Deciphering Complex Interactions in Bioactive Lipid Signaling
Abstract
1. Introduction
2. Bioactive Lipids
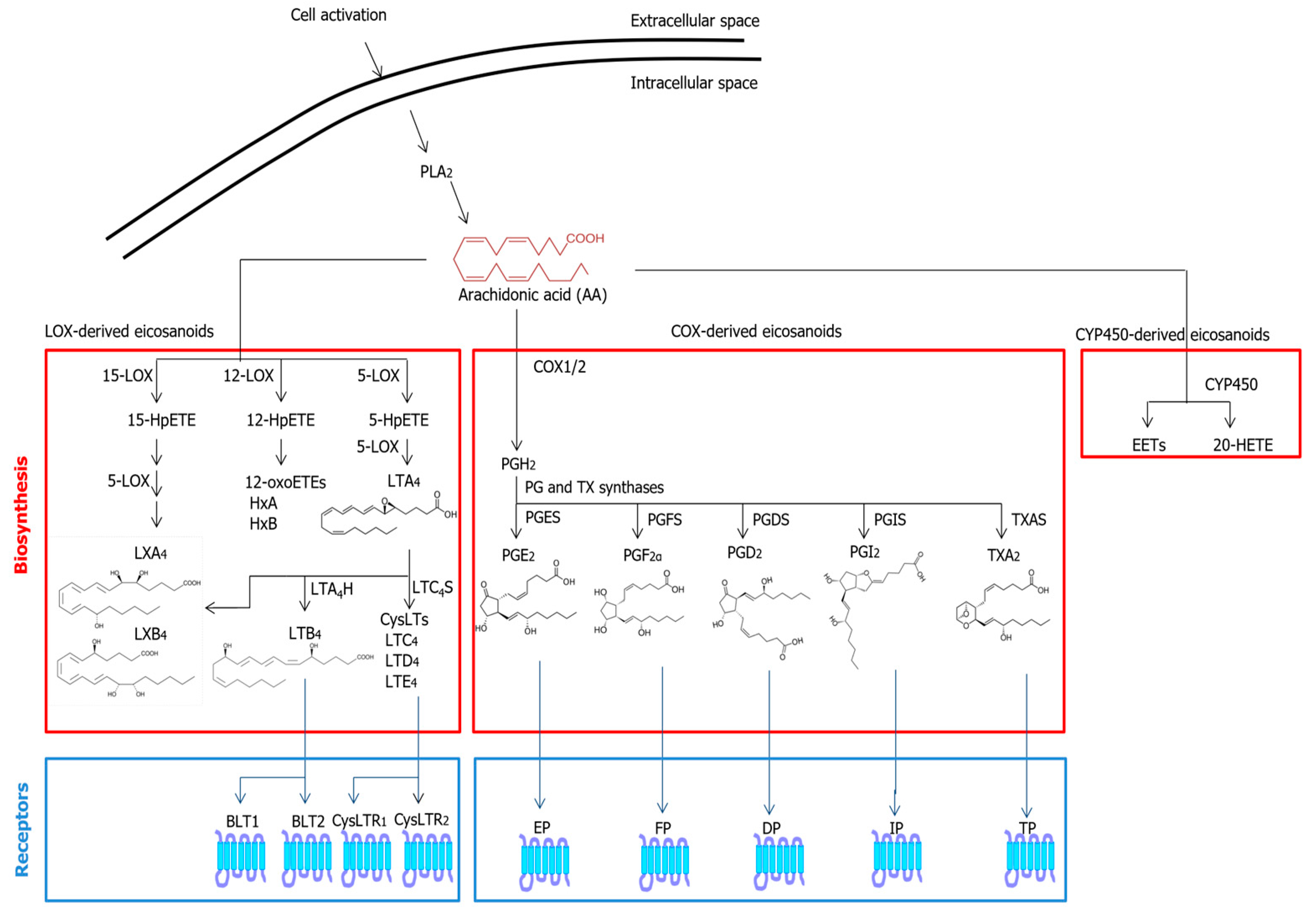
2.1. Eicosanoids
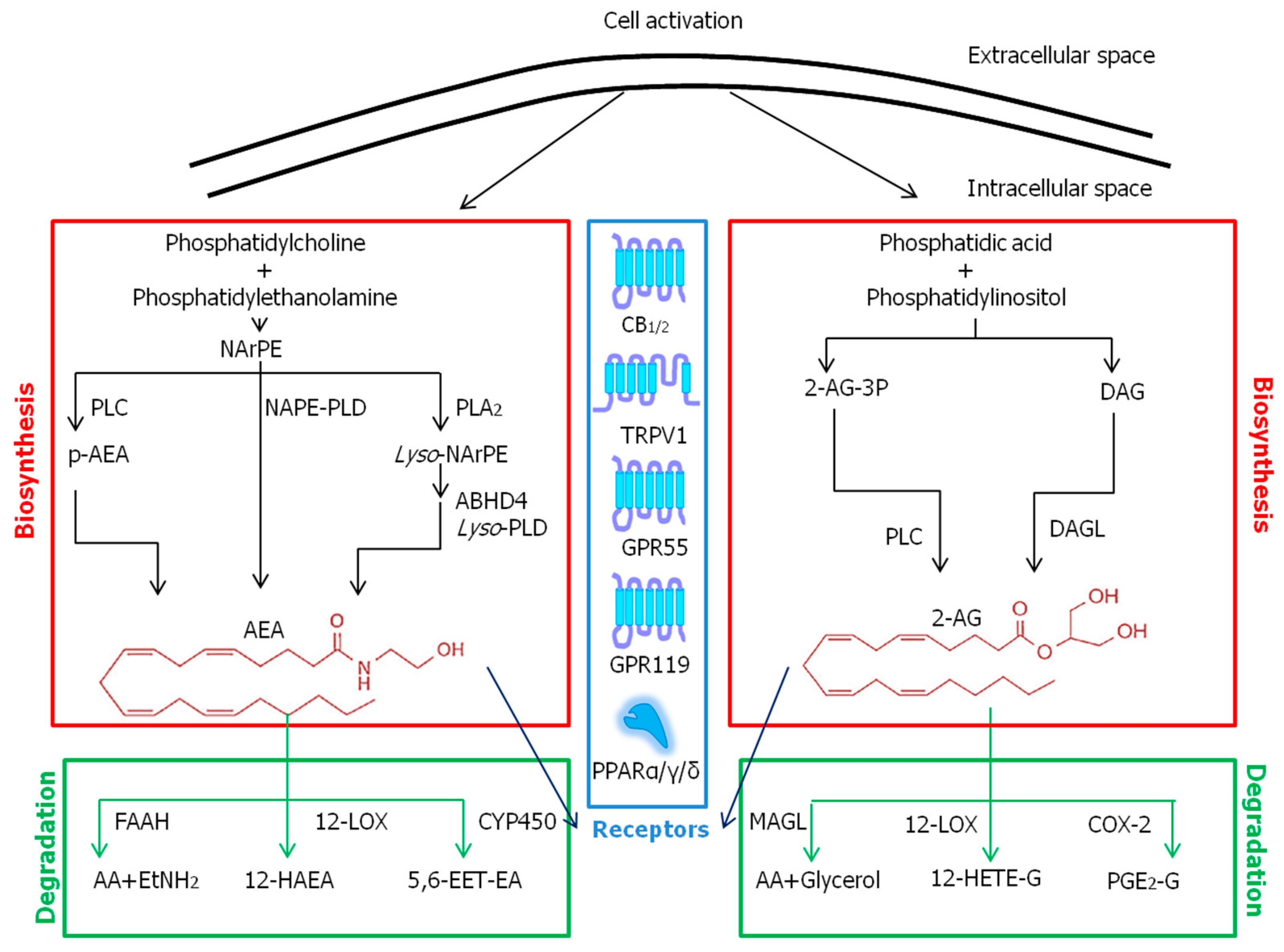
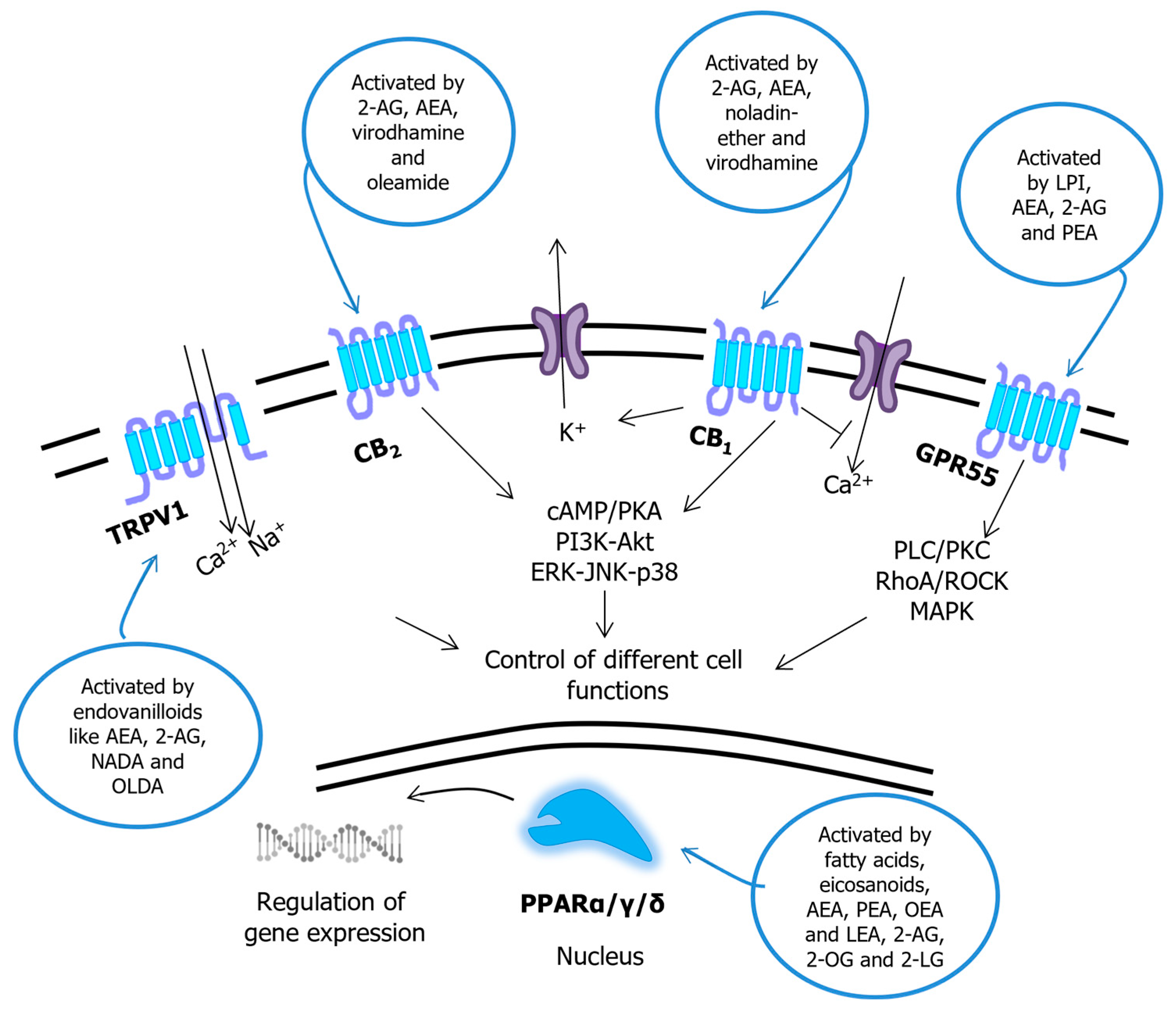
2.2. Endocannabinoids
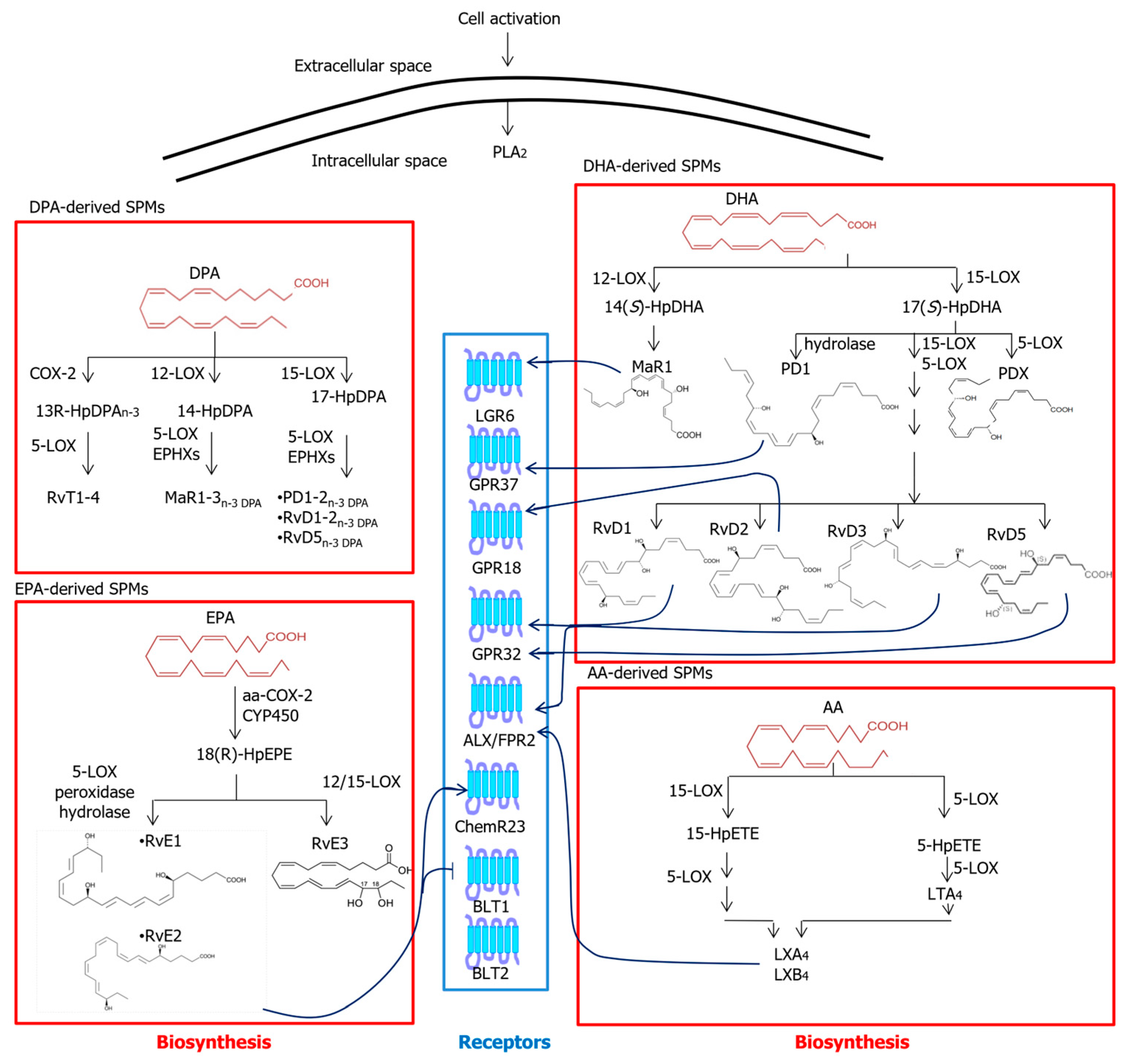
2.3. Specialized Pro-Resolving Mediators
3. Interactions in Lipid Signaling Pathways
3.1. Common Metabolic Pathways
3.2. Common Receptor Targets
3.3. Interaction of EICs, ECBs and SPMs with AA/PUFA-Unrelated Bioactive Lipids
4. Conclusions and Future Directions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abd-Nikfarjam, B.; Dolati-Somarin, A.; Baradaran Rahimi, V.; Askari, V.R. Cannabinoids in neuroinflammatory disorders: Focusing on multiple sclerosis, Parkinson’s, and Alzheimer’s diseases. Biofactors, 2023; in press. [Google Scholar] [CrossRef] [PubMed]
- Ang, S.P.; Sidharthan, S.; Lai, W.; Hussain, N.; Patel, K.V.; Gulati, A.; Henry, O.; Kaye, A.D.; Orhurhu, V. Cannabinoids as a potential alternative to opioids in the management of various pain subtypes: Benefits, limitations, and risks. Pain Ther. 2023; in press. [Google Scholar] [CrossRef]
- Bhaswant, M.; Shanmugam, D.K.; Miyazawa, T.; Abe, C.; Miyazawa, T. Microgreens-A comprehensive review of bioactive molecules and health benefits. Molecules 2023, 28, 867. [Google Scholar] [CrossRef] [PubMed]
- Mulyani, Y.; Sinaga, S.E.; Supratman, U. Phytochemistry and biological activities of endophytic fungi from the meliaceae family. Molecules 2023, 28, 778. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Hu, X.; Zhou, R.; Li, Y.; Wu, W.; Liu, N. A review on dietary flavonoids as modulators of the tumor microenvironment. Mol. Nutr. Food Res. 2023, 2023, e2200435. [Google Scholar] [CrossRef]
- Kumar, A.P.N.; Kumar, M.; Jose, A.; Tomer, V.; Oz, E.; Proestos, C.; Zeng, M.; Elobeid, T.K.S.; Oz, F. Major phytochemicals: Recent advances in health benefits and extraction method. Molecules 2023, 28, 887. [Google Scholar] [CrossRef]
- Leuti, A.; Fazio, D.; Fava, M.; Piccoli, A.; Oddi, S.; Maccarrone, M. Bioactive lipids, inflammation and chronic diseases. Adv. Drug Deliv. Rev. 2020, 159, 133–169. [Google Scholar] [CrossRef]
- Cecconi, S.; Rapino, C.; Di Nisio, V.; Rossi, G.; Maccarrone, M. The (endo)cannabinoid signaling in female reproduction: What are the latest advances? Prog. Lipid Res. 2020, 77, 101019. [Google Scholar] [CrossRef]
- Maccarrone, M.; Rapino, C.; Francavilla, F.; Barbonetti, A. Cannabinoid signalling and effects of cannabis on the male reproductive system. Nat. Rev. Urol. 2021, 18, 19–32. [Google Scholar] [CrossRef]
