Impact of Plastic-Related Compounds on P-Glycoprotein and Breast Cancer Resistance Protein In Vitro
Abstract
1. Introduction
2. Results
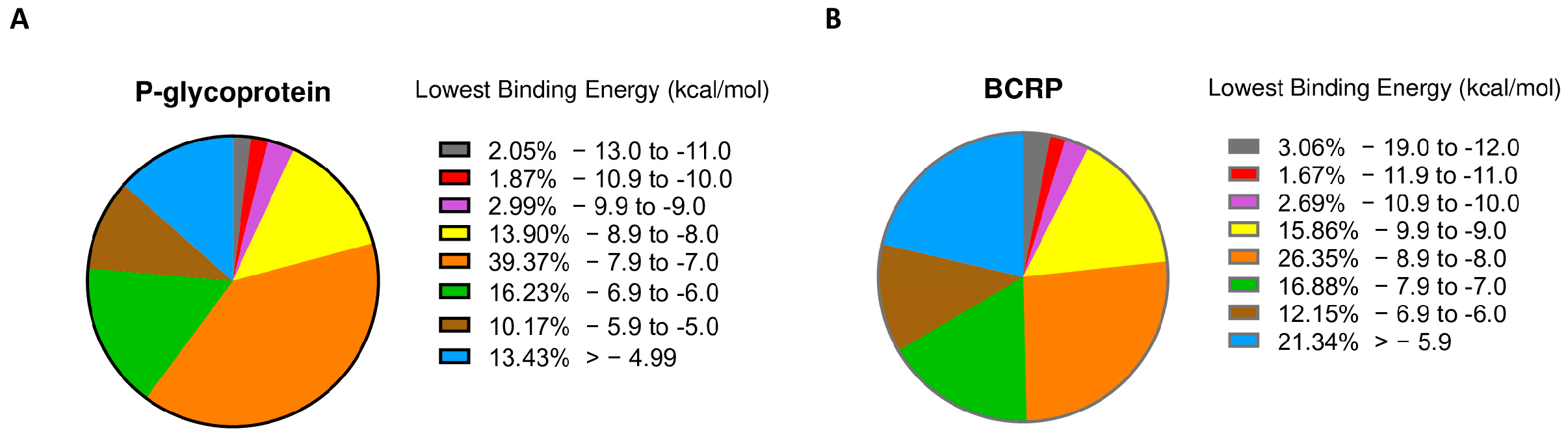
2.1. PyRx Screening
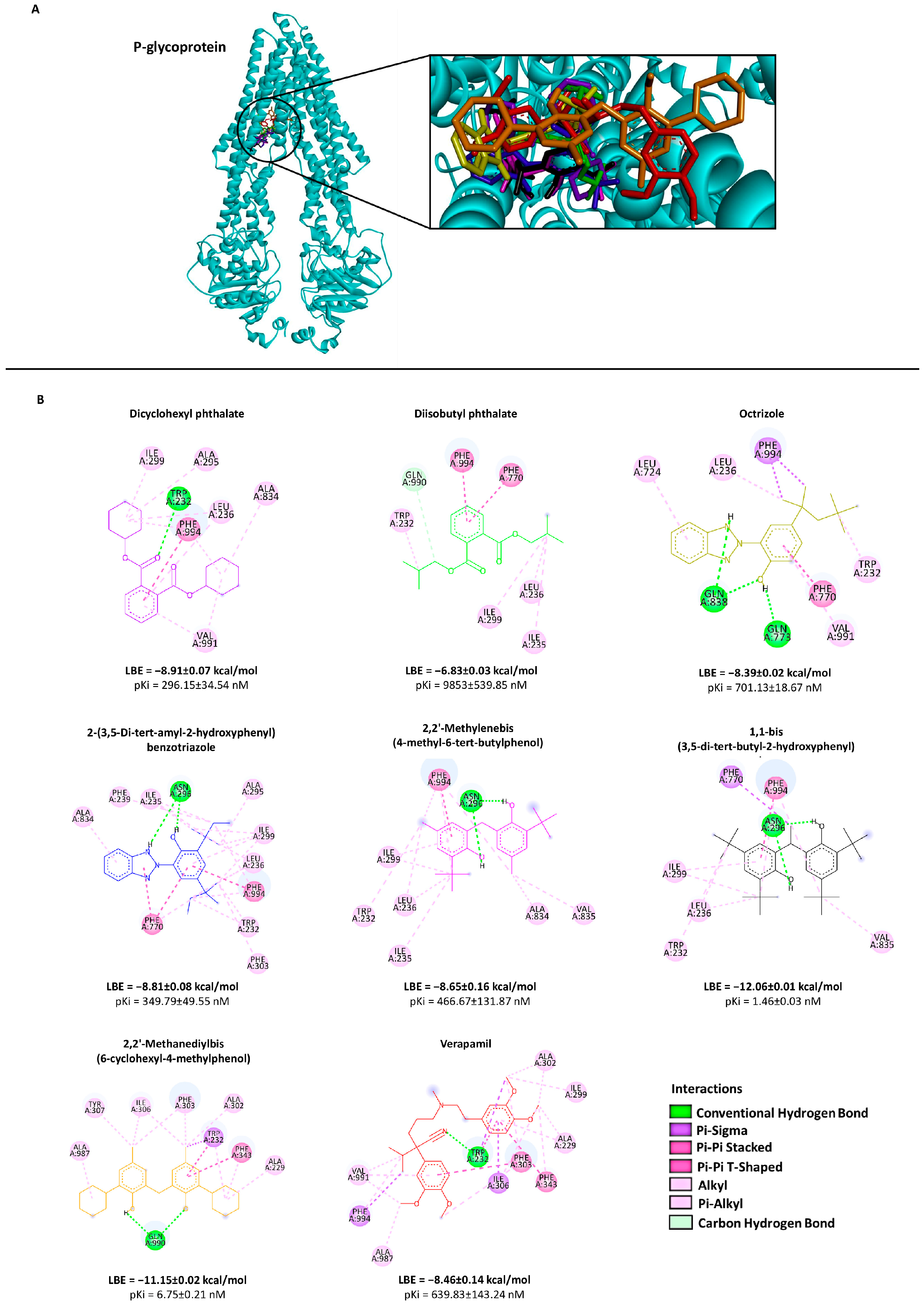
2.2. Molecular Docking
2.3. Cytotoxicity Assay
2.4. P-Glycoprotein Transport Assay
2.5. BCRP Transport Assay
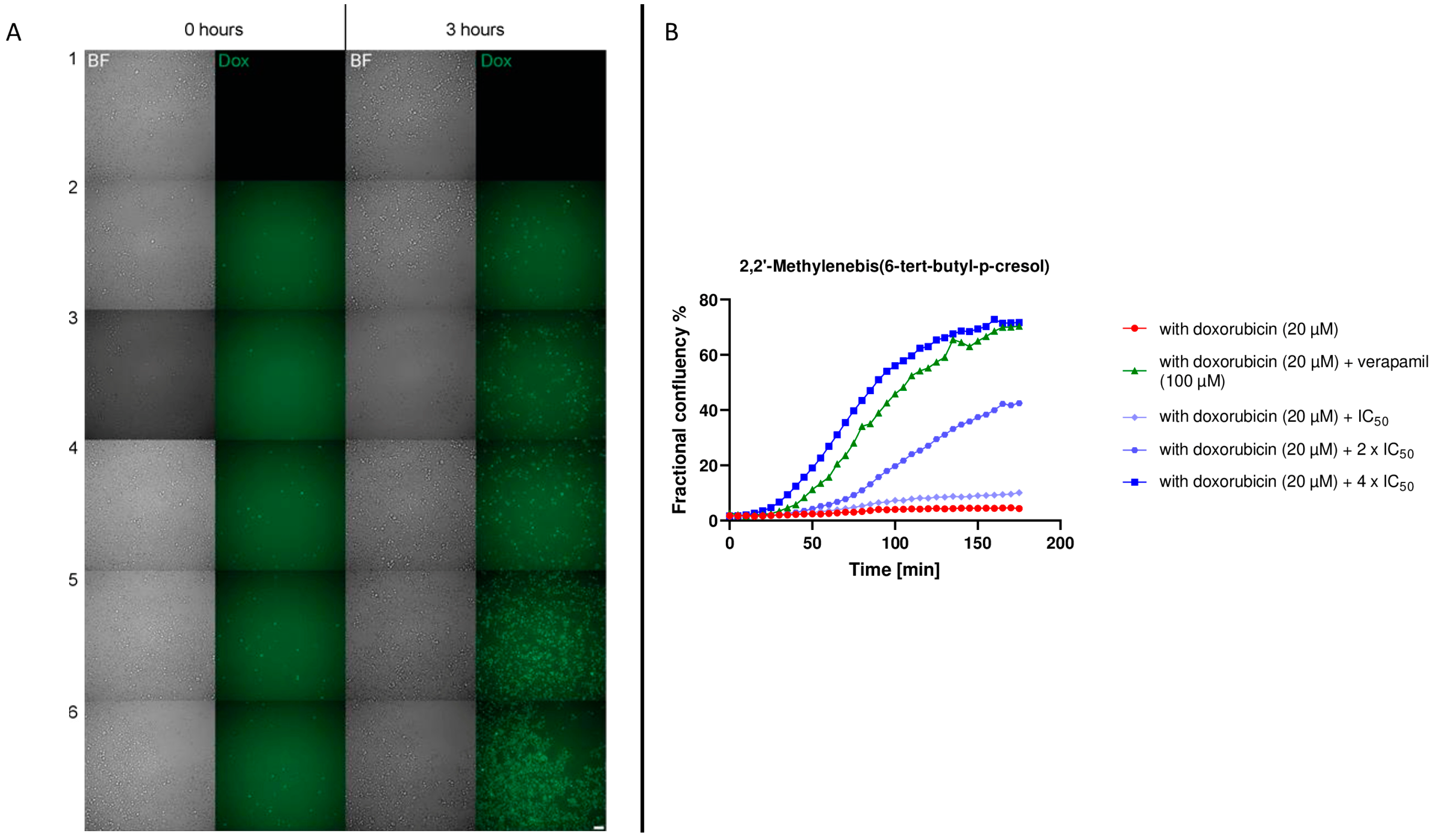
2.6. Live Cell Time-Lapse Microscopy
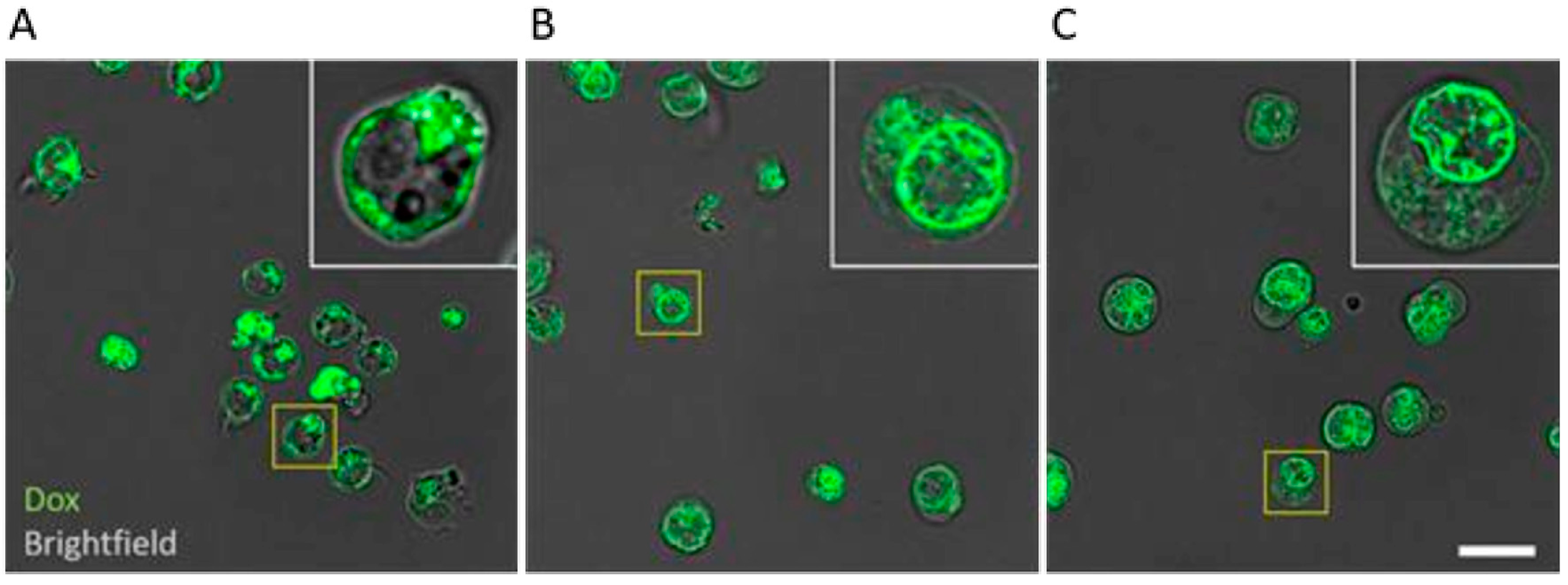
2.7. Confocal Fluorescence Microscopy
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Lines
4.3. PyRx Screening
4.4. Molecular Docking
4.5. Cytotoxicity Assay
4.6. P-Glycoprotein Transport Assay
4.7. BCRP Transport Assay
4.8. Live Cell Time-Lapse Microscopy
4.9. Confocal Fluorescence Microscopy
4.10. Data Analysis
4.10.1. Quantification of Cell Area (Cell Confluency)
4.10.2. Quantification of the Doxorubicin Plus Cells Area (Dox + Cells Confluency)
4.10.3. Quantification of Fractional Confluency of Doxorubicin Plus Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Efferth, T.; Paul, N.W. Threats to Human Health by Great Ocean Garbage Patches. Lancet Planet. Health 2017, 1, e301–e303. [Google Scholar] [CrossRef] [PubMed]
- Cózar, A.; Echevarría, F.; González-Gordillo, J.I.; Irigoien, X.; Úbeda, B.; Hernández-León, S.; Palma, Á.T.; Navarro, S.; García-de-Lomas, J.; Ruiz, A.; et al. Plastic Debris in the Open Ocean. Proc. Natl. Acad. Sci. USA 2014, 111, 10239–10244. [Google Scholar] [CrossRef]
- Vince, J.; Hardesty, B.D. Plastic Pollution Challenges in Marine and Coastal Environments: From Local to Global Governance. Restor. Ecol. 2016, 25, 123–128. [Google Scholar] [CrossRef]
- Prata, J.C. Airborne Microplastics: Consequences to Human Health? Environ. Pollut. 2018, 234, 115–126. [Google Scholar] [CrossRef] [PubMed]
