Study of MDM2 as Prognostic Biomarker in Brain-LGG Cancer and Bioactive Phytochemicals Inhibit the p53-MDM2 Pathway: A Computational Drug Development Approach
Abstract
1. Introduction
2. Results
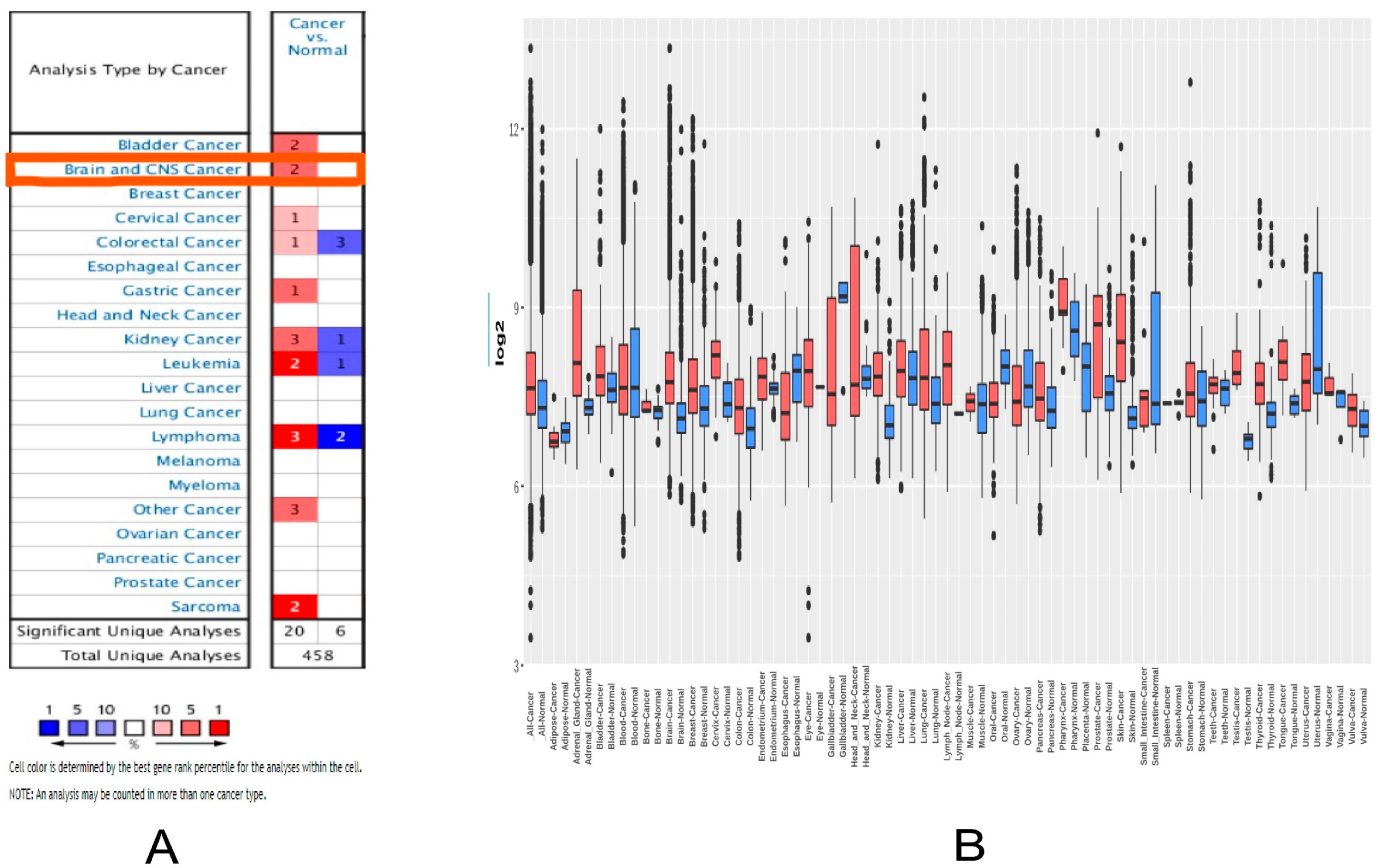
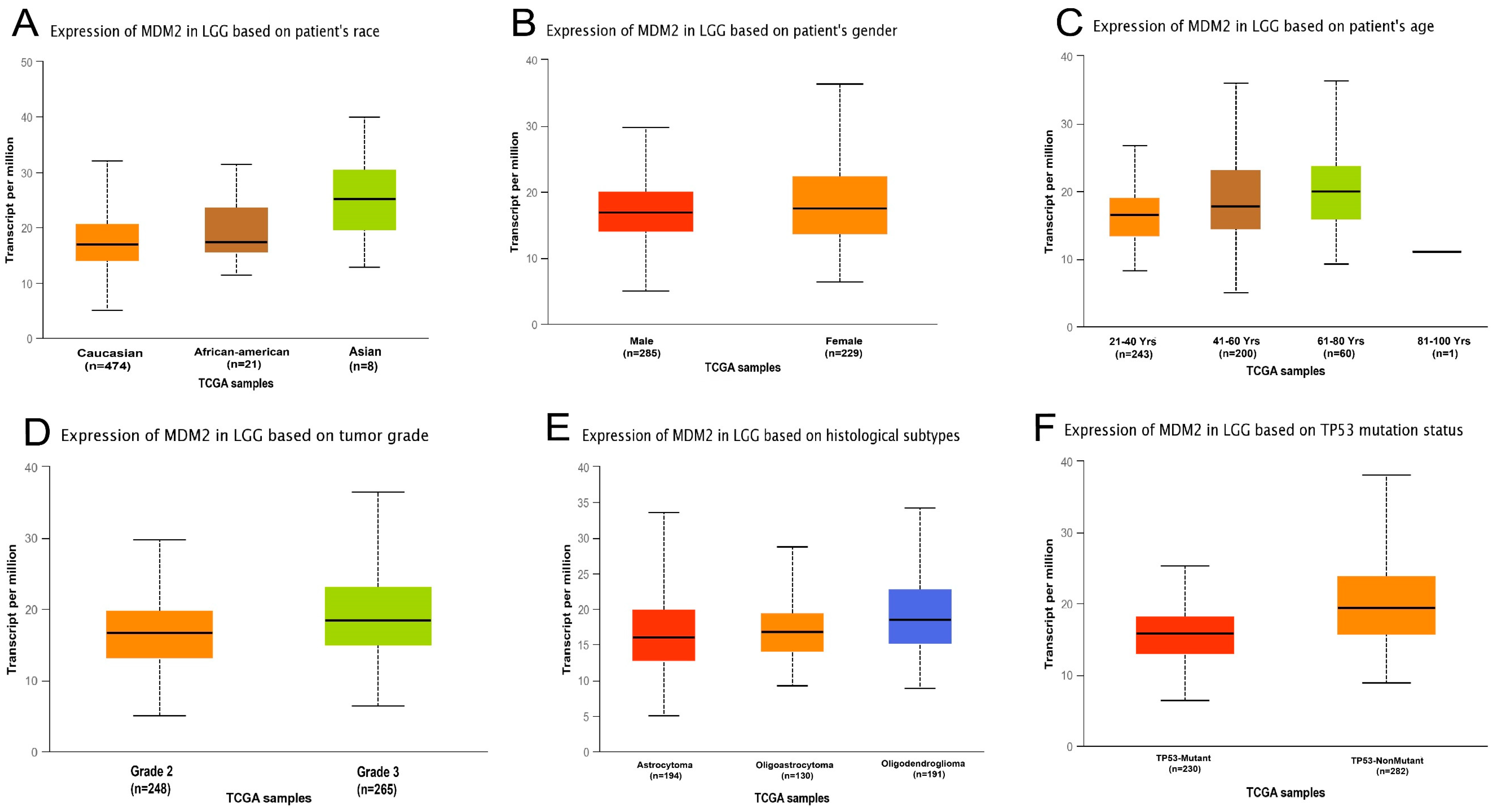
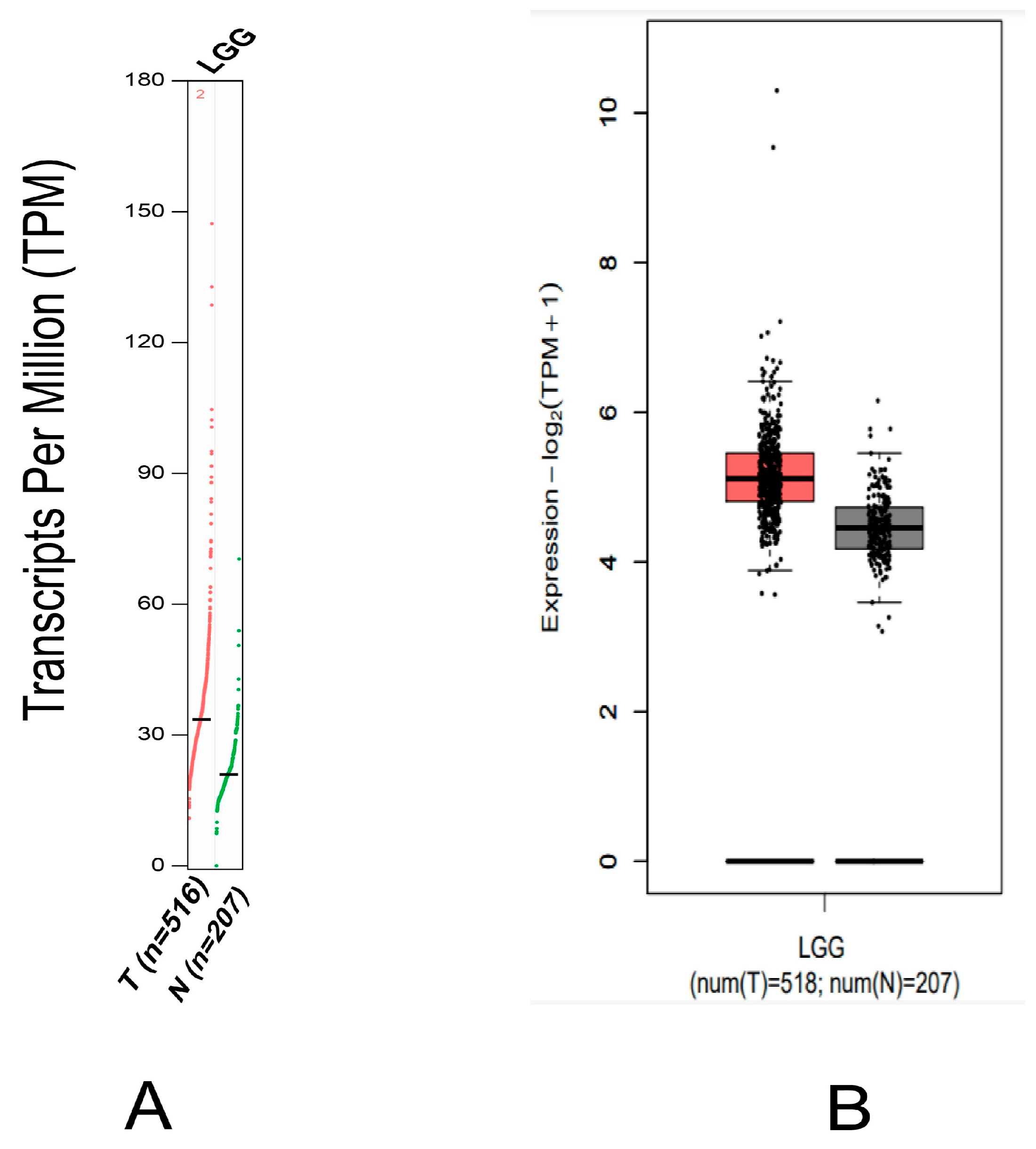
2.1. MDM2 Gene Expression Analysis
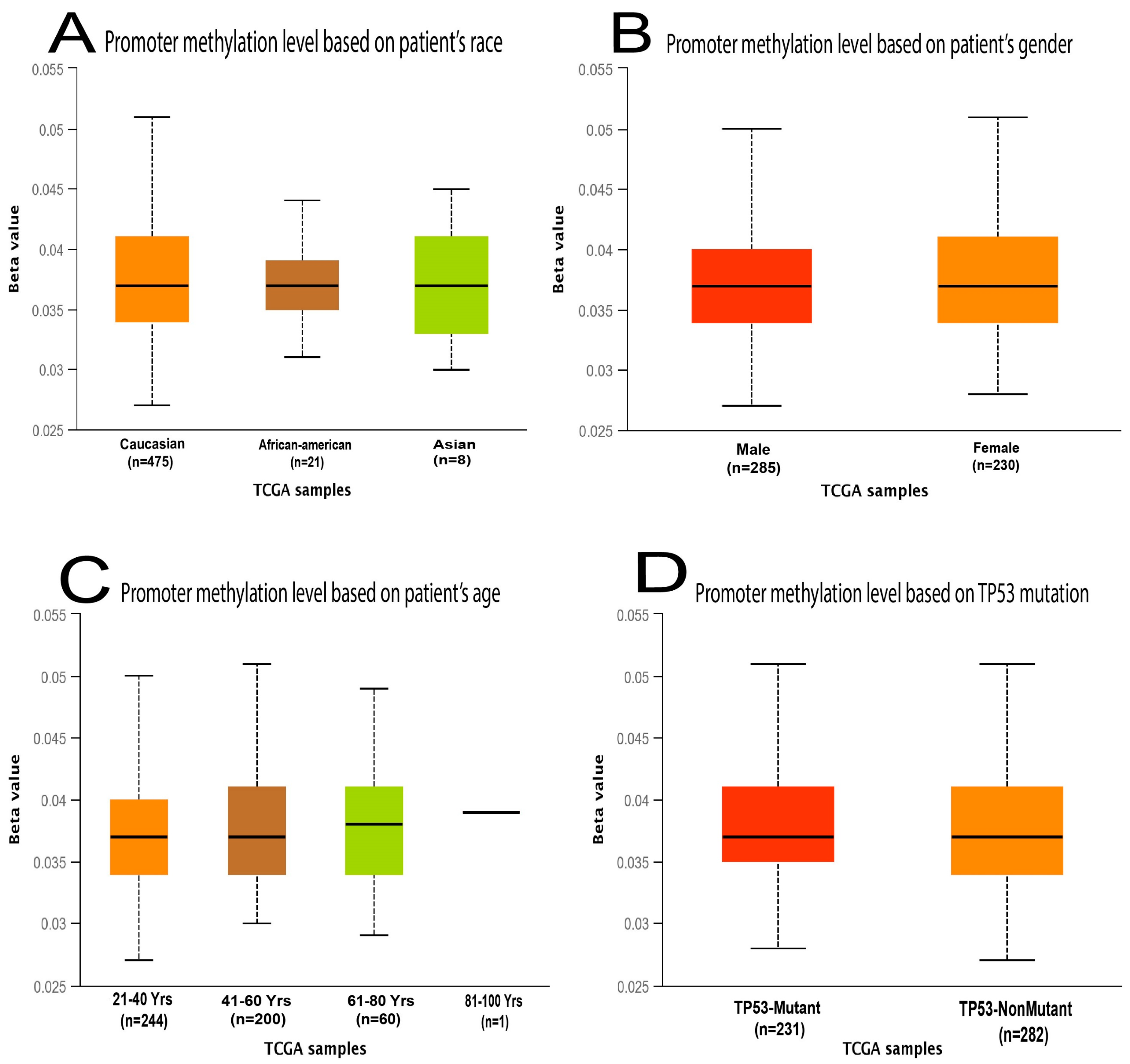
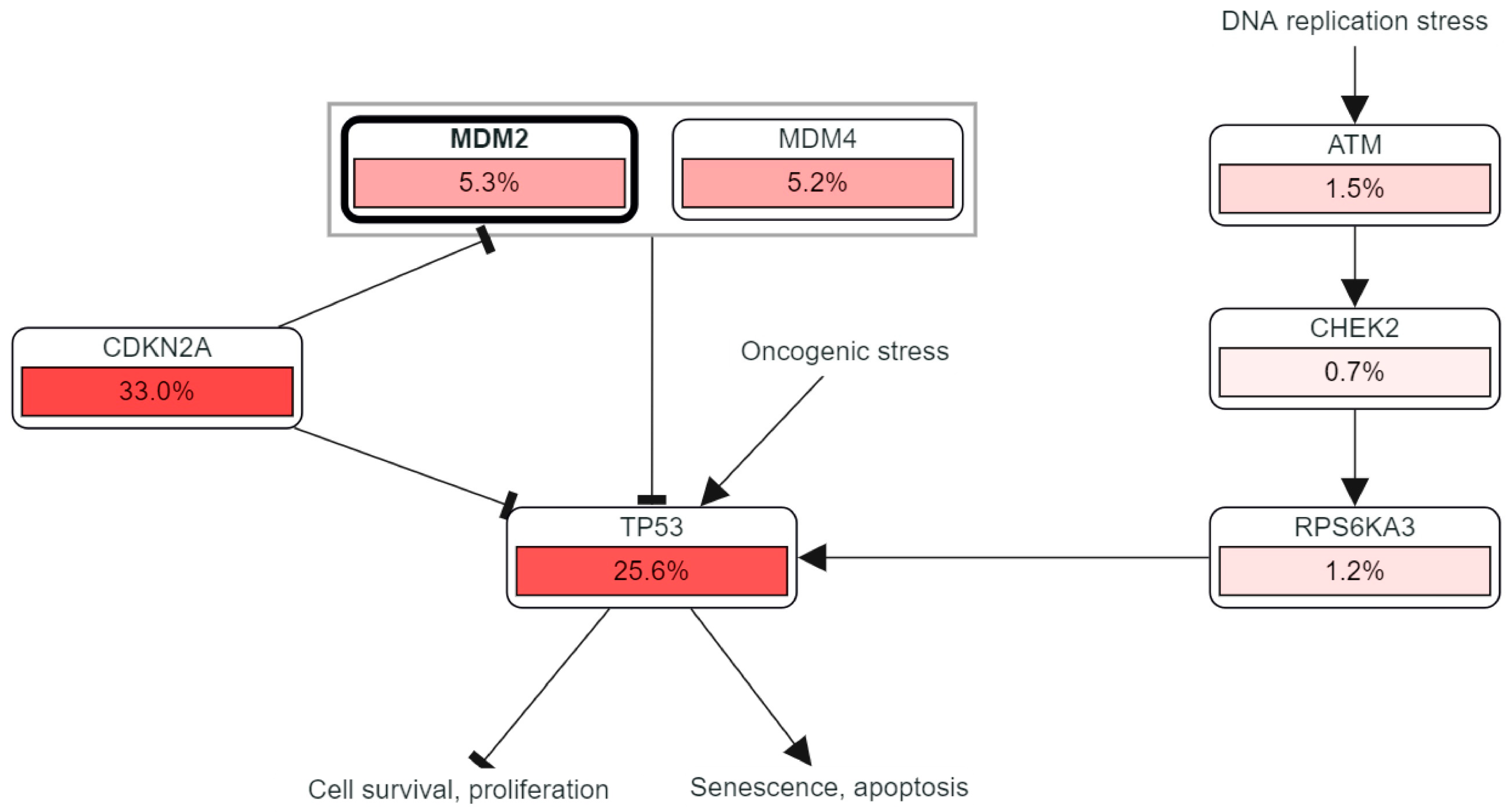
2.2. Genetic Mutations and Copy Number Alterations (CNAs) Analysis of MDM2 Genomic Sequences Correlated with Brain Cancer Development
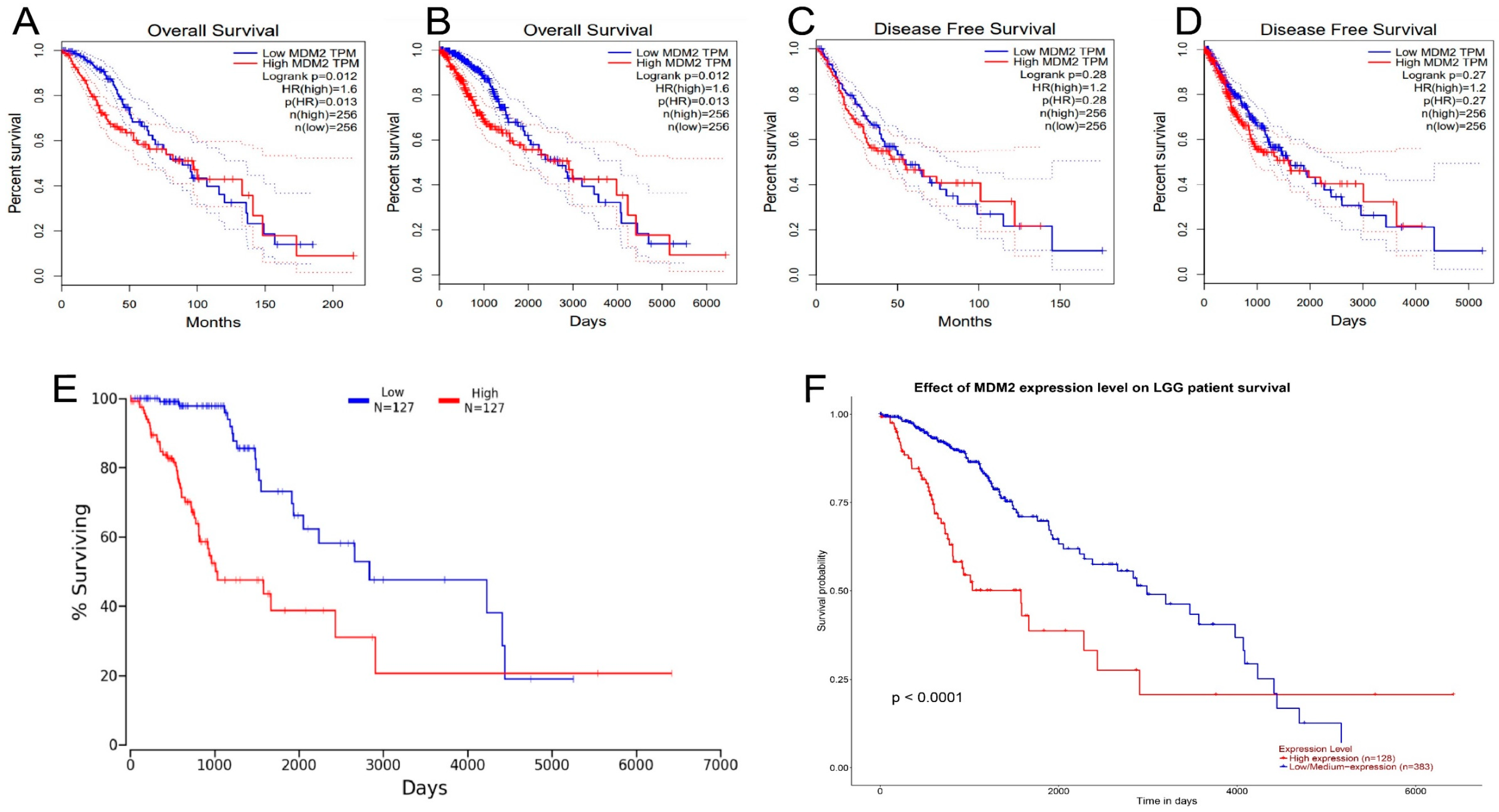
2.3. The Analysis of Prognostic Value and Survival Assay of MDM2 Gene
2.4. Study of Correlated Genes, and PIP Network
2.5. ADMET Profiling
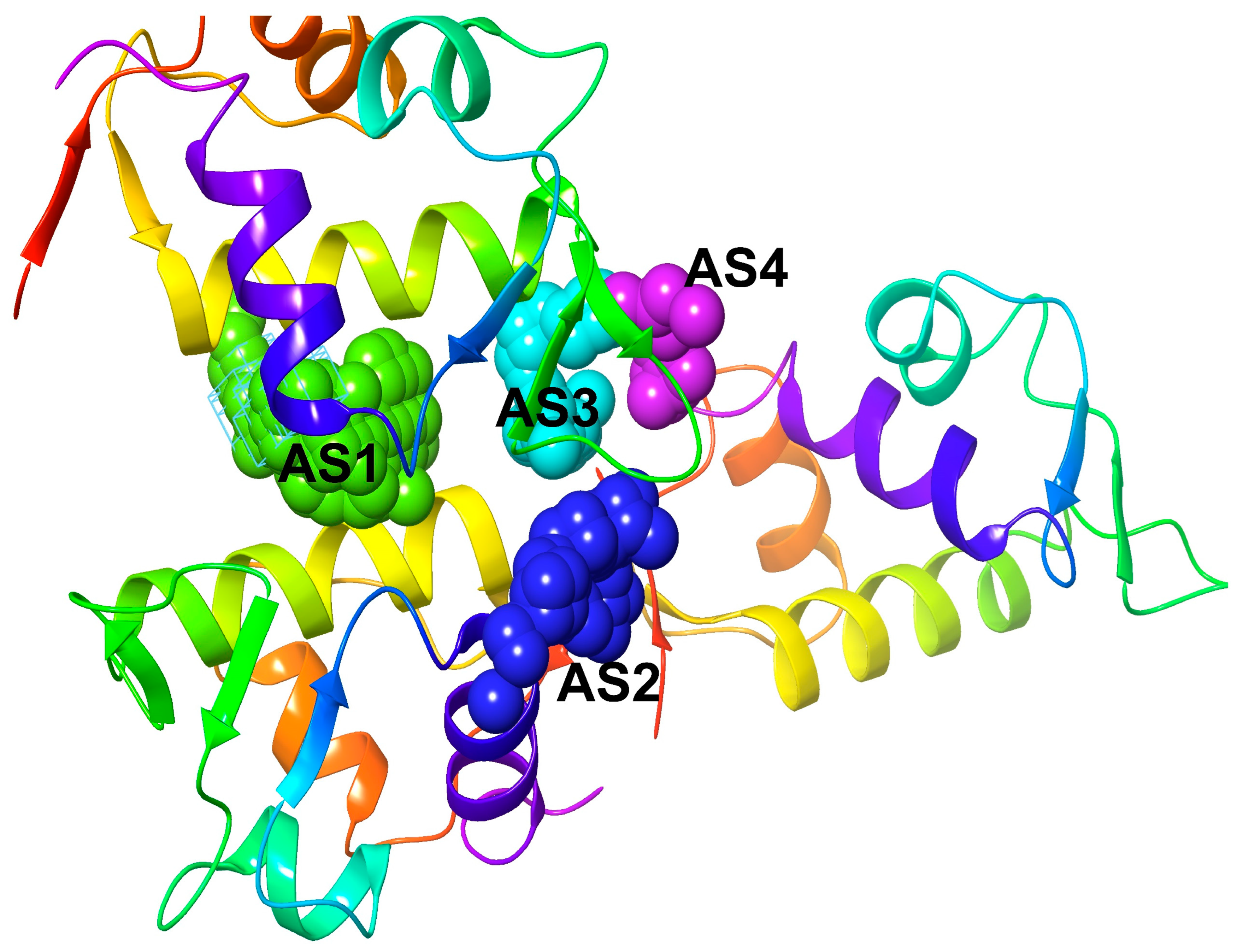
2.6. Active Site Identification and Generation of Receptor Grid
2.7. Interpretation of Molecular Docking
2.8. Visualization of Post-Docking Protein-Ligands Interactions
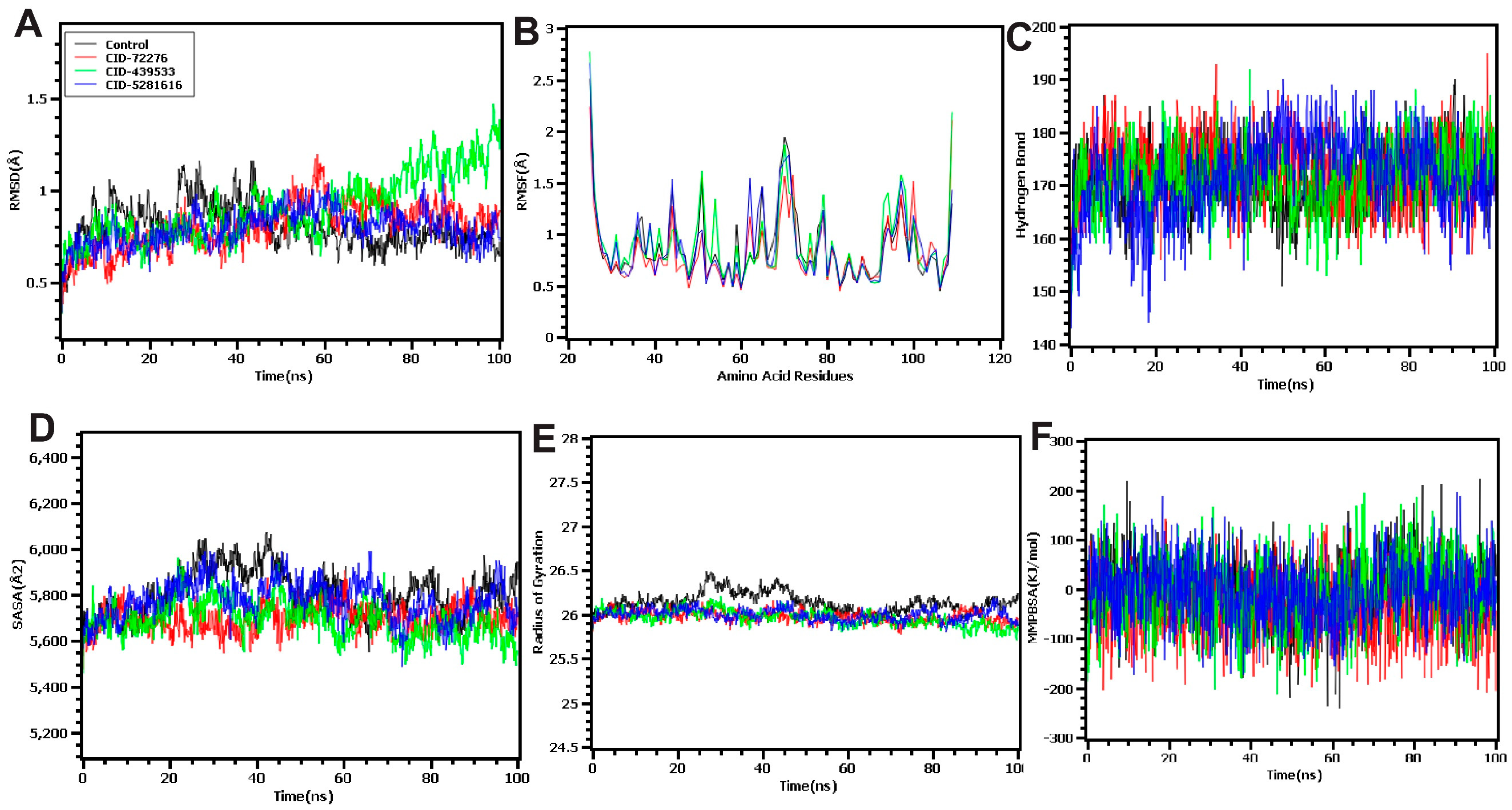
2.9. Molecular Dynamics Simulation (MDS) Analysis
2.9.1. RMSD Analysis
2.9.2. RMSF Study
2.9.3. Hydrogen Bond Analysis
2.9.4. Analysis of SASA Value
2.9.5. Study of Rg
2.9.6. Analysis of MM-PBSA Value
3. Discussion
4. Materials and Methods
4.1. MDM2 Gene Expression in Brain Cancer Research
4.2. Determination Copy Number Alterations and Mutation of MDM2 Gene
4.3. Survival Data Analysis
4.4. Analysis of Correlation and Interaction Networks
4.5. Compounds Library Preparation and ADMET Screening for Selection of the Lead Compounds
4.6. Retrieval and Preparation of Compounds
4.7. Protein Retrieval and Preparation
4.8. Active Site Prediction and Generation of Receptor Grid
4.9. Site Specific Super Molecular Docking
4.10. Post-Docking Protein-Ligands Interactions Visualization
4.11. Molecular Dynamics Simulation (MDS)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hayman, L.; Chaudhry, W.R.; Revin, V.V.; Zhelev, N.; Bourdon, J.C. What is the potential of p53 isoforms as a predictive biomarker in the treatment of cancer? Expert Rev. Mol. Diagn. 2019, 19, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Lee, S.H.; Kim, D.H.; Ahn, M.J.; Kim, J.S.; Woo, J.Y.; Torizawa, T.; Kainosho, M.; Han, K.H. Structural details on mdm2-p53 interaction. J. Biol. Chem. 2005, 280, 38795–38802. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Zilani, M.N.H.; Islam, M.A.; Biswas, P.; Anisuzzman, M.; Hossain, H.; Shilpi, J.A.; Hasan, M.N.; Hossain, M.G. Metabolite profiling, anti-inflammatory, analgesic potentials of edible herb Colocasia gigantea and molecular docking study against COX-II enzyme. J. Ethnopharmacol. 2021, 281, 114577. [Google Scholar] [CrossRef]
- Bouchet, B.P.; Caron de Fromentel, C.; Puisieux, A.; Galmarini, C.M. p53 as a target for anti-cancer drug development. Crit. Rev. Oncol. Hematol. 2006, 58, 190–207. [Google Scholar] [CrossRef]
- Paul, P.; Biswas, P.; Dey, D.; Saikat, A.S.M.; Islam, M.A.; Sohel, M.; Hossain, R.; Mamun, A.A.; Rahman, M.A.; Hasan, M.N.; et al. Exhaustive Plant Profile of “Dimocarpus longan Lour” with Significant Phytomedicinal Properties: A Literature Based-Review. Processes 2021, 9, 1803. [Google Scholar] [CrossRef]
- Nayak, S.K.; Panesar, P.S.; Kumar, H. p53-Induced apoptosis and inhibitors of p53. Curr. Med. Chem. 2009, 16, 2627–2640. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, M.D.H.; Hossain, M.S.; Biswas, P.; Islam, R.; Uddin, M.J.; Rahman, M.H.; Rhim, H. Molecular Insights into the Multifunctional Role of Natural Compounds: Autophagy Modulation and Cancer Prevention. Biomedicines 2020, 8, 517. [Google Scholar] [CrossRef]
- Wang, W.; Hu, Y. Small molecule agents targeting the p53-MDM2 pathway for cancer therapy. Med. Res. Rev. 2012, 32, 1159–1196. [Google Scholar] [CrossRef]