- Baker, R.R. The eicosanoids: A historical overview. Clin. Biochem. 1990, 23, 455–458. [Google Scholar] [CrossRef]
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N. Systems approach to inflammation resolution: Identification of novel anti-inflammatory and pro-resolving mediators. J. Thromb. Haemost. 2009, 7, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 158, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, H.; Kohno, D.; Koga, T.; Sasaki, T.; Fukunaka, A.; Okuno, T.; Jo-Watanabe, A.; Kazuno, S.; Miyatsuka, T.; Kitamura, T. Leukotriene A4 hydrolase deficiency protects mice from diet-induced obesity by increasing energy expenditure through neuroendocrine axis. FASEB J. 2020, 34, 13949–13958. [Google Scholar] [CrossRef]
- Félix-Soriano, E.; Sáinz, N.; Gil-Iturbe, E.; Collantes, M.; Fernández-Galilea, M.; Castilla-Madrigal, R.; Ly, L.; Dalli, J.; Moreno-Aliaga, M.J. Changes in brown adipose tissue lipid mediator signatures with aging, obesity, and DHA supplementation in female mice. FASEB J. 2021, 35, e21592. [Google Scholar] [CrossRef]
- Fujimori, K.; Uno, S.; Kuroda, K.; Matsumoto, C.; Maehara, T. Leukotriene C4 synthase is a novel PPARγ target gene, and leukotriene C4 and D4 activate adipogenesis through cysteinyl LT1 receptors in adipocytes. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119203. [Google Scholar] [CrossRef]
- Eckenstaler, R.; Ripperger, A.; Hauke, M.; Petermann, M.; Hemkemeyer, S.A.; Schwedhelm, E.; Ergün, S.; Frye, M.; Werz, O.; Koeberle, A. A thromboxane A2 receptor-driven COX-2-dependent feedback loop that affects endothelial homeostasis and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 444–461. [Google Scholar] [CrossRef]
- Leslie, C.C. Cytosolic phospholipase A2: Physiological function and role in disease. J. Lipid Res. 2015, 56, 1386–1402. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Dennis, E.A. Phospholipase A2 catalysis and lipid mediator lipidomics. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 766–771. [Google Scholar] [CrossRef]
- Smith, W.L.; Urade, Y.; Jakobsson, P. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 2011, 111, 5821–5865. [Google Scholar] [CrossRef]
- Clark, S.R.; Guy, C.J.; Scurr, M.J.; Taylor, P.R.; Kift-Morgan, A.P.; Hammond, V.J.; Thomas, C.P.; Coles, B.; Roberts, G.W.; Eberl, M.; et al. Esterified eicosanoids are acutely generated by 5-lipoxygenase in primary human neutrophils and in human and murine infection. Blood 2011, 117, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Roman, R.J. Effect of cytochrome P450 metabolites of arachidonic acid in nephrology. J. Am. Soc. Nephrol. 2017, 28, 2845–2855. [Google Scholar] [CrossRef]