- da Costa, J.P.; Santos, P.S.M.; Duarte, A.C.; Rocha-Santos, T. (Nano) Plastics in the Environment—Sources, Fates and Effects. Sci. Total Environ. 2016, 566–567, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Anbumani, S.; Kakkar, P. Ecotoxicological Effects of Microplastics on Biota: A Review. Environ. Sci. Pollut. Res. 2018, 25, 14373–14396. [Google Scholar] [CrossRef]
- de Sá, L.C.; Oliveira, M.; Ribeiro, F.; Rocha, T.L.; Futter, M.N. Studies of the Effects of Microplastics on Aquatic Organisms: What Do We Know and Where Should We Focus Our Efforts in the Future? Sci. Total Environ. 2018, 645, 1029–1039. [Google Scholar] [CrossRef]
- Alomar, C.; Sureda, A.; Capó, X.; Guijarro, B.; Tejada, S.; Deudero, S. Microplastic Ingestion by Mullus Surmuletus Linnaeus, 1758 Fish and Its Potential for Causing Oxidative Stress. Environ. Res. 2017, 159, 135–142. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain (CONTAM). Presence of Microplastics and Nanoplastics in Food, with Particular Focus on Seafood. EFSA J. 2016, 14, e04501. [Google Scholar] [CrossRef]
- Lithner, D.; Larsson, Å.; Dave, G. Science of the Total Environment Environmental and Health Hazard Ranking and Assessment of Plastic Polymers Based on Chemical Composition. Sci. Total Environ. 2011, 409, 3309–3324. [Google Scholar] [CrossRef]
- Lehner, R.; Weder, C.; Petri-Fink, A.; Rothen-Rutishauser, B. Emergence of Nanoplastic in the Environment and Possible Impact on Human Health. Environ. Sci. Technol. 2019, 53, 1748–1765. [Google Scholar] [CrossRef]
- Danopoulos, E.; Twiddy, M.; Rotchell, J.M. Microplastic Contamination of Drinking Water: A Systematic Review. PLoS ONE 2020, 15, e0236838. [Google Scholar] [CrossRef]
- Oliveri Conti, G.; Ferrante, M.; Banni, M.; Favara, C.; Nicolosi, I.; Cristaldi, A.; Fiore, M.; Zuccarello, P. Micro- and Nano-Plastics in Edible Fruit and Vegetables. The First Diet Risks Assessment for the General Population. Environ. Res. 2020, 187, 109677. [Google Scholar] [CrossRef]