- Zhang, Q.; Zeng, S.X.; Lu, H. Targeting p53-MDM2-MDMX loop for cancer therapy. Subcell. Biochem. 2014, 85, 281–319. [Google Scholar] [CrossRef]
- Shvarts, A.; Steegenga, W.T.; Riteco, N.; van Laar, T.; Dekker, P.; Bazuine, M.; van Ham, R.C.; van der Houven van Oordt, W.; Hateboer, G.; van der Eb, A.J.; et al. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996, 15, 5349–5357. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, M.H.; Biswas, P.; Hossain, M.S.; Islam, R.; Hannan, M.A.; Uddin, M.J.; Rhim, H. Potential Therapeutic Role of Phytochemicals to Mitigate Mitochondrial Dysfunctions in Alzheimer’s Disease. Antioxidants 2020, 10, 23. [Google Scholar] [CrossRef]
- Fu, T.; Min, H.; Xu, Y.; Chen, J.; Li, G. Molecular dynamic simulation insights into the normal state and restoration of p53 function. Int. J. Mol. Sci. 2012, 13, 9709–9740. [Google Scholar] [CrossRef]
- Arefin, A.; Ismail Ema, T.; Islam, T.; Hossen, S.; Islam, T.; Al Azad, S.; Uddin Badal, N.; Islam, A.; Biswas, P.; Alam, N.U.; et al. Target specificity of selective bioactive compounds in blocking α-dystroglycan receptor to suppress Lassa virus infection: An in silico approach. J. Biomed. Res. 2021, 35, 459–473. [Google Scholar] [CrossRef]
- Bibi, S.; Hasan, M.M.; Biswas, P.; Shkodina, A.; Shah, M.A.; Shah, G.M.; Khan, A.; Al-Harrasi, A. Chapter 7—Phytonutrients in the management of lipids metabolism. In The Role of Phytonutrients in Metabolic Disorders; Khan, H., Akkol, E.K., Daglia, M., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 195–236. [Google Scholar]
- Pinzi, L.; Caporuscio, F.; Rastelli, G. Selection of protein conformations for structure-based polypharmacology studies. Drug Discov. Today 2018, 23, 1889–1896. [Google Scholar] [CrossRef]
- Pei, D.; Zhang, Y.; Zheng, J. Regulation of p53: A collaboration between Mdm2 and Mdmx. Oncotarget 2012, 3, 228–235. [Google Scholar] [CrossRef]
- Biswas, P.; Dey, D.; Rahman, A.; Islam, M.A.; Susmi, T.F.; Kaium, M.A.; Hasan, M.N.; Rahman, M.D.H.; Mahmud, S.; Saleh, M.A.; et al. Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation. J. Pers. Med. 2021, 11, 888. [Google Scholar] [CrossRef]
- Dey, D.; Hossain, R.; Biswas, P.; Paul, P.; Islam, M.A.; Ema, T.I.; Gain, B.K.; Hasan, M.M.; Bibi, S.; Islam, M.T.; et al. Amentoflavone derivatives significantly act towards the main protease (3CL(PRO)/M(PRO)) of SARS-CoV-2: In silico admet profiling, molecular docking, molecular dynamics simulation, network pharmacology. Mol. Divers. 2022, 1–15. [Google Scholar] [CrossRef]
- Liu, E.T.; Lemberger, T. Higher order structure in the cancer transcriptome and systems medicine. Mol. Syst. Biol. 2007, 3, 94. [Google Scholar] [CrossRef]
- McMurray, H.R.; Sampson, E.R.; Compitello, G.; Kinsey, C.; Newman, L.; Smith, B.; Chen, S.R.; Klebanov, L.; Salzman, P.; Yakovlev, A.; et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 2008, 453, 1112–1116. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef] [PubMed]
- Laird, P.W.; Jaenisch, R. The role of DNA methylation in cancer genetic and epigenetics. Annu. Rev. Genet. 1996, 30, 441–464. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv. Exp. Med. Biol. 2003, 532, 39–49. [Google Scholar] [CrossRef]
- Schiebe, M.; Ohneseit, P.; Hoffmann, W.; Meyermann, R.; Rodemann, H.P.; Bamberg, M. Analysis of mdm2 and p53 gene alterations in glioblastomas and its correlation with clinical factors. J. Neurooncol. 2000, 49, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.R.; Lee, M. Estimating the Prognosis of Low-Grade Glioma with Gene Attention Using Multi-Omics and Multi-Modal Schemes. Biology 2022, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Rayan, A.; Raiyn, J.; Falah, M. Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef]
