Antitumor and Antibacterial Activity of Ni(II), Cu(II), Ag(I), and Hg(II) Complexes with Ligand Derived from Thiosemicarbazones: Characterization and Theoretical Studies
Abstract
1. Introduction
2. Results
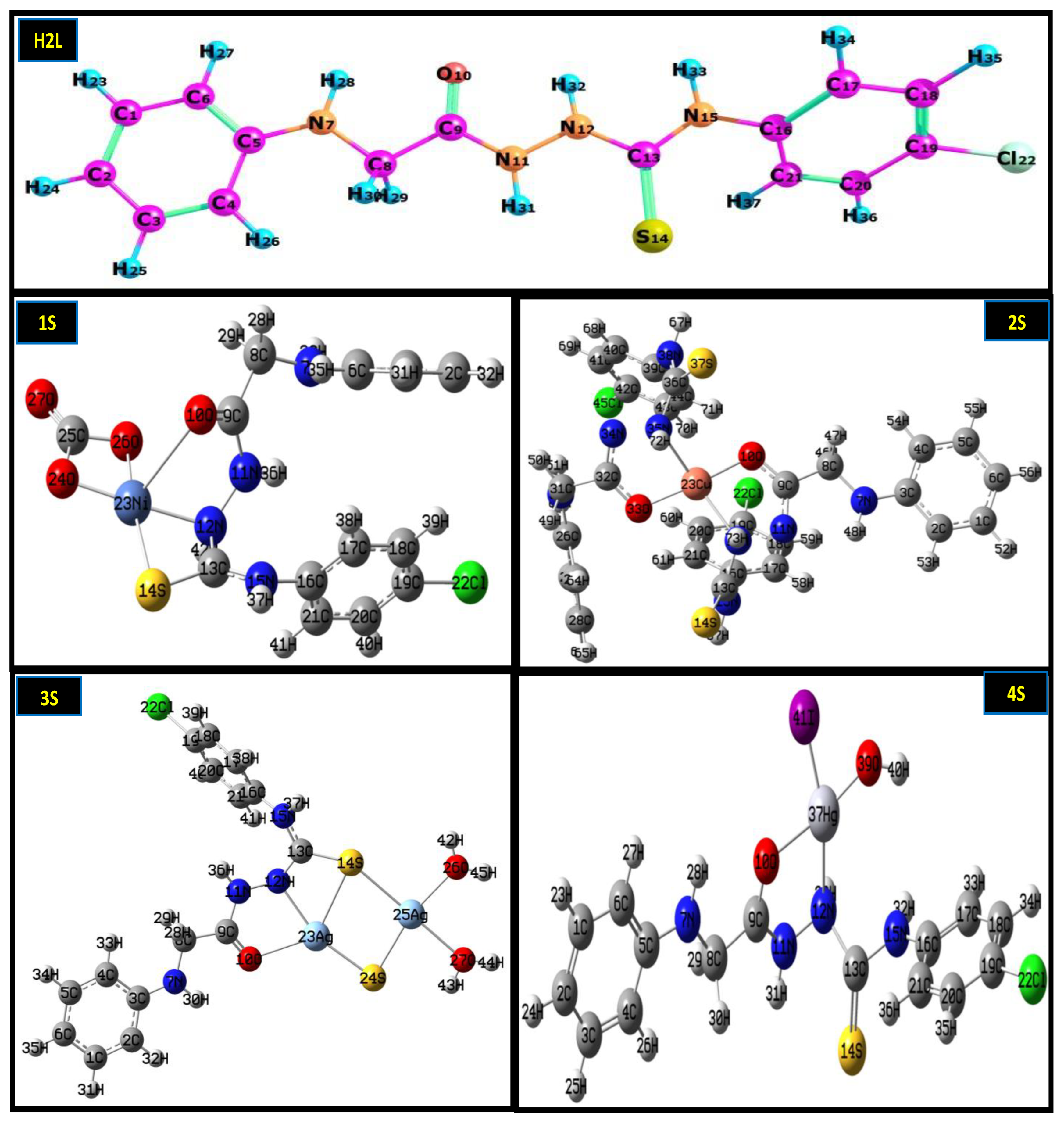
2.1. Physicochemical Properties
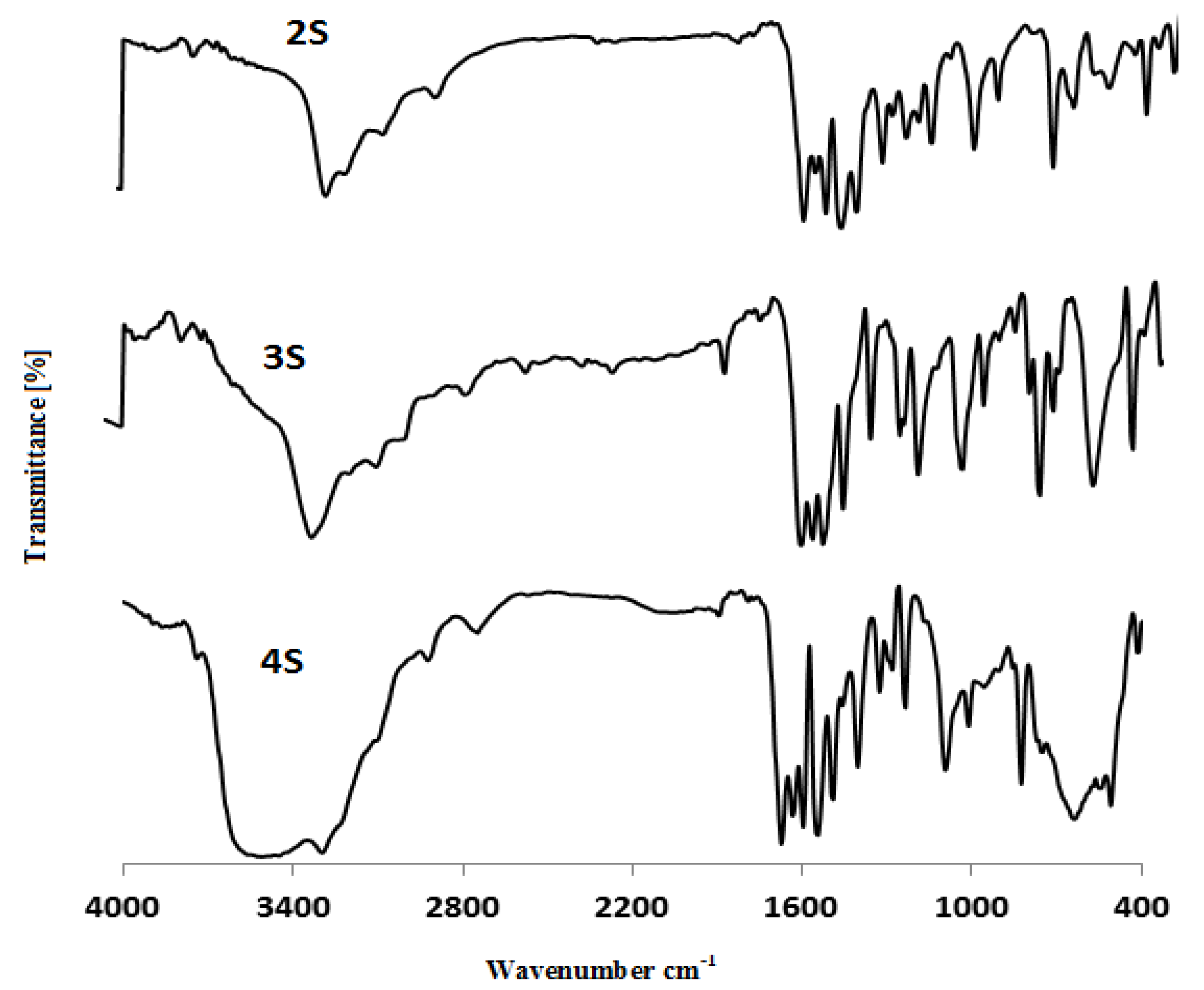
2.2. FT-IR
2.3. ESI-MS Spectra
2.4. Electronic Spectral Bands
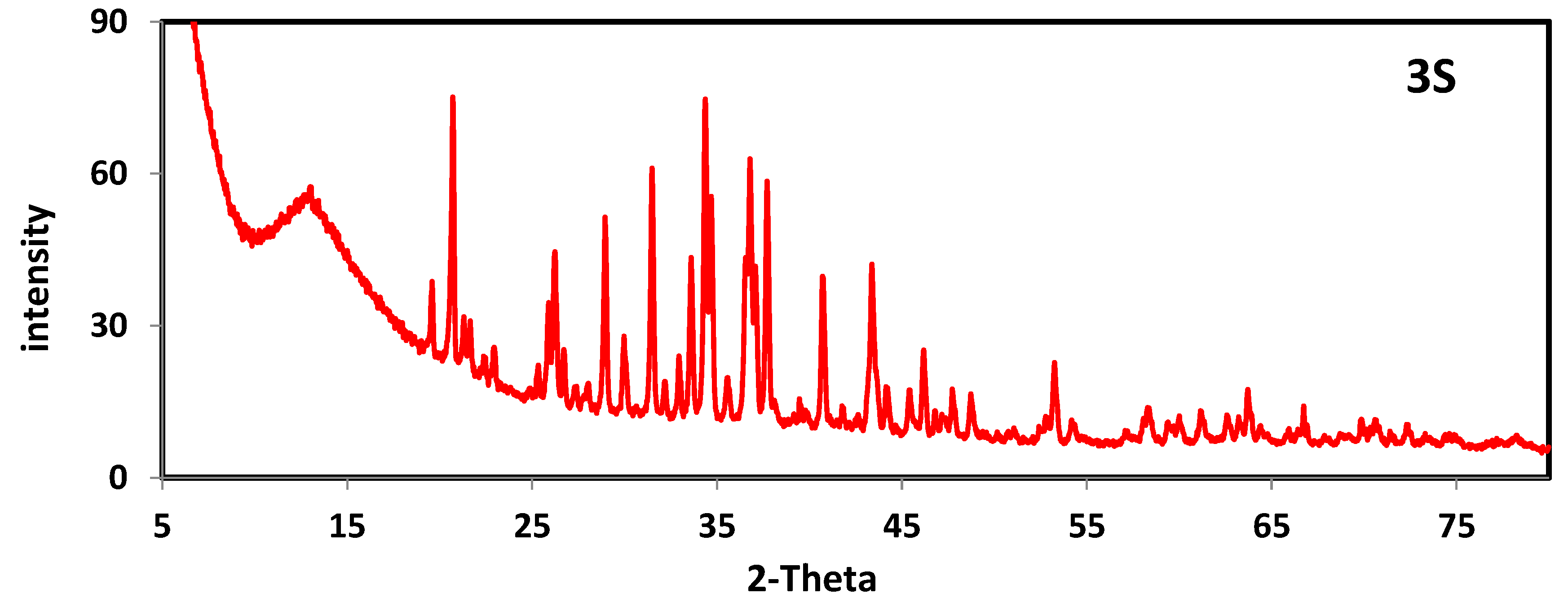
2.5. PXRD of Ligand and Metal Complexes
2.6. Thermal Analysis
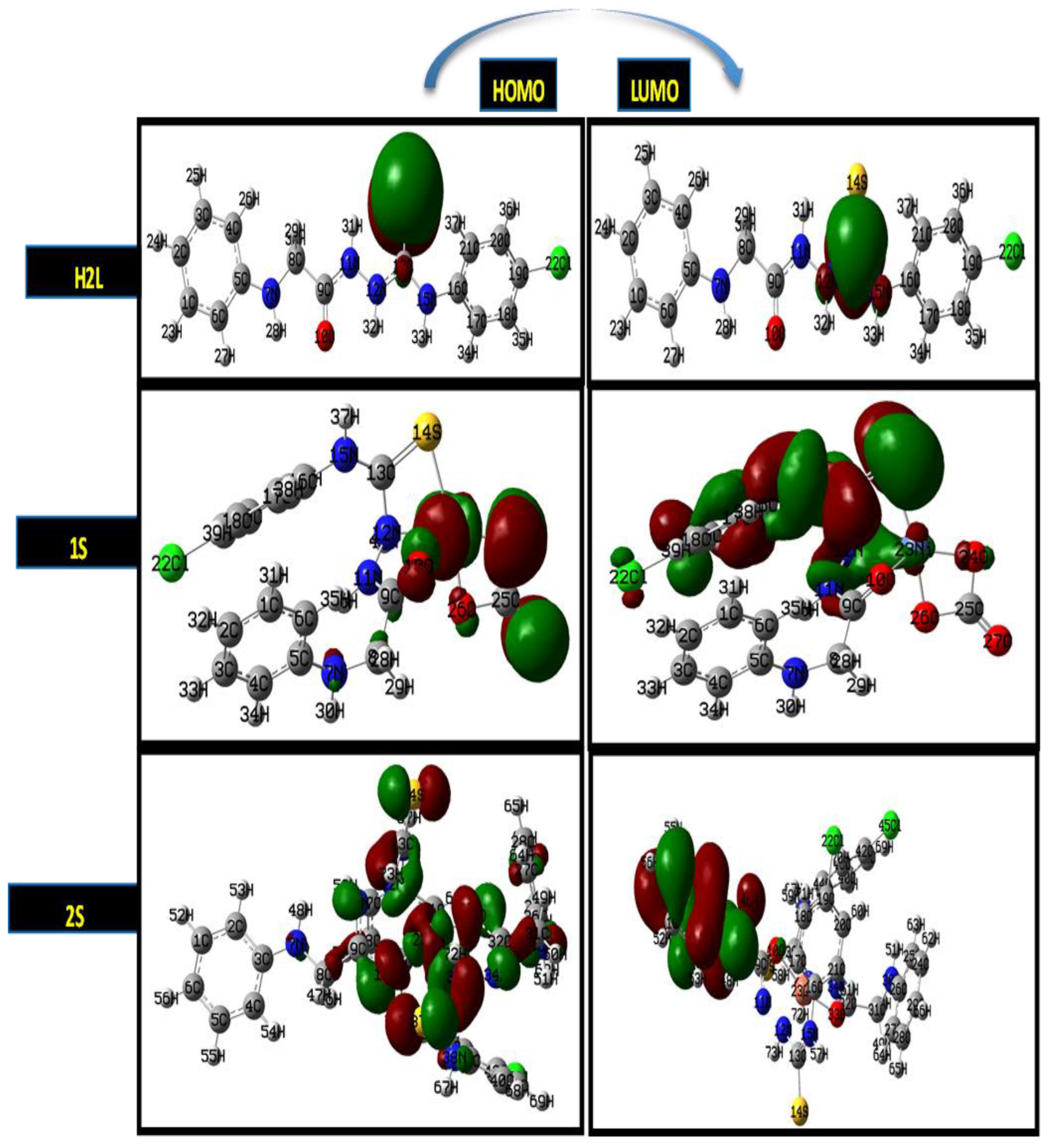
2.7. DFT Calculations of the Ligand and Metal Complexes
2.8. Biological Applications
2.8.1. Antibacterial Activity
2.8.2. Cytotoxicity
2.8.3. Molecular Docking Studies
3. Experimental Section
3.1. Material and Methods
3.2. Preparation of Ligand and Metal Complexes
3.3. Computational Study
3.4. Antibacterial Assay
3.5. Cytotoxicity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Gamasy, S.M.; Ebrahim, S. 4N donor atoms moiety of transition metal complexes of a Schiff base ligand: Synthesis, characterization and biological activities study. Egypt. J. Chem. 2021, 64, 3–4. [Google Scholar] [CrossRef]
- Kyhoiesh, H.A.K.; Al-Adilee, K.J. Synthesis, spectral characterization, antimicrobial evaluation studies and cytotoxic activity of some transition metal complexes with tridentate (N, N, O) donor azo dye ligand. Results Chem. 2021, 3, 100245. [Google Scholar] [CrossRef]