- Wright, S.L.; Kelly, F.J. Plastic and Human Health: A Micro Issue? Environ. Sci. Technol. 2017, 51, 6634–6647. [Google Scholar] [CrossRef]
- Leslie, H.A.; van Velzen, M.J.M.; Brandsma, S.H.; Vethaak, A.D.; Garcia-Vallejo, J.J.; Lamoree, M.H. Discovery and Quantification of Plastic Particle Pollution in Human Blood. Environ. Int. 2022, 163, 107199. [Google Scholar] [CrossRef]
- Ragusa, A.; Svelato, A.; Santacroce, C.; Catalano, P.; Notarstefano, V.; Carnevali, O.; Papa, F.; Rongioletti, M.C.A.; Baiocco, F.; Draghi, S.; et al. Plasticenta: First Evidence of Microplastics in Human Placenta. Environ. Int. 2021, 146, 106274. [Google Scholar] [CrossRef]
- Meeker, J.D.; Sathyanarayana, S.; Swan, S.H. Phthalates and Other Additives in Plastics: Human Exposure and Associated Health Outcomes. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2097–2113. [Google Scholar] [CrossRef]
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovidou, E.; Purnell, P. An Overview of Chemical Additives Present in Plastics: Migration, Release, Fate and Environmental Impact during Their Use, Disposal and Recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, T.; Zhang, L.; Zhu, P.; Deng, M.; Huang, C.; Hu, T.; Jiang, L.; Li, J. Mammalian Drug Efflux Transporters of the ATP Binding Cassette (ABC) Family in Multidrug Resistance: A Review of the Past Decade. Cancer Lett. 2016, 370, 153–164. [Google Scholar] [CrossRef]
- Efferth, T.; Volm, M. Multiple Resistance to Carcinogens and Xenobiotics: P-Glycoproteins as Universal Detoxifiers. Arch. Toxicol. 2017, 91, 2515–2538. [Google Scholar] [CrossRef]