- Khan, M.S.; Mehmood, B.; Yousafi, Q.; Bibi, S.; Fazal, S.; Saleem, S.; Sajid, M.W.; Ihsan, A.; Azhar, M.; Kamal, M.A. Molecular Docking Studies Reveal Rhein from rhubarb (Rheum rhabarbarum) as a Putative Inhibitor of ATP-binding Cassette Super-family G member 2. Med. Chem. 2021, 17, 273–288. [Google Scholar] [CrossRef]
- Fridlender, M.; Kapulnik, Y.; Koltai, H. Plant derived substances with anti-cancer activity: From folklore to practice. Front. Plant Sci. 2015, 6, 799. [Google Scholar] [CrossRef]
- Caputi, L.; Franke, J.; Farrow, S.C.; Chung, K.; Payne, R.M.E.; Nguyen, T.D.; Dang, T.T.; Soares Teto Carqueijeiro, I.; Koudounas, K.; Dugé de Bernonville, T.; et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360, 1235–1239. [Google Scholar] [CrossRef]
- Slichenmyer, W.J.; Von Hoff, D.D. Taxol: A new and effective anti-cancer drug. Anticancer Drugs 1991, 2, 519–530. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Ettinger, D.S. The Role of Topotecan in the Treatment of Small Cell Lung Cancer. Oncologist 1998, 3, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.A.; Sayed, S.; Bello, M.; Hussain, N.; Chando, R.K.; Alam, S.; Hasan, M.K. CDK4 as a phytochemical based anticancer drug target. Inform. Med. Unlocked 2022, 28, 100826. [Google Scholar] [CrossRef]
- Singh, J.; Sangwan, N.; Chauhan, A.; Avti, P.K. Integrative network and computational simulation of clinical and genomic data for the identification of mutated EGFR in breast cancer patients for therapeutic targeting using purine analogues. Mol. Simul. 2022, 48, 1548–1560. [Google Scholar] [CrossRef]
- Malik, S. Biotechnology and Production of Anti-Cancer Compounds; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; Volume 919. [Google Scholar]
- Hamedi, A.; Bayat, M.; Asemani, Y.; Amirghofran, Z. A review of potential anti-cancer properties of some selected medicinal plants grown in Iran. J. Herb. Med. 2022, 33, 100557. [Google Scholar] [CrossRef]
- Ferdausi, N.; Islam, S.; Rimti, F.H.; Quayum, S.T.; Arshad, E.M.; Ibnat, A.; Islam, T.; Arefin, A.; Ema, T.I.; Biswas, P.; et al. Point-specific interactions of isovitexin with the neighboring amino acid residues of the hACE2 receptor as a targeted therapeutic agent in suppressing the SARS-CoV-2 influx mechanism. J. Adv. Vet. Anim. Res. 2022, 9, 230–240. [Google Scholar] [CrossRef]
- Siddiqui, A.J.; Jahan, S.; Singh, R.; Saxena, J.; Ashraf, S.A.; Khan, A.; Choudhary, R.K.; Balakrishnan, S.; Badraoui, R.; Bardakci, F.; et al. Plants in Anticancer Drug Discovery: From Molecular Mechanism to Chemoprevention. Biomed Res. Int. 2022, 2022, 5425485. [Google Scholar] [CrossRef]
- Hasan, A.; Biswas, P.; Bondhon, T.A.; Jannat, K.; Paul, T.K.; Paul, A.K.; Jahan, R.; Nissapatorn, V.; Mahboob, T.; Wilairatana, P.; et al. Can Artemisia herba-alba Be Useful for Managing COVID-19 and Comorbidities? Molecules 2022, 27, 492. [Google Scholar] [CrossRef]
- Hossain, R.; Dey, D.; Biswas, P.; Paul, P.; Ahmed, S.Z.; Khan, A.A.; Ema, T.I.; Islam, M.T. Chlorophytum borivilianum (Musli) and Cimicifuga racemosa (Black Cohosh). In Herbs, Shrubs, and Trees of Potential Medicinal Benefits; CRC Press: Boca Raton, FL, USA, 2022; pp. 45–82. [Google Scholar]
- Halgren, T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Fu, C.; Deng, S.; Koneski, I.; Awad, M.M.; Akram, Z.; Matinlinna, J.; Pichika, M.R.; Daood, U.; Fawzy, A.S. Multiscale in-vitro analysis of photo-activated riboflavin incorporated in an experimental universal adhesive. J. Mech. Behav. Biomed. Mater. 2020, 112, 104082. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Ojo, O.A.; Ojo, A.B.; Okolie, C.; Nwakama, M.C.; Iyobhebhe, M.; Evbuomwan, I.O.; Nwonuma, C.O.; Maimako, R.F.; Adegboyega, A.E.; Taiwo, O.A.; et al. Deciphering the Interactions of Bioactive Compounds in Selected Traditional Medicinal Plants against Alzheimer’s Diseases via Pharmacophore Modeling, Auto-QSAR, and Molecular Docking Approaches. Molecules 2021, 26, 1996. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Dubey, A.; Yadava, U.; Mishra, S.K.; Kang, S.G.; Dwivedi, V.D. Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 Mpro. Brief. Bioinform. 2021, 22, 1361–1377. [Google Scholar] [CrossRef]