- Abdalla, E.M.; Hassan, S.S.; Elganzory, H.H.; Aly, S.A.; Alshater, H. Molecular Docking, DFT Calculations, Effect of High Energetic Ionizing Radiation, and Biological Evaluation of Some Novel Metal (II) Heteroleptic Complexes Bearing the Thiosemicarbazone Ligand. Molecules 2021, 26, 5851. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, L.H.; Abdelghani, A.A.; AlObaid, A.A.; El-ezz, D.A.; Warad, I.; Shehata, M.R.; Abdalla, E.M. Novel Bromo and methoxy substituted Schiff base complexes of Mn (II), Fe (III), and Cr (III) for anticancer, antimicrobial, docking, and ADMET studies. Sci. Rep. 2023, 13, 3199. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, E.M.; Abd-Allah, M. Synthesis, Characterization, Antimicrobial/Antitumor Activity of Binary and Ternary Neodymium (III) Complex with 2, 2’-((1E, 1’E)-(ethane-1, 2-diylbis (azaneylylidene)) bis (methaneylylidene)) diphenol and Imidazole. Egypt. J. Chem. 2022, 65, 735–744. [Google Scholar] [CrossRef]
- Aly, S.A.; Eldourghamy, A.; El-Fiky, B.A.; Megahed, A.A.; El-Sayed, W.A.; Abdalla, E.M.; Elganzory, H.H. Synthesis, spectroscopic characterization, thermal studies, and molecular docking of novel Cr (III), Fe (III), and Co (II) complexes based on Schiff base: In vitro antibacterial and antitumor activities. J. Appl. Pharm. Sci. 2023, 13, 196–210. [Google Scholar] [CrossRef]