- Schinkel, A.H.; Jonker, J.W. Mammalian Drug Efflux Transporters of the ATP Binding Cassette (ABC) Family: An Overview. Adv. Drug Deliv. Rev. 2003, 55, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F. Structure, Function and Regulation of P-Glycoprotein and Its Clinical Relevance in Drug Disposition. Xenobiotica 2008, 38, 802–832. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Klaassen, C. Structure, Function, Expression, Genomic Organization, and Single Nucleotide Polymorphisms of Human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) Efflux Transporters. Int. J. Toxicol. 2006, 25, 231–259. [Google Scholar] [CrossRef]
- Mao, Q.; Unadkat, J.D. Role of the Breast Cancer Resistance Protein (BCRP/ABCG2) in Drug Transport—An Update. AAPS J. 2015, 17, 65–82. [Google Scholar] [CrossRef]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Fan, J.; Fu, A.; Zhang, L. Progress in Molecular Docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Ferreira, R.J.; Ferreira, M.J.U.; Dos Santos, D.J.V.A. Molecular Docking Characterizes Substrate-Binding Sites and Efflux Modulation Mechanisms within P-Glycoprotein. J. Chem. Inf. Model. 2013, 53, 1747–1760. [Google Scholar] [CrossRef]
- Zeino, M.; Saeed, M.E.M.; Kadioglu, O.; Efferth, T. The Ability of Molecular Docking to Unravel the Controversy and Challenges Related to P-Glycoprotein—A Well-Known, yet Poorly Understood Drug Transporter. Investig. New Drugs 2014, 32, 618–625. [Google Scholar] [CrossRef]
- Benson, R. Hazard to the Developing Male Reproductive System from Cumulative Exposure to Phthalate Esters-Dibutyl Phthalate, Diisobutyl Phthalate, Butylbenzyl Phthalate, Diethylhexyl Phthalate, Dipentyl Phthalate, and Diisononyl Phthalate. Regul. Toxicol. Pharmacol. 2009, 53, 90–101. [Google Scholar] [CrossRef]