- Yokomizo, T.; Nakamura, M.; Shimizu, T. Leukotriene receptors as potential therapeutic targets. J. Clin. Invest. 2018, 128, 2691–2701. [Google Scholar] [CrossRef]
- El Sohly, M.A.; Radwan, M.M.; Gul, W. Phytochemistry of Cannabis sativa L. Prog. Chem. Org. Nat. Prod. 2017, 103, 1–36. [Google Scholar] [PubMed]
- Gómez-Cañas, M.; Rodríguez-Cueto, C.; Satta, V.; Hernández-Fisac, I.; Navarro, E.; Fernández-Ruiz, J. Endocannabinoid-binding receptors as drug targets. Methods Mol. Biol. 2023, 2576, 67–94. [Google Scholar] [PubMed]
- Maccarrone, M. Missing pieces to the endocannabinoid puzzle. Trends Mol. Med. 2020, 26, 263–272. [Google Scholar] [CrossRef]
- Mechoulam, R.; Ben-Shabat, S.; Hanus, L.; Ligumsky, M.; Kaminski, N.E.; Schatz, A.R.; Gopher, A.; Almog, S.; Martin, B.R.; Compton, D.R. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995, 50, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Morishita, J.; Tsuboi, K.; Tonai, T.; Ueda, N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 2004, 279, 5298–5305. [Google Scholar] [CrossRef]
- Bisogno, T.; Howell, F.; Williams, G.; Minassi, A.; Cascio, M.G.; Ligresti, A.; Matias, I.; Schiano-Moriello, A.; Paul, P.; Williams, E.J.; et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 2003, 163, 463–468. [Google Scholar] [CrossRef]
- Guo, Y.; Uyama, T.; Rahman, S.M.K.; Sikder, M.M.; Hussain, Z.; Tsuboi, K.; Miyake, M.; Ueda, N. Involvement of the γ isoform of cPLA2 in the biosynthesis of bioactive N-acylethanolamines. Molecules 2021, 26, 5213. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Harvey-White, J.; Osei-Hyiaman, D.; Razdan, R.; Gong, Q.; Chan, A.C.; Zhou, Z.; Huang, B.X.; Kim, H.Y. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. USA 2006, 103, 13345–13350. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.M.; Cravatt, B.F. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J. Biol. Chem. 2006, 281, 26465–26472. [Google Scholar] [CrossRef] [PubMed]
- Cravatt, B.F.; Giang, D.K.; Mayfield, S.P.; Boger, D.L.; Lerner, R.A.; Gilula, N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature 1996, 384, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.P.; Carpenter, D.; Leslie, F.M.; Freund, T.F.; Katona, I.; Sensi, S.L.; Kathuria, S.; Piomelli, D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci. USA 2002, 99, 10819–10824. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Structural and chemical biology of the interaction of cyclooxygenase with substrates and non-steroidal anti-inflammatory drugs. Chem. Rev. 2020, 120, 7592–7641. [Google Scholar] [CrossRef]
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat. Rev. Neurol. 2020, 16, 9–29. [Google Scholar] [CrossRef]
