.; Qiu, J.; Schutze, K.; Yachdav, G.; et al. PredictProtein-Predicting Protein Structure and Function for 29 Years. Nucleic Acids Res. 2021, 49, W535–W540. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A Brief History of Protein Sorting Prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pauleta, S.R.; Lu, Y.; Goodhew, C.F.; Moura, I.; Pettigrew, G.W.; Shelnutt, J.A. Calcium-dependent conformation of a heme and fingerprint peptide of the diheme cytochrome c peroxidase from Paracoccus pantotrophus. Biochemistry 2001, 40, 6570–6579. [Google Scholar] [CrossRef]
- Pauleta, S.R.; Lu, Y.; Goodhew, C.F.; Moura, I.; Pettigrew, G.W.; Shelnutt, J.A. Calcium-dependent heme structure in the reduced forms of the bacterial cytochrome c peroxidase from Paracoccus pantotrophus. Biochemistry 2008, 47, 5841–5850. [Google Scholar] [CrossRef]
- Arslan, E.; Schulz, H.; Zufferey, R.; Kunzler, P.; Thony-Meyer, L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 1998, 251, 744–747. [Google Scholar] [CrossRef]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Barr, I.; Guo, F. Pyridine hemochromagen assay for determining the concentration of heme in purified protein solutions. Bio-Protocol 2015, 5, e1594. [Google Scholar] [CrossRef] [PubMed]
- Anthis, N.J.; Clore, G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013, 22, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Giordani, R.; Buc, J. Evidence for two different electron transfer pathways in the same enzyme, nitrate reductase A from Escherichia coli. Eur. J. Biochem. 2004, 271, 2400–2407. [Google Scholar] [CrossRef] [PubMed]

| YhjA | YhjA M82A | YhjA M125A | YhjA H134A | |
|---|---|---|---|---|
| ABTS2− | 110 ± 3 | 111 ± 12 | 68 ± 14 | 110 ± 18 |
| Hydroquinone | 54.6 ± 0.1 | 54 ± 13 | 32 ± 11 | 56 ± 13 |
| Data from BeStSel/DichroWeb or 2Struc | |||
|---|---|---|---|
| α-Helix (%) | β-Sheet | Other | |
| YhjA WT | 31/36 | 14/14 | 55/50 |
| YhjA structural model | 40–50 | 1–14 | 46–50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, R.N.S.; de Aguiar, S.R.M.M.; Pauleta, S.R. Coordination of the N-Terminal Heme in the Non-Classical Peroxidase from Escherichia coli. Molecules 2023, 28, 4598. https://doi.org/10.3390/molecules28124598
Oliveira RNS, de Aguiar SRMM, Pauleta SR. Coordination of the N-Terminal Heme in the Non-Classical Peroxidase from Escherichia coli. Molecules. 2023; 28(12):4598. https://doi.org/10.3390/molecules28124598
Chicago/Turabian StyleOliveira, Ricardo N. S., Sara R. M. M. de Aguiar, and Sofia R. Pauleta. 2023. "Coordination of the N-Terminal Heme in the Non-Classical Peroxidase from Escherichia coli" Molecules 28, no. 12: 4598. https://doi.org/10.3390/molecules28124598
APA StyleOliveira, R. N. S., de Aguiar, S. R. M. M., & Pauleta, S. R. (2023). Coordination of the N-Terminal Heme in the Non-Classical Peroxidase from Escherichia coli. Molecules, 28(12), 4598. https://doi.org/10.3390/molecules28124598