- Yost, E.E.; Euling, S.Y.; Weaver, J.A.; Beverly, B.E.J.; Keshava, N.; Mudipalli, A.; Arzuaga, X.; Blessinger, T.; Dishaw, L.; Hotchkiss, A.; et al. Hazards of Diisobutyl Phthalate (DIBP) Exposure: A Systematic Review of Animal Toxicology Studies. Environ. Int. 2019, 125, 579–594. [Google Scholar] [CrossRef]
- Hall, M.D.; Handley, M.D.; Gottesman, M.M. Is Resistance Useless? Multidrug Resistance and Collateral Sensitivity. Trends Pharmacol. Sci. 2009, 30, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Saeed, M.E.M.; Kadioglu, O.; Seo, E.J.; Shirooie, S.; Mbaveng, A.T.; Nabavi, S.M.; Kuete, V. Collateral Sensitivity of Natural Products in Drug-Resistant Cancer Cells. Biotechnol. Adv. 2020, 38, 107342. [Google Scholar] [CrossRef] [PubMed]
- De Lange, E.C.M. Potential Role of ABC Transporters as a Detoxification System at the Blood-CSF Barrier. Adv. Drug Deliv. Rev. 2004, 56, 1793–1809. [Google Scholar] [CrossRef]
- Nicklisch, S.C.T.; Rees, S.D.; McGrath, A.P.; Gökirmak, T.; Bonito, L.T.; Vermeer, L.M.; Cregger, C.; Loewen, G.; Sandin, S.; Chang, G.; et al. Global Marine Pollutants Inhibit P-Glycoprotein: Environmental Levels, Inhibitory Effects, and Cocrystal Structure. Sci. Adv. 2016, 2, e1600001. [Google Scholar] [CrossRef]
- Fardel, O.; Kolasa, E.; Le Vee, M. Environmental Chemicals as Substrates, Inhibitors or Inducers of Drug Transporters: Implication for Toxicokinetics, Toxicity and Pharmacokinetics. Expert Opin. Drug Metab. Toxicol. 2012, 8, 29–46. [Google Scholar] [CrossRef]
- MacArthur, D.E. Rethinking the Future of Plastics Rethinking the Future of Plastics the New Plastics Economy; Ellen MacArthur Foundation: Cowes, UK, 2014; pp. 1–120. [Google Scholar]
- Liu, L.; Fokkink, R.; Koelmans, A.A. Sorption of Polycyclic Aromatic Hydrocarbons to Polystyrene Nanoplastic. Environ. Toxicol. Chem. 2016, 35, 1650–1655. [Google Scholar] [CrossRef]