- Opo, F.; Rahman, M.M.; Ahammad, F.; Ahmed, I.; Bhuiyan, M.A.; Asiri, A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021, 11, 4049. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Carr-Wilkinson, J.; O’Toole, K.; Wood, K.M.; Challen, C.C.; Baker, A.G.; Board, J.R.; Evans, L.; Cole, M.; Cheung, N.K.; Boos, J.; et al. High Frequency of p53/MDM2/p14ARF Pathway Abnormalities in Relapsed Neuroblastoma. Clin. Cancer Res. 2010, 16, 1108–1118. [Google Scholar] [CrossRef]
- Biswas, P.; Dey, D.; Biswas, P.K.; Rahaman, T.I.; Saha, S.; Parvez, A.; Khan, D.A.; Lily, N.J.; Saha, K.; Sohel, M.; et al. A Comprehensive Analysis and Anti-Cancer Activities of Quercetin in ROS-Mediated Cancer and Cancer Stem Cells. Int. J. Mol. Sci. 2022, 23, 1746. [Google Scholar] [CrossRef]
- Onel, K.; Cordon-Cardo, C. MDM2 and prognosis. Mol. Cancer Res. 2004, 2, 1–8. [Google Scholar] [CrossRef]
- Deben, C.; Deschoolmeester, V.; Lardon, F.; Rolfo, C.; Pauwels, P. TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit. Rev. Oncol. Hematol. 2016, 99, 63–73. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Zeng, Z.; Yang, Y.; Qing, C.; Hu, Z.; Huang, Y.; Zhou, C.; Li, D.; Jiang, Y. Distinct expression and prognostic value of members of SMAD family in non-small cell lung cancer. Medicine 2020, 99, e19451. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lánczky, A.; Szállási, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Z.; Jin, J.; Long, Z.; Liu, X.; Cai, H.; Zhou, Y.; Huang, H.; Wang, Y. MDM2 binding protein as a predictor of metastasis and a novel prognostic biomarker in patients with gastric cancer. Oncol. Lett. 2017, 14, 6409–6416. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, L.; Jiang, Y.; Hu, T.; Zhang, D.; Qiao, Q.; Wang, R.; Wang, M.; Han, S. MTBP regulates cell survival and therapeutic sensitivity in TP53 wildtype glioblastomas. Theranostics 2019, 9, 6019–6030. [Google Scholar] [CrossRef]
- Inoue, R.; Moghaddam, K.A.; Ranasinghe, M.; Saeki, Y.; Chiocca, E.A.; Wade-Martins, R. Infectious delivery of the 132 kb CDKN2A/CDKN2B genomic DNA region results in correctly spliced gene expression and growth suppression in glioma cells. Gene Ther. 2004, 11, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, M.J.; Büschges, R.; Wolter, M.; Reifenberger, J.; Boström, J.; Kraus, J.A.; Schlegel, U.; Reifenberger, G. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999, 59, 6091–6096. [Google Scholar] [PubMed]
- Xiong, Y.; Zhang, Y.; Xiong, S.; Williams-Villalobo, A.E. A Glance of p53 Functions in Brain Development, Neural Stem Cells, and Brain Cancer. Biology 2020, 9, 285. [Google Scholar] [CrossRef]
- Bibi, S.; Sakata, K. Current Status of Computer-Aided Drug Design for Type 2 Diabetes. Curr. Comput. Aided Drug Des. 2016, 12, 167–177. [Google Scholar] [CrossRef]
- Dey, D.; Biswas, P.; Paul, P.; Mahmud, S.; Ema, T.I.; Khan, A.A.; Ahmed, S.Z.; Hasan, M.M.; Saikat, A.S.M.; Fatema, B.; et al. Natural flavonoids effectively block the CD81 receptor of hepatocytes and inhibit HCV infection: A computational drug development approach. Mol. Divers. 2022. [Google Scholar] [CrossRef]
- Sohel, M.; Biswas, P.; Al Amin, M.; Hossain, M.A.; Sultana, H.; Dey, D.; Aktar, S.; Setu, A.; Khan, M.S.; Paul, P.; et al. Genistein, a Potential Phytochemical against Breast Cancer Treatment-Insight into the Molecular Mechanisms. Processes 2022, 10, 415. [Google Scholar] [CrossRef]