- Matsuda, L.A.; Lolait, S.J.; Brownstein, M.J.; Young, A.C.; Bonner, T.I. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 1990, 346, 561–564. [Google Scholar] [CrossRef]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Joffre, J.; Wong, E.; Lawton, S.; Lloyd, E.; Nguyen, N.; Xu, F.; Sempio, C.; Kobzik, L.; Zlatanova, I.; Schumacher, M.; et al. N-Oleoyl dopamine induces IL-10 via central nervous system TRPV1 and improves endotoxemia and sepsis outcomes. J. Neuroinflamm. 2022, 19, 118. [Google Scholar] [CrossRef]
- Serhan, C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014, 510, 92–101. [Google Scholar] [CrossRef]
- Dalli, J.; Colas, R.A.; Serhan, C.N. Novel n-3 immunoresolvents: Structures and actions. Sci. Rep. 2013, 3, 1940. [Google Scholar] [CrossRef] [PubMed]
- Dalli, J.; Chiang, N.; Serhan, C.N. Elucidation of novel 13-series resolvins that increase with atorvastatin and clear infections. Nat. Med. 2015, 21, 1071–1075. [Google Scholar] [CrossRef]
- López-Vicario, C.; Rius, B.; Alcaraz-Quiles, J.; García-Alonso, V.; Lopategi, A.; Titos, E.; Clària, J. Pro-resolving mediators produced from EPA and DHA: Overview of the pathways involved and their mechanisms in metabolic syndrome and related liver diseases. Eur. J. Pharmacol. 2016, 785, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.; Serhan, C.N. Structural elucidation and physiologic functions of specialized pro-resolving mediators and their receptors. Mol. Aspects Med. 2017, 58, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Taketomi, Y.; Murakami, M. Regulatory roles of phospholipase A2 enzymes and bioactive lipids in mast cell biology. Front. Immunol. 2022, 13, 923265. [Google Scholar] [CrossRef]
- Pantazi, D.; Tellis, C.; Tselepis, A.D. Oxidized phospholipids and lipoprotein-associated phospholipase A2 (Lp-PLA2) in atherosclerotic cardiovascular disease: An update. Biofactors 2022, 48, 1257–1270. [Google Scholar] [CrossRef]
- Wu, W.; Li, W.X.; Huang, C.H. Phospholipase A2, a nonnegligible enzyme superfamily in gastrointestinal diseases. Biochimie 2022, 194, 79–95. [Google Scholar] [CrossRef]
- Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Compartmentalized regulation of lipid signaling in oxidative stress and inflammation: Plasmalogens, oxidized lipids and ferroptosis as new paradigms of bioactive lipid research. Prog. Lipid Res. 2023, 89, 101207. [Google Scholar] [CrossRef]
- Mitchener, M.M.; Hermanson, D.J.; Shockley, E.M.; Brown, H.A.; Lindsley, C.W.; Reese, J.; Rouzer, C.A.; Lopez, C.F.; Marnett, L.J. Competition and allostery govern substrate selectivity of cyclooxygenase-2. Proc. Natl. Acad. Sci. USA 2015, 112, 12366–12371. [Google Scholar] [CrossRef]