- El-ezz, A.; Abdel-Rahman, L.H.; Al-Farhan, B.S.; Mostafa, D.A.; Ayad, E.G.; Basha, M.T.; Abdelaziz, M.; Abdalla, E.M. Enhanced In Vivo Wound Healing Efficacy of a Novel Hydrogel Loaded with Copper (II) Schiff Base Quinoline Complex (CuSQ) Solid Lipid Nanoparticles. Pharmaceuticals 2022, 15, 978. [Google Scholar] [CrossRef]
- Abdel Rahman, L.H.; Al-Zaqri, N.; Abdelghani, A.A.; Abdalla, E.M. Physicochemical, in vitro therapeutic activity, DNA-binding, and in silico molecular docking studies of samarium (III) complexes bearing N, O-chelated Schiff base ligands. J. Coord. Chem. 2022, 75, 994–1018. [Google Scholar] [CrossRef]
- Al-Farhan, B.S.; Basha, M.T.; Abdel Rahman, L.H.; El-Saghier, A.M.; Abou El-Ezz, D.; Marzouk, A.A.; Shehata, M.R.; Abdalla, E.M. Synthesis, DFT Calculations, Antiproliferative, Bactericidal Activity and Molecular Docking of Novel Mixed-Ligand Salen/8-Hydroxyquinoline Metal Complexes. Molecules 2021, 26, 4725. [Google Scholar] [CrossRef] [PubMed]
- Gaber, M.; Fathalla, S.K.; El-Ghamry, H.A. 2, 4-Dihydroxy-5-[(5-mercapto-1H-1, 2, 4-triazole-3-yl) diazenyl] benzaldehyde acetato, chloro and nitrato Cu (II) complexes: Synthesis, structural characterization, DNA binding and anticancer and antimicrobial activity. Appl. Organomet. Chem. 2019, 33, e4707. [Google Scholar] [CrossRef]