- Ahmed, H.; Mahmud, A.R.; Siddiquee, M.F.R.; Shahriar, A.; Biswas, P.; Shimul, M.E.K.; Ahmed, S.Z.; Ema, T.I.; Rahman, N.; Khan, M.A.; et al. Role of T cells in cancer immunotherapy: Opportunities and challenges. Cancer Pathog. Ther. 2022, 1, E003. [Google Scholar] [CrossRef]
- Al Azad, S.; Ahmed, S.; Biswas, P.; Mia, M.A.R.; Farjana, M.; Arshe, F.A.; Mily, S.J.; Ankhi, A.B.; Shaikat, M.M.; Sultana10, S. Quantitative analysis of the factors influencing IDA and TSH downregulation in correlation to the fluctuation of activated vitamin D3 in women. J. Adv. Biotechnol. Exp. Ther. 2022, 5, 320–333. [Google Scholar] [CrossRef]
- Paul, P.K.; Al Azad, S.; Rahman, M.H.; Farjana, M.; Uddin, M.R.; Dey, D.; Mahmud, S.; Ema, T.I.; Biswas, P.; Anjum, M.; et al. Catabolic profiling of selective enzymes in the saccharification of non-food lignocellulose parts of biomass into functional edible sugars and bioenergy: An in silico bioprospecting. J. Adv. Vet. Anim. Res. 2022, 9, 19–32. [Google Scholar] [CrossRef]
- Mannhold, R.; Kubinyi, H.; Folkers, G. Pharmacokinetics and Metabolism in Drug Design; John Wiley & Sons: Hoboken, NJ, USA, 2012; Volume 51. [Google Scholar]
- Notari, R.E. Pharmacokinetics and molecular modification: Implications in drug design and evaluation. J. Pharm. Sci. 1973, 62, 865–881. [Google Scholar] [CrossRef]
- Biswas, P.; Hasan, M.M.; Dey, D.; Dos Santos Costa, A.C.; Polash, S.A.; Bibi, S.; Ferdous, N.; Kaium, M.A.; Rahman, M.D.H.; Jeet, F.K.; et al. Candidate antiviral drugs for COVID-19 and their environmental implications: A comprehensive analysis. Environ. Sci. Pollut. Res. Int. 2021, 28, 59570–59593. [Google Scholar] [CrossRef]
- Matin, M.M.; Roshid, M.H.; Bhattacharjee, S.C.; Azad, A.K. PASS predication, antiviral, in vitro Antimicrobial, and ADMET studies of rhamnopyranoside esters. Med. Res. Arch. 2020, 8. [Google Scholar] [CrossRef]
- Rahman, M.S.; Zilani, M.N.H.; Islam, M.A.; Hasan, M.M.; Islam, M.M.; Yasmin, F.; Biswas, P.; Hirashima, A.; Rahman, M.A.; Hasan, M.N.; et al. In Vivo Neuropharmacological Potential of Gomphandra tetrandra (Wall.) Sleumer and In-Silico Study against β-Amyloid Precursor Protein. Processes 2021, 9, 1449. [Google Scholar] [CrossRef]
- Al Saber, M.; Biswas, P.; Dey, D.; Kaium, M.A.; Islam, M.A.; Tripty, M.I.A.; Rahman, M.H.; Rahaman, T.I.; Biswas, M.Y.; Paul, P.; et al. A Comprehensive Review of Recent Advancements in Cancer Immunotherapy and Generation of CAR T Cell by CRISPR-Cas9. Processes 2022, 10, 16. [Google Scholar] [CrossRef]
- Munshi, M.; Zilani, M.N.H.; Islam, M.A.; Biswas, P.; Das, A.; Afroz, F.; Hasan, M.N. Novel compounds from endophytic fungi of Ceriops decandra inhibit breast cancer cell growth through estrogen receptor alpha in in-silico study. Inform. Med. Unlocked 2022, 32, 101046. [Google Scholar] [CrossRef]
- Aziz, S.; Bibi, S.; Hasan, M.M.; Biswas, P.; Ali, M.I.; Bilal, M.; Chopra, H.; Mukerjee, N.; Maitra, S. A review on influence of biochar amendment on soil processes and environmental remediation. Biotechnol. Genet. Eng. Rev. 2023, 1–35. [Google Scholar] [CrossRef]
- Biswas, P.; Polash, S.A.; Dey, D.; Kaium, M.A.; Mahmud, A.R.; Yasmin, F.; Baral, S.K.; Islam, M.A.; Rahaman, T.I.; Abdullah, A.; et al. Advanced implications of nanotechnology in disease control and environmental perspectives. Biomed. Pharmacother. 2023, 158, 114172. [Google Scholar] [CrossRef]
- Hasan, M.M.; Zilani, M.N.H.; Akter, S.; Nasrin, P.; Shajib, G.M.A.; Islam, M.A.; Biswas, P.; Mahmud, S.; Saleh, M.A.; Hasan, M.N.; et al. UHPLC-Q/Orbitrap/MS based chemical fingerprinting and hepatoprotective potential of a medicinal plant, Morinda angustifolia Roxb. S. Afr. J. Bot. 2022, 148, 561–572. [Google Scholar] [CrossRef]
- Sarker, M.T.; Saha, S.; Biswas, P.; Islam, M.T.; Sheikh, M.A.; Hasan, M.N.; Islam, N.; Rabbe, M.M.I.; Rafi, M.O. Identification of blood-based inflammatory biomarkers for the early-stage detection of acute myocardial infarction. Netw. Model. Anal. Health Inform. Bioinform. 2022, 11, 28. [Google Scholar] [CrossRef]