- Verderio, C.; Gabrielli, M.; Giussani, P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018, 59, 1325–1340. [Google Scholar] [CrossRef]
- Gabrielli, M.; Battista, N.; Riganti, L.; Prada, I.; Antonucci, F.; Cantone, L.; Matteoli, M.; Maccarrone, M.; Verderio, C. Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep. 2015, 16, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, H.; Tang, C.K. Sterol carrier protein 2 in lipid metabolism and non-alcoholic fatty liver disease: Pathophysiology, molecular biology, and potential clinical implications. Metabolism 2022, 131, 155180. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Dryanovski, D.I.; Kimura, Y.; Jackson, S.N.; Woods, A.S.; Yasui, Y.; Tsai, S.Y.; Patel, S.; Covey, D.P.; Su, T.P.; et al. Cocaine-induced endocannabinoid signaling mediated by sigma-1 receptors and extracellular vesicle secretion. eLife 2019, 8, e47209. [Google Scholar] [CrossRef]
- Marion, M.; Hamilton, J.; Richardson, B.; Roeder, N.; Figueiredo, A.; Nubelo, A.; Hetelekides, E.; Penman, S.; Owada, Y.; Kagawa, Y.; et al. Environmental enrichment sex-dependently rescues memory impairment in FABP5 KO mice not mediated by brain-derived neurotrophic factor. Behav. Brain Res. 2022, 425, 113814. [Google Scholar] [CrossRef] [PubMed]
- Oddi, S.; Fezza, F.; Pasquariello, N.; D’Agostino, A.; Catanzaro, G.; De Simone, C.; Rapino, C.; Finazzi-Agrò, A.; Maccarrone, M. Molecular identification of albumin and Hsp70 as cytosolic anandamide-binding proteins. Chem. Biol. 2009, 16, 624–632. [Google Scholar] [CrossRef]
- Kaczocha, M.; Glaser, S.T.; Deutsch, D.G. Identification of intracellular carriers for the endocannabinoid anandamide. Proc. Natl. Acad. Sci. USA 2009, 106, 6375–6380. [Google Scholar] [CrossRef]
- Elmes, M.W.; Prentis, L.E.; McGoldrick, L.L.; Giuliano, C.J.; Sweeney, J.M.; Joseph, O.M.; Che, J.; Carbonetti, G.S.; Studholme, K.; Deutsch, D.G.; et al. FABP1 controls hepatic transport and biotransformation of D9-THC. Sci. Rep. 2019, 9, 7588. [Google Scholar] [CrossRef]
- Fu, J.; Bottegoni, G.; Sasso, O.; Bertorelli, R.; Rocchia, W.; Masetti, M.; Guijarro, A.; Lodola, A.; Armirotti, A.; Garau, G.; et al. A catalytically silent FAAH-1 variant drives anandamide transport in neurons. Nat. Neurosci. 2011, 15, 64–69. [Google Scholar] [CrossRef]
- Hillard, C.J.; Huang, H.; Vogt, C.D.; Rodrigues, B.E.; Neumann, T.S.; Sem, D.S.; Schroeder, F.; Cunningham, C.W. Endocannabinoid transport proteins: Discovery of tools to study sterol carrier protein-2. Methods Enzymol. 2017, 593, 99–121. [Google Scholar]