- Du, H.; Williams, C.T.; Ebner, A.D.; Ritter, J.A. In situ FTIR spectroscopic analysis of carbonate transformations during adsorption and desorption of CO2 in K-promoted HTlc. Chem. Mater. 2010, 22, 3519–3526. [Google Scholar] [CrossRef]
- Qin, L.; Liang, F.; Li, Y.; Wu, J.; Guan, S.; Wu, M.; Xie, S.; Luo, M.; Ma, D. A 2D porous zinc-organic framework platform for loading of 5-fluorouracil. Inorganics 2022, 10, 202. [Google Scholar] [CrossRef]
- Qin, L.; Li, Y.; Liang, F.; Li, L.; Lan, Y.; Li, Z.; Lu, X.; Yang, M.; Ma, D. A microporous 2D cobalt-based MOF with pyridyl sites and open metal sites for selective adsorption of CO2. Microporous Mesoporous Mater. 2022, 341, 112098. [Google Scholar] [CrossRef]
- Young, A.G.; Hanton, L.R. Square planar silver (I) complexes: A rare but increasingly observed stereochemistry for silver (I). Coord. Chem. Rev. 2008, 252, 1346–1386. [Google Scholar] [CrossRef]
- Soliman, S.M.; Mabkhot, Y.N.; Albering, J.H. X-ray Structure and DFT Studies of a New Square Planar Silver (I) Complex of Ketene S, S-Dithioacetal Ligand. J. Chem. Crystallogr. 2020, 50, 52–61. [Google Scholar] [CrossRef]
- Pointillart, F.; Herson, P.; Boubekeur, K.; Train, C. Square-planar and trigonal prismatic silver (I) in bipyrimidine and oxalate bridged tetranuclear complexes and one-dimensional compounds: Synthesis and crystal structures. Inorg. Chim. Acta 2008, 361, 373–379. [Google Scholar] [CrossRef]