- Singh, R.; Khalid, M.; Batra, N.; Biswas, P.; Singh, L.; Bhatti, R. Exploring the Anticonvulsant Activity of Aqueous Extracts of Ficus benjamina L. Figs in Experimentally Induced Convulsions. J. Chem. 2023, 2023, 6298366. [Google Scholar] [CrossRef]
- Dey, D.; Paul, P.K.; Al Azad, S.; Al Mazid, M.F.; Khan, A.M.; Sharif, M.A.; Rahman, M.H. Molecular optimization, docking, and dynamic simulation profiling of selective aromatic phytochemical ligands in blocking the SARS-CoV-2 S protein attachment to ACE2 receptor: An in silico approach of targeted drug designing. J. Adv. Vet. Anim. Res. 2021, 8, 24–35. [Google Scholar] [CrossRef]
- Dipta, D.; Tanzila Ismail, E.; Partha BISWAS, S.A.; Shoeba ISLAM, U.R.R.; FIROZ, M.; AHMED, S.Z.; Salauddin, A.; RAHMAN, A.; AFRIN10, S.; MAHEDI10, R.A.J.F.A.S.E. Antiviral effects of bacteriocin against animal-to-human transmittable mutated SARS-CoV-2: A systematic review. Front. Agric. Sci. Eng. 2021, 8, 603–622. [Google Scholar]
- Aljahdali, M.O.; Molla, M.H.R.; Ahammad, F. Compounds Identified from Marine Mangrove Plant (Avicennia alba) as Potential Antiviral Drug Candidates against WDSV, an In-Silico Approach. Mar. Drugs 2021, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A.; Elshemey, W.M. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J. Med. Virol. 2018, 90, 13–18. [Google Scholar] [CrossRef]
- Mahmud, S.; Rahman, E.; Nain, Z.; Billah, M.; Karmakar, S.; Mohanto, S.C.; Paul, G.K.; Amin, A.; Acharjee, U.K.; Saleh, M.A. Computational discovery of plant-based inhibitors against human carbonic anhydrase IX and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2021, 39, 2754–2770. [Google Scholar] [CrossRef]
- Alamri, M.A.; Altharawi, A.; Alabbas, A.B.; Alossaimi, M.A.; Alqahtani, S.M. Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab. J. Chem. 2020, 13, 7224–7234. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Goodford, P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar] [CrossRef]
- Weber, A.E.; Halgren, T.A.; Doyle, J.J.; Lynch, R.J.; Siegl, P.K.; Parsons, W.H.; Greenlee, W.J.; Patchett, A.A. Design and synthesis of P2-P1′-linked macrocyclic human renin inhibitors. J. Med. Chem. 1991, 34, 2692–2701. [Google Scholar] [CrossRef]
- Khan, R.A.; Hossain, R.; Siyadatpanah, A.; Al-Khafaji, K.; Khalipha, A.B.R.; Dey, D.; Asha, U.H.; Biswas, P.; Saikat, A.S.M.; Chenari, H.A.; et al. Diterpenes/Diterpenoids and Their Derivatives as Potential Bioactive Leads against Dengue Virus: A Computational and Network Pharmacology Study. Molecules 2021, 26, 6821. [Google Scholar] [CrossRef]
- Baral, S.K.; Biswas, P.; Kaium, M.A.; Islam, M.A.; Dey, D.; Al Saber, M.; Rahaman, T.I.; Emran, T.B.; Hasan, M.N.; Jeong, M.K.; et al. A Comprehensive Discussion in Vaginal Cancer Based on Mechanisms, Treatments, Risk Factors and Prevention. Front. Oncol. 2022, 12, 883805. [Google Scholar] [CrossRef]
- Thérien, M.; Skorey, K.; Zamboni, R.; Li, C.S.; Lau, C.K.; LeRiche, T.; Linh Truong, V.; Waddleton, D.; Ramachandran, C. Synthesis of a novel peptidic photoaffinity probe for the PTP-1B enzyme. Bioorg. Med. Chem. Lett. 2004, 14, 2319–2322. [Google Scholar] [CrossRef]
- Morshed, A.; Al Azad, S.; Mia, M.A.R.; Uddin, M.F.; Ema, T.I.; Yeasin, R.B.; Srishti, S.A.; Sarker, P.; Aurthi, R.Y.; Jamil, F.; et al. Oncoinformatic screening of the gene clusters involved in the HER2-positive breast cancer formation along with the in silico pharmacodynamic profiling of selective long-chain omega-3 fatty acids as the metastatic antagonists. Mol. Divers. 2022. [Google Scholar] [CrossRef]
- Bibi, S.; Sakata, K. An Integrated Computational Approach for Plant-Based Protein Tyrosine Phosphatase Non-Receptor Type 1 Inhibitors. Curr. Comput. Aided Drug. Des. 2017, 13, 319–335. [Google Scholar] [CrossRef]