- Cooper, J.E.; Kendig, E.L.; Belcher, S.M. Assessment of Bisphenol A Released from Reusable Plastic, Aluminium and Stainless Steel Water Bottles. Chemosphere 2011, 85, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Kwan, C.S.; Takada, H. Release of Additives and Monomers from Plastic Wastes. In Handbook of Environmental Chemistry; NY Research Press: New York, NY, USA, 2019; Volume 78, pp. 51–70. [Google Scholar]
- Takahashi, O.; Oishi, S. Male Reproductive Toxicity of Four Bisphenol Antioxidants in Mice and Rats and Their Estrogenic Effect. Arch. Toxicol. 2006, 80, 225–241. [Google Scholar] [CrossRef]
- Yang, X.; Sun, Z.; Wang, W.; Zhou, Q.; Shi, G.; Wei, F.; Jiang, G. Developmental Toxicity of Synthetic Phenolic Antioxidants to the Early Life Stage of Zebrafish. Sci. Total Environ. 2018, 643, 559–568. [Google Scholar] [CrossRef]
- Fromm, M.F. Importance of P-Glycoprotein at Blood-Tissue Barriers. Trends Pharmacol. Sci. 2004, 25, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Chu, S.; Bence, A.K.; Bailey, B.; Xue, X.; Erickson, P.A.; Montrose, M.H.; Beck, W.T.; Erickson, L.C. Quantitation of Doxorubicin Uptake, Efflux, and Modulation of Multidrug Resistance (MDR) in MDR Human Cancer Cells. J. Pharmacol. Exp. Ther. 2008, 324, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Abdelfatah, S.; Böckers, M.; Asensio, M.; Kadioglu, O.; Klinger, A.; Fleischer, E.; Efferth, T. Isopetasin and S-Isopetasin as Novel P-Glycoprotein Inhibitors against Multidrug-Resistant Cancer Cells. Phytomedicine 2020, 86, 153196. [Google Scholar] [CrossRef]
- Kimmig, A.; Gekeler, V.; Neumann, M.; Frese, G.; Handgretinger, R.; Kardos, G.; Diddens, H.; Niethammer, D. Susceptibility of Multidrug-Resistant Human Leukemia Cell Lines to Human Interleukin 2-Activated Killer Cells. Cancer Res. 1990, 50, 6793–6799. [Google Scholar]