- Aly, S.A.; Elganzory, H.H.; Mahross, M.H.; Abdalla, E.M. Quantum chemical studies and effect of gamma irradiation on the spectral, thermal, X-ray diffraction and DNA interaction with Pd (II), Cu (I), and Cd (II) of hydrazone derivatives. Appl. Organomet. Chem. 2021, 35, e6153. [Google Scholar] [CrossRef]
- Elganzory, H.H.; Hassan, S.S.; Aly, S.A.; Abdalla, E.M. Synthesis, Characterization, PXRD Studies, Theoretical Calculation, and Antitumor Potency Studies of a Novel N, O-Multidentate Chelating Ligand and Its Zr (IV), V (IV), Ru (III), and Cd (II) Complexes. Bioinorg. Chem. Appl. 2022, 2022, 2006451. [Google Scholar] [CrossRef]
- Abu-Khadra, A.S.; Farag, R.S.; Abdel-Hady, A.E.-D.M. Synthesis, characterization and antimicrobial activity of Schiff base (E)-N-(4-(2-hydroxybenzylideneamino) phenylsulfonyl) acetamide metal complexes. Am. J. Anal. Chem. 2016, 7, 233. [Google Scholar] [CrossRef]
- Abdalla, E.M.; Abdel Rahman, L.H.; Abdelhamid, A.A.; Shehata, M.R.; Alothman, A.A.; Nafady, A. Synthesis, Characterization, Theoretical Studies, and Antimicrobial/Antitumor Potencies of Salen and Salen/Imidazole Complexes of Co (II), Ni (II), Cu (II), Cd (II), Al (III) and La (III). Appl. Organomet. Chem. 2020, 34, e5912. [Google Scholar] [CrossRef]