- Rahman, M.D.H.; Biswas, P.; Dey, D.; Hannan, M.A.; Sahabuddin, M.; Araf, Y.; Kwon, Y.; Emran, T.B.; Ali, M.S.; Uddin, M.J. An In-Silico Identification of Potential Flavonoids against Kidney Fibrosis Targeting TGFβR-1. Life 2022, 12, 1764. [Google Scholar] [CrossRef]
- Studio, D. Discovery Studio; [2.1]; Accelrys: San Diego, CA, USA, 2008. [Google Scholar]
- Saleem, U.; Iman, S.; Ahmad, B.; Shah, M.A.; Bibi, S.; Alqarni, M.; Khan, M.S.; Shah, G.M.; Khan, H.; Alhasani, R.H.; et al. Antidepressant activity of phytochemicals of Mangifera indica seeds assisted by integrated computational analysis. Metab. Brain Dis. 2023, 38, 483–505. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Margolis, K.G. Building community in the gut: A role for mucosal serotonin. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 6–8. [Google Scholar] [CrossRef]
- Bibi, S.; Hasan, M.M.; Wang, Y.B.; Papadakos, S.P.; Yu, H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). Curr. Med. Chem. 2022, 29, 152–162. [Google Scholar] [CrossRef]
- Harrach, M.F.; Drossel, B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014, 140, 174501. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef]
- Dash, R.; Ali, M.C.; Dash, N.; Azad, M.A.K.; Hosen, S.M.Z.; Hannan, M.A.; Moon, I.S. Structural and Dynamic Characterizations Highlight the Deleterious Role of SULT1A1 R213H Polymorphism in Substrate Binding. Int. J. Mol. Sci. 2019, 20, 6256. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Zilani, M.N.H.; Biswas, P.; Khan, D.A.; Rahman, M.H.; Nahid, R.; Nahar, N.; Samad, A.; Ahammad, F.; Hasan, M.N. Evaluation of in vitro and in silico anti-inflammatory potential of some selected medicinal plants of Bangladesh against cyclooxygenase-II enzyme. J. Ethnopharmacol. 2022, 285, 114900. [Google Scholar] [CrossRef] [PubMed]
- Bibi, S.; Khan, M.S.; El-Kafrawy, S.A.; Alandijany, T.A.; El-Daly, M.M.; Yousafi, Q.; Fatima, D.; Faizo, A.A.; Bajrai, L.H.; Azhar, E.I. Virtual screening and molecular dynamics simulation analysis of Forsythoside A as a plant-derived inhibitor of SARS-CoV-2 3CLpro. Saudi Pharm. J. 2022, 30, 979–1002. [Google Scholar] [CrossRef]
- Dey, D.; Hasan, M.M.; Biswas, P.; Papadakos, S.P.; Rayan, R.A.; Tasnim, S.; Bilal, M.; Islam, M.J.; Arshe, F.A.; Arshad, E.M.; et al. Investigating the Anticancer Potential of Salvicine as a Modulator of Topoisomerase II and ROS Signaling Cascade. Front. Oncol. 2022, 12, 899009. [Google Scholar] [CrossRef]
- Biswas, P.; Hany Rumi, O.; Ahmed Khan, D.; Ahmed, M.N.; Nahar, N.; Jahan, R.; Hasan Zilani, M.N.; Paul, T.K.; Hasan, A.; Bondhon, T.A.; et al. Evaluation of Melongosides as Potential Inhibitors of NS2B-NS3 Activator-Protease of Dengue Virus (Serotype 2) by Using Molecular Docking and Dynamics Simulation Approach. J. Trop. Med. 2022, 2022, 7111786. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Z.; Ren, J.; Wei, Z.; Li, J. Potential inhibitors for the novel coronavirus (SARS-CoV-2). Brief. Bioinform. 2021, 22, 1225–1231. [Google Scholar] [CrossRef]



| Cancer Study | Sample Size | Protein Change | Mutation Type | Sample ID |
|---|---|---|---|---|
| Brain Lower-Grade Glioma (TCGA Firehose Legacy) | 530 | S304P | Missense | TCGA-FG-8185-01 |
| Brain Lower-Grade Glioma (TCGA PanCancer Atlas) | 514 | S304P | Missense | TCGA-FG-8185-01 |
| V207E | Missense | TCGA-KT-A7W1-01 | ||
| Glioma (MSK, Nature 2019) | 91 | N334K | Missense | Patient-19-CSF |
| N334K | Missense | Patient-19-T | ||
| A351V | Missense | Patient-34-CSF-VP | ||
| Glioma (MSKCC, Clin Cancer Res 2019) | 1004 | I195V | Missense | P-0010402-T01-IM5 |
| S235N | Missense | P-0003900-T01-IM5 | ||
| R332G | Missense | P-0008166-T01-IM5 | ||