- Plau, J.; Golczak, M.; Paik, J.; Calderon, R.M.; Blaner, W.S. Retinol-binding protein 2 (RBP2): More than just dietary retinoid uptake. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159179. [Google Scholar] [CrossRef]
- Khasabova, I.A.; Golovko, M.Y.; Golovko, S.A.; Simone, D.A.; Khasabov, S.G. Intrathecal administration of resolvin D1 and E1 decreases hyperalgesia in mice with bone cancer pain: Involvement of endocannabinoid signaling. Prostaglandins Other Lipid Mediat. 2020, 151, 106479. [Google Scholar] [CrossRef] [PubMed]
- Hermes, D.J.; Yadav-Samudrala, B.J.; Xu, C.; Paniccia, J.E.; Meeker, R.B.; Armstrong, M.L.; Reisdorph, N.; Cravatt, B.F.; Mackie, K.; Lichtman, A.H.; et al. GPR18 drives FAAH inhibition-induced neuroprotection against HIV-1 Tat-induced neurodegeneration. Exp. Neurol. 2021, 341, 113699. [Google Scholar] [CrossRef]
- McHugh, D. GPR18 in microglia: Implications for the CNS and endocannabinoid system signalling. Br. J. Pharmacol. 2012, 167, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Maccarrone, M.; Fiorucci, L.; Erba, F.; Bari, M.; Finazzi-Agrò, M.; Ascoli, F. Human mast cells take up and hydrolyze anandamide under the control of 5-lipoxygenase and do not express cannabinoid receptors. FEBS Lett. 2000, 468, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Marti-Solano, M. A multi-dimensional view of context-dependent G protein-coupled receptor function. Biochem. Soc. Trans. 2023; in press. [Google Scholar] [CrossRef]
- Chen, Q.; Tesmer, J.J.G. G protein-coupled receptor interactions with arrestins and GPCR kinases: The unresolved issue of signal bias. J. Biol. Chem. 2022, 298, 102279. [Google Scholar] [CrossRef]
- Grabiec, U.; Hohmann, T.; Ghadban, C.; Rothgänger, C.; Wong, D.; Antonietti, A.; Growth, T.; Mackie, K.; Dehghani, F. Protective effect of N-arachidonoyl glycine-GPR18 signaling after excitotoxical lesion in murine organotypic hippocampal slice cultures. Int. J. Mol. Sci. 2019, 20, 1266. [Google Scholar] [CrossRef]
- Patricio, F.; Morales-Andrade, A.A.; Patricio-Martínez, A.; Limón, I.D. Cannabidiol as a therapeutic target: Evidence of its neuroprotective and neuromodulatory function in Parkinson’s disease. Front. Pharmacol. 2020, 11, 595635. [Google Scholar] [CrossRef]
- Rinne, P.; Guillamat-Prats, R.; Rami, M.; Bindila, L.; Ring, L.; Lyytikäinen, L.P.; Raitoharju, E.; Oksala, N.; Lehtimäki, T.; Weber, C.; et al. Palmitoylethanolamide promotes a proresolving macrophage phenotype and attenuates atherosclerotic plaque formation. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2562–2575. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, Y.; Huang, X.; Yuan, A.; Shao, Q.; Pu, J.; He, B. Selective activation of CB2 receptor improves efferocytosis in cultured macrophages. Life Sci. 2016, 161, 10–18. [Google Scholar] [CrossRef]