- Abdel-Rahman, L.H.; Basha, M.T.; Al-Farhan, B.S.; Shehata, M.R.; Abdalla, E.M. Synthesis, characterization, potential antimicrobial, antioxidant, anticancer, DNA binding, and molecular docking activities and DFT on novel Co (II), Ni (II), VO (II), Cr (III), and La (III) Schiff base complexes. Appl. Organomet. Chem. 2022, 36, e6484. [Google Scholar] [CrossRef]
- Adly, O.M.; Taha, A.; Ibrahim, M.A. New nickel (II), cobalt (III), and iron (III) complexes with N′-[(2-aminochromon-3-yl) methylidene] benzohydrazide: Synthesis, characterization, solvatochromic shift, dipole moment, and DFT calculations. Appl. Organomet. Chem. 2022, 36, e6558. [Google Scholar] [CrossRef]
- El Alfy, H.; Hassan, A.; Khattab, E.S.A.; Heakal, B.H. Synthesis, Characterization and Biological Evaluation Studies of 4-((3-Formyl-4-hydroxyphenyl) diazinyl)-N-(4-methyloxazol-2-yl) Benzene Sulfonamide with Cu (II), Ni (II), Zn (II) and Ag (I) Using a Microwave Irradiation. Egypt. J. Chem. 2018, 61, 569–580. [Google Scholar] [CrossRef]
- Mandal, S.; Mondal, M.; Biswas, J.K.; Cordes, D.B.; Slawin, A.M.; Butcher, R.J.; Saha, M.; Saha, N.C. Synthesis, characterization and antimicrobial activity of some nickel, cadmium and mercury complexes of 5-methyl pyrazole-3yl-N-(2′-methylthiophenyl) methyleneimine,(MPzOATA) ligand. J. Mol. Struct. 2018, 1152, 189–198. [Google Scholar] [CrossRef]
- Nasiri Sovari, S.; Zobi, F. Recent studies on the antimicrobial activity of transition metal complexes of groups 6–12. Chemistry 2020, 2, 418–452. [Google Scholar] [CrossRef]