- Efferth, T.; Sauerbrey, A.; Olbrich, A.; Gebhart, E.; Rauch, P.; Weber, H.O.; Hengstler, J.G.; Halatsch, M.E.; Volm, M.; Tew, K.D.; et al. Molecular Modes of Action of Artesunate in Tumor Cell Lines. Mol. Pharmacol. 2003, 64, 382–394. [Google Scholar] [CrossRef]
- Efferth, T.; Konkimalla, V.B.; Wang, Y.F.; Sauerbrey, A.; Meinhardt, S.; Zintl, F.; Mattern, J.; Volm, M. Prediction of Broad Spectrum Resistance of Tumors towards Anticancer Drugs. Clin. Cancer Res. 2008, 14, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Yang, W.; Abruzzo, L.V.; Krogmann, T.; Gao, Y.; Rishi, A.K.; Ross, D.D. A Multidrug Resistance Transporter from Human MCF-7 Breast Cancer Cells. Med. Sci. 1998, 95, 15665–15670. [Google Scholar]
- Böckers, M.; Paul, N.W.; Efferth, T. Indeno [1,2,3-Cd] Pyrene and Picene Mediate Actions via Estrogen Receptor α Signaling Pathway in in Vitro Cell Systems, Altering Gene Expression. Toxicol. Appl. Pharmacol. 2020, 396, 114995. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank: A Historical Perspective. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 88–95. [Google Scholar] [CrossRef]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural Insight into Substrate and Inhibitor Discrimination by Human P-Glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef]
- Jackson, S.M.; Manolaridis, I.; Kowal, J.; Zechner, M.; Taylor, N.M.I.; Bause, M.; Bauer, S.; Bartholomaeus, R.; Bernhardt, G.; Koenig, B.; et al. Structural Basis of Small-Molecule Inhibition of Human Multidrug Transporter ABCG2. Nat. Struct. Mol. Biol. 2018, 25, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]



| Compound | Structure | CEM/ADR5000 IC50 (µM) | CCRF-CEM IC50 (µM) | Resistance Ratio P-gp | MDA-MB-231-BCRP IC50 (µM) | MDA-MB_231-pcDNA IC50 (µM) | Resistance Ratio BCRP |
|---|---|---|---|---|---|---|---|
| (1) Dicyclohexyl phthalate |  | 45.76 ± 7.38 | 73.03 ± 10.10 | 0.63 | 80.66 ± 3.00 | 81.58 ± 7.41 | 0.99 |