- Reyes-Resina, I.; Navarro, G.; Aguinaga, D.; Canela, E.I.; Schoeder, C.T.; Załuski, M.; Kieć-Kononowicz, K.; Saura, C.A.; Müller, C.E.; Franco, R. Molecular and functional interaction between GPR18 and cannabinoid CB2 G-protein-coupled receptors. Relevance in neurodegenerative diseases. Biochem. Pharmacol. 2018, 157, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Langmead, C.J.; Riddy, D.M. New advances in targeting the resolution of inflammation: Implications for specialized pro-resolving mediator GPCR drug discovery. ACS Pharmacol. Transl. Sci. 2020, 3, 88–106. [Google Scholar] [CrossRef]
- Pamplona, F.A.; Ferreira, J.; Menezes de Lima, O., Jr.; Duarte, F.S.; Bento, A.F.; Forner, S.; Villarinho, J.G.; Bellocchio, L.; Wotjak, C.T.; Lerner, R.; et al. Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor. Proc. Natl. Acad. Sci. USA 2012, 109, 21134–21139. [Google Scholar] [CrossRef] [PubMed]
- Pertwee, R.G. Lipoxin A4 is an allosteric endocannabinoid that strengthens anandamide-induced CB1 receptor activation. Proc. Natl. Acad. Sci. USA 2012, 109, 20781–206782. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Bucher, C.; Liu, W.; Müller, M.; Schmidt, T.; Kardell, M.; Driessen, M.N.; Rossaint, J.; Gross, E.R.; Wagner, N.M. 12(S)-HETE mediates diabetes-induced endothelial dysfunction by activating intracellular endothelial cell TRPV1. J. Clin. Investig. 2020, 130, 4999–5010. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xiong, M.; Zong, X.; Ge, Y.; Hui Zhang, H.; Wang, M.; Han, G.W.; Yi, C.; Ma, L.; Ye, R.D.; et al. Structural basis of ligand binding modes at the human formyl peptide receptor 2. Nat. Commun. 2020, 11, 1208. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; He, X.; Zhao, J.; Jiang, H.; Cheng, X.; Xia, Y.; Xu, H.E.; He, Y. Structural basis of leukotriene B4 receptor 1 activation. Nat. Commun. 2022, 13, 1156. [Google Scholar] [CrossRef]
- Nakamura, H.; Murayama, T. Role of sphingolipids in arachidonic acid metabolism. J. Pharmacol. Sci. 2014, 124, 307–312. [Google Scholar] [CrossRef]
- Standoli, S.; Pecchioli, S.; Tortolani, D.; Di Meo, C.; Fanti, F.; Sergi, M.; Bacci, M.; Seidita, I.; Bernacchioni, C.; Donati, C.; et al. The TRPV1 receptor is up-regulated by sphingosine 1-phosphate and is implicated in the anandamide-dependent regulation of mitochondrial activity in C2C12 myoblasts. Int. J. Mol. Sci. 2022, 23, 11103. [Google Scholar] [CrossRef]