- Sangwan, V.; Singh, D. Macrocyclic Schiff base complexes as potent antimicrobial agents: Synthesis, characterization and biological studies. Mater. Sci. Eng. C 2019, 105, 110119. [Google Scholar] [CrossRef] [PubMed]
- Focaccetti, C.; Bruno, A.; Magnani, E.; Bartolini, D.; Principi, E.; Dallaglio, K.; Bucci, E.O.; Finzi, G.; Sessa, F.; Noonan, D.M. Effects of 5-fluorouracil on morphology, cell cycle, proliferation, apoptosis, autophagy and ROS production in endothelial cells and cardiomyocytes. PLoS ONE 2015, 10, e0115686. [Google Scholar] [CrossRef]
- Yan, X.; Chen, J.-Q.; Hu, M.-L.; Sakiyama, H.; Muddassir, M.; Liu, J.-Q. Syntheses, structures and mechanisms of interactions with DNA of two new 20-core silver (I) complexes with different ligands. Inorg. Chim. Acta 2023, 546, 121297. [Google Scholar] [CrossRef]
- Zhang, W.; Ye, G.; Liao, D.; Chen, X.; Lu, C.; Nezamzadeh-Ejhieh, A.; Khan, M.S.; Liu, J.; Pan, Y.; Dai, Z. Recent Advances of Silver-Based Coordination Polymers on Antibacterial Applications. Molecules 2022, 27, 7166. [Google Scholar] [CrossRef]
- Khedr, A.M.; Gouda, A.A.; ElGhamry, H.A. Nano-synthesis approach, elaborated spectral, biological activity and in silico assessment of novel nano-metal complexes based on sulfamerazine azo dye. J. Mol. Liq. 2022, 352, 118737. [Google Scholar] [CrossRef]