| (2) Diisobutyl phthalate | 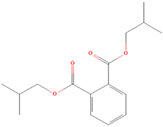 | 35.13 ± 4.77 | 75.15 ± 8.57 | 0.47 | >100 | >100 | / |
| (3) Octrizole |  | 54.73 ± 1.42 | 58.82 ± 2.69 | 0.93 | 66.30 ± 1.38 | 60.33 ± 2.58 | 1.10 |
| (4) 2-(3,5-Di-tert-amyl-2-hydroxyphenyl)benzotriazole | 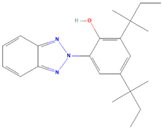 | 31.86 ± 3.35 | 40.20 ± 3.05 | 0.79 | >100 | >100 | / |
| (5) 2,2′-Methylenebis(4-methyl-6-tert-butylphenol) | 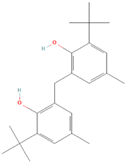 | 14.42 ± 2.81 | 15.34 ± 0.21 | 0.94 | 17.12 ± 0.46 | 16.35 ± 0.47 | 1.05 |
| (6) 1,1-bis(3,5-di-tert-butyl-2-hydroxyphenyl)ethane | 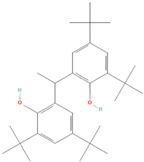 | 36.25 ± 6.79 | 17.09 ± 1.77 | 2.12 | 19.08 ± 0.24 | 16.46 ± 0.49 | 1.16 |
| (7) 2,2′-Methanediylbis(6-cyclohexyl-4-methylphenol) |  | 16.48 ± 1.74 | 20.43 ± 1.08 | 0.81 | 18.12 ± 0.21 | 20.21 ± 1.45 | 0.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosellini, M.; Turunen, P.; Efferth, T. Impact of Plastic-Related Compounds on P-Glycoprotein and Breast Cancer Resistance Protein In Vitro. Molecules 2023, 28, 2710. https://doi.org/10.3390/molecules28062710
Rosellini M, Turunen P, Efferth T. Impact of Plastic-Related Compounds on P-Glycoprotein and Breast Cancer Resistance Protein In Vitro. Molecules. 2023; 28(6):2710. https://doi.org/10.3390/molecules28062710
Chicago/Turabian StyleRosellini, Matteo, Petri Turunen, and Thomas Efferth. 2023. "Impact of Plastic-Related Compounds on P-Glycoprotein and Breast Cancer Resistance Protein In Vitro" Molecules 28, no. 6: 2710. https://doi.org/10.3390/molecules28062710
APA StyleRosellini, M., Turunen, P., & Efferth, T. (2023). Impact of Plastic-Related Compounds on P-Glycoprotein and Breast Cancer Resistance Protein In Vitro. Molecules, 28(6), 2710. https://doi.org/10.3390/molecules28062710