- Kang, J.W.; Choi, H.S.; Shin, J.K.; Lee, S.M. Resolvin D1 activates the sphingosine-1-phosphate signaling pathway in murine livers with ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2019, 514, 1058–1065. [Google Scholar] [CrossRef]
- Grimaldi, P.; Pucci, M.; Di Siena, S.; Di Giacomo, D.; Pirazzi, V.; Geremia, R.; Maccarrone, M. The faah gene is the first direct target of estrogen in the testis: Role of histone demethylase LSD1. Cell. Mol. Life Sci. 2012, 69, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Maccarrone, M.; Bari, M.; Di Rienzo, M.; Finazzi-Agrò, A.; Rossi, A. Progesterone activates fatty acid amide hydrolase (FAAH) promoter in human T lymphocytes through the transcription factor Ikaros. Evidence for a synergistic effect of leptin. J. Biol. Chem. 2003, 278, 32726–32732. [Google Scholar] [CrossRef] [PubMed]
- Castro-Muñoz, L.J.; Ulloa, E.V.; Sahlgren, C.; Lizano, M.; De La Cruz-Hernández, E.; Contreras-Paredes, A. Modulating epigenetic modifications for cancer therapy. Oncol. Rep. 2023, 49, 59. [Google Scholar] [CrossRef] [PubMed]
- Punt, J.M.; van der Vliet, D.; van der Stelt, M. Chemical probes to control and visualize lipid metabolism in the brain. Acc. Chem. Res. 2022, 55, 3205–3217. [Google Scholar] [CrossRef]
- Gazzi, T.; Brennecke, B.; Atz, K.; Korn, C.; Sykes, D.; Forn-Cuni, G.; Pfaff, P.; Sarott, R.C.; Westphal, M.V.; Mostinski, Y.; et al. Detection of cannabinoid receptor type 2 in native cells and zebrafish with a highly potent, cell-permeable fluorescent probe. Chem. Sci. 2022, 13, 5539–5545. [Google Scholar] [CrossRef]
| Time Range | EICs | ECBs | SPMs |
|---|---|---|---|
| 1992–1997 | 22,258 | 200 | 3 |
| 1998–2002 | 16,534 | 648 | 5 |
| 2003–2007 | 17,065 | 1791 | 18 |
| 2008–2012 | 16,128 | 2866 | 85 |
| 2013–2017 | 14,402 | 3271 | 316 |
| 2018–2022 | 10,713 | 3949 | 847 |
| Enzyme/Receptor | EICs | ECBs | SPMs |
|---|---|---|---|
| PLA2 | + | + | + |
| CYP450 | + | + | + |
| 12-LOX | + | + | + |
| COX-2 | + | + | + |
| CB1 | + | + | + |
| CB2 | + | + | |
| GPR18 | + | + | |
| TRPV1 | + | + | + |
| BLT1 | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maccarrone, M. Deciphering Complex Interactions in Bioactive Lipid Signaling. Molecules 2023, 28, 2622. https://doi.org/10.3390/molecules28062622
Maccarrone M. Deciphering Complex Interactions in Bioactive Lipid Signaling. Molecules. 2023; 28(6):2622. https://doi.org/10.3390/molecules28062622
Chicago/Turabian StyleMaccarrone, Mauro. 2023. "Deciphering Complex Interactions in Bioactive Lipid Signaling" Molecules 28, no. 6: 2622. https://doi.org/10.3390/molecules28062622
APA StyleMaccarrone, M. (2023). Deciphering Complex Interactions in Bioactive Lipid Signaling. Molecules, 28(6), 2622. https://doi.org/10.3390/molecules28062622