| G462E | Missense | P-0000500-T01-IM3 | ||
| E210K | Missense | P-0003900-T01-IM5 | ||
| E263K | Missense | P-0003900-T01-IM5 | ||
| G183D | Missense | P-0004400-T01-IM5 | ||
| V207A | Missense | P-0013506-T01-IM5 | ||
| G449d | Missense | TRF047202 | ||
| I208T | Missense | P-0019164-T01-IM6 | ||
| Merged Cohort of LGG and GBM (TCGA, Cell 2016) | 1102 | S304P | Missense | TCGA-FG-8185-01 |
| V207E | Missense | TCGA-KT-A7W1-01 | ||
| Glioblastoma (TCGA, Cell 2013) | 543 | V94M | Missense | TCGA-06-0155-01 |
| X229_splice | Splice | TCGA-12-0618-01 | ||
| Glioblastoma (TCGA, Nature 2008) | 206 | Y287H | Missense | TCGA-02-0085-01 |
| Glioblastoma Multiforme (TCGA, Firehose Legacy) | 604 | V94M | Missense | TCGA-06-0155-01 |
| X229_splice | Splice | TCGA-12-0618-01 | ||
| Glioblastoma Multiforme (TCGA, PanCancer Atlas) | 592 | D86Y | Missense | TCGA-06-2566-01 |
| S127F | Missense | TCGA-06-5416-01 | ||
| I303M | Missense | TCGA-19-5956-01 | ||
| MDM2 CACNA1C | Fusion | TCGA-06-A7TK-01 | ||
| CTDSP2-MDM2 | Fusion | TCGA-06-5856-06 |
| Compds | MW (g/mol) | HBA | HBD | Num rot. | ToPoSA (Å2) | Log P | B.S. | LD50 | BBB | HpT | AT | MToD | ToC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temozolomide (control) | 194.15 | 5 | 1 | 1 | 108.17 | −0.92 | 0.55 | 2.178 | −1.142 | yes | yes | 1.226 | 0.153 |
| Taxifolin | 304.25 | 7 | 5 | 1 | 127.45 | 0.63 | 0.55 | 2.261 | −0.725 | no | no | 0.345 | −0.078 |
| (-)-Epicatechin | 290.27 | 6 | 5 | 1 | 110.38 | 0.85 | 0.55 | 2.428 | −1.00 | no | no | 0.438 | 0.183 |
| Galangin | 270.24 | 5 | 3 | 1 | 90.90 | 1.99 | 0.55 | 2.450 | −0.748 | no | no | 0.333 | 0.256 |
| Compounds | Docking Score (Kcal/mol) | Amino Acid Participation in Bonding Interaction | |
|---|---|---|---|
| Interaction of Hydrogen Bond | Interaction of Hydrophobic Bond | ||
| Temozolomide (Reference Drug) | −5.0 | Gln59 (3.04 Å) | Phe55, Phe55, Lys51, Gly58 |
| Imidazoline (The native ligand of 1RV1) | −2.5 | Leu54, Phe55, Gly59 | |
| Taxifolin | −10.0 | Gln59 (2.81 Å), Gln59 (3.01 Å) | Lys51, Phe55, Gly58, Lys51, Phe55, Gln59 |
| (-)-Epicatechin | −8.8 | Lys51 (2.89 Å) | Leu54, Phe55, Gln59, Phe55, Leu54, Lys51 |
| Galangin | −7.4 | Lys51, Phe55, Gln59, Phe55, Lys51, Leu54 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, P.; Bibi, S.; Yousafi, Q.; Mehmood, A.; Saleem, S.; Ihsan, A.; Dey, D.; Hasan Zilani, M.N.; Hasan, M.N.; Saleem, R.; et al. Study of MDM2 as Prognostic Biomarker in Brain-LGG Cancer and Bioactive Phytochemicals Inhibit the p53-MDM2 Pathway: A Computational Drug Development Approach. Molecules 2023, 28, 2977. https://doi.org/10.3390/molecules28072977
Biswas P, Bibi S, Yousafi Q, Mehmood A, Saleem S, Ihsan A, Dey D, Hasan Zilani MN, Hasan MN, Saleem R, et al. Study of MDM2 as Prognostic Biomarker in Brain-LGG Cancer and Bioactive Phytochemicals Inhibit the p53-MDM2 Pathway: A Computational Drug Development Approach. Molecules. 2023; 28(7):2977. https://doi.org/10.3390/molecules28072977
Chicago/Turabian StyleBiswas, Partha, Shabana Bibi, Qudsia Yousafi, Asim Mehmood, Shahzad Saleem, Awais Ihsan, Dipta Dey, Md. Nazmul Hasan Zilani, Md. Nazmul Hasan, Rasha Saleem, and et al. 2023. "Study of MDM2 as Prognostic Biomarker in Brain-LGG Cancer and Bioactive Phytochemicals Inhibit the p53-MDM2 Pathway: A Computational Drug Development Approach" Molecules 28, no. 7: 2977. https://doi.org/10.3390/molecules28072977
APA StyleBiswas, P., Bibi, S., Yousafi, Q., Mehmood, A., Saleem, S., Ihsan, A., Dey, D., Hasan Zilani, M. N., Hasan, M. N., Saleem, R., Awaji, A. A., Fahmy, U. A., & Abdel-Daim, M. M. (2023). Study of MDM2 as Prognostic Biomarker in Brain-LGG Cancer and Bioactive Phytochemicals Inhibit the p53-MDM2 Pathway: A Computational Drug Development Approach. Molecules, 28(7), 2977. https://doi.org/10.3390/molecules28072977