- Frisch, A.; Hratchian, H.P.; I, R.D.I.; Keith, T.; Millam, J.; Nielsen, B.; Holder, A.; Hiscocks, J. GaussView Version 5.0.8; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Jahangirian, H.; Haron, M.J.; Shah, M.H.; Abdollahi, Y.; Rezayi, M.; Vafaei, N. Well diffusion method for evaluation of antibacterial activity of copper phenyl fatty hydroxamate synthesized from canola and palm kernel oils. Dig. J. Nanomater. Biostruct. 2013, 8, 1263–1270. [Google Scholar]
- Ikotun, A.; Ojo, Y.; Obafemi, C.; Egharevba, G. Synthesis and antibacterial activity of metal complexes of barbituric acid. Afr. J. Pure Appl. Chem. 2011, 5, 97–103. [Google Scholar]
- Stockert, J.C.; Horobin, R.W.; Colombo, L.L.; Blázquez-Castro, A. Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018, 120, 159–167. [Google Scholar] [CrossRef]
- Tolosa, L.; Donato, M.T.; Gómez-Lechón, M.J. General cytotoxicity assessment by means of the MTT assay. In Protocols in In Vitro Hepatocyte Research; Springer: Berlin/Heidelberg, Germany, 2015; pp. 333–348. [Google Scholar]
- Meerloo, J.V.; Kaspers, G.J.; Cloos, J. Cell sensitivity assays: The MTT assay. In Cancer Cell Culture; Springer: Berlin/Heidelberg, Germany, 2011; pp. 237–245. [Google Scholar]
- Van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 47. [Google Scholar] [CrossRef] [PubMed]



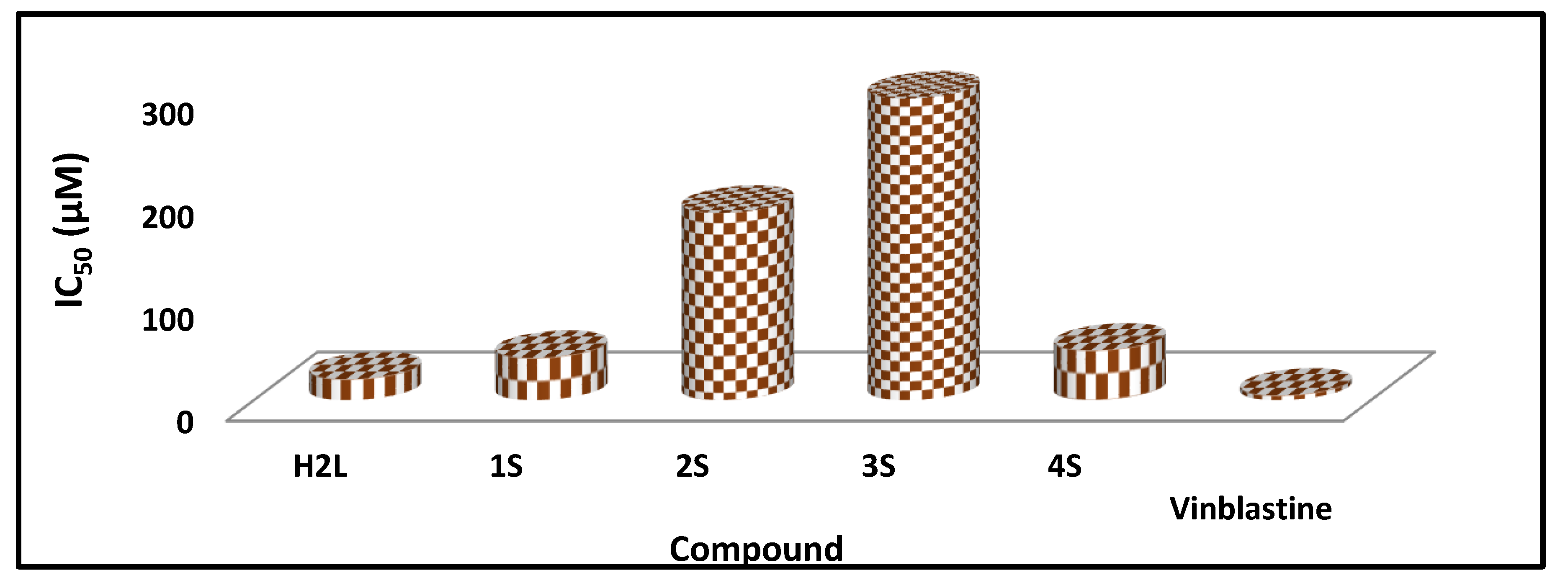

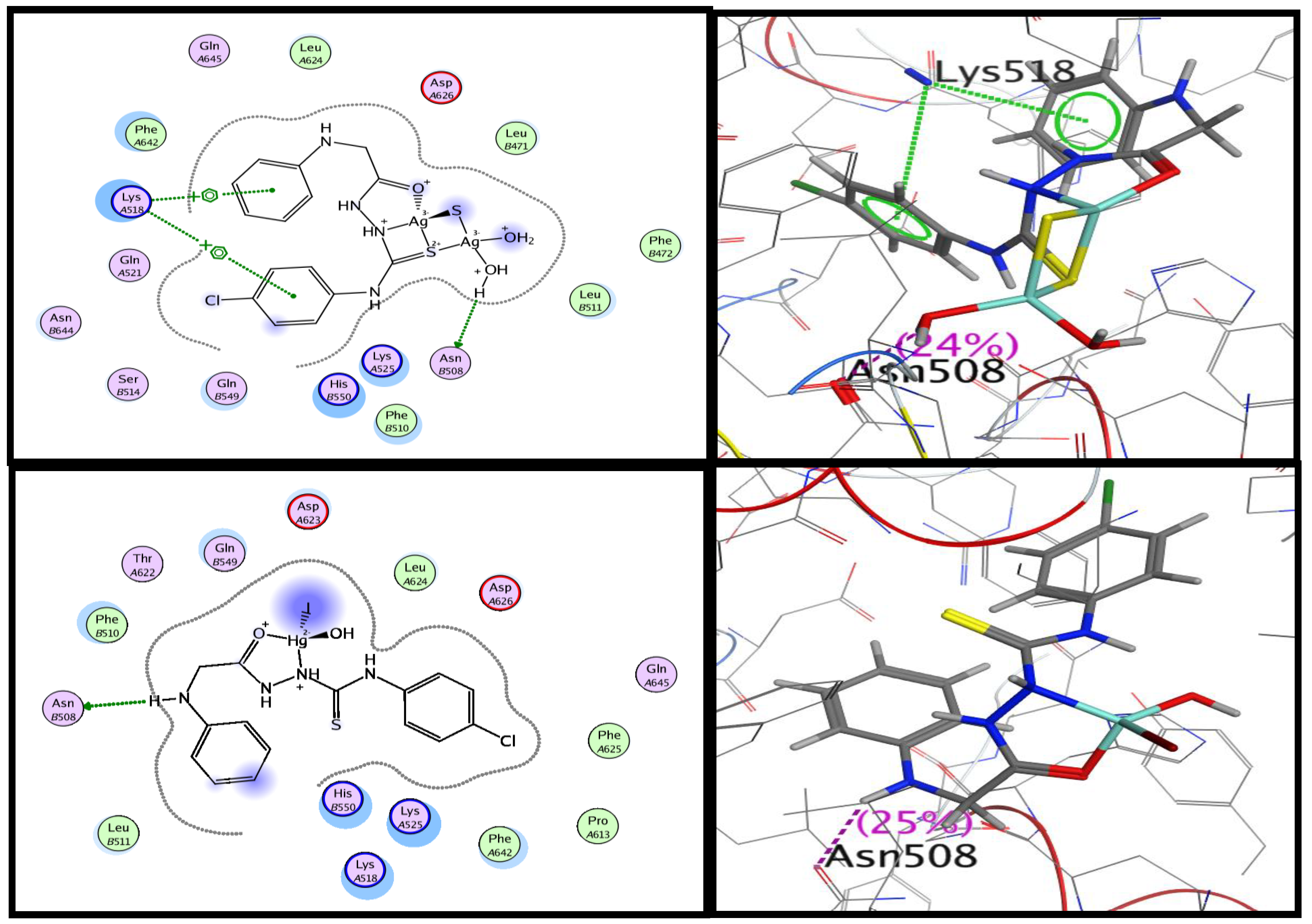
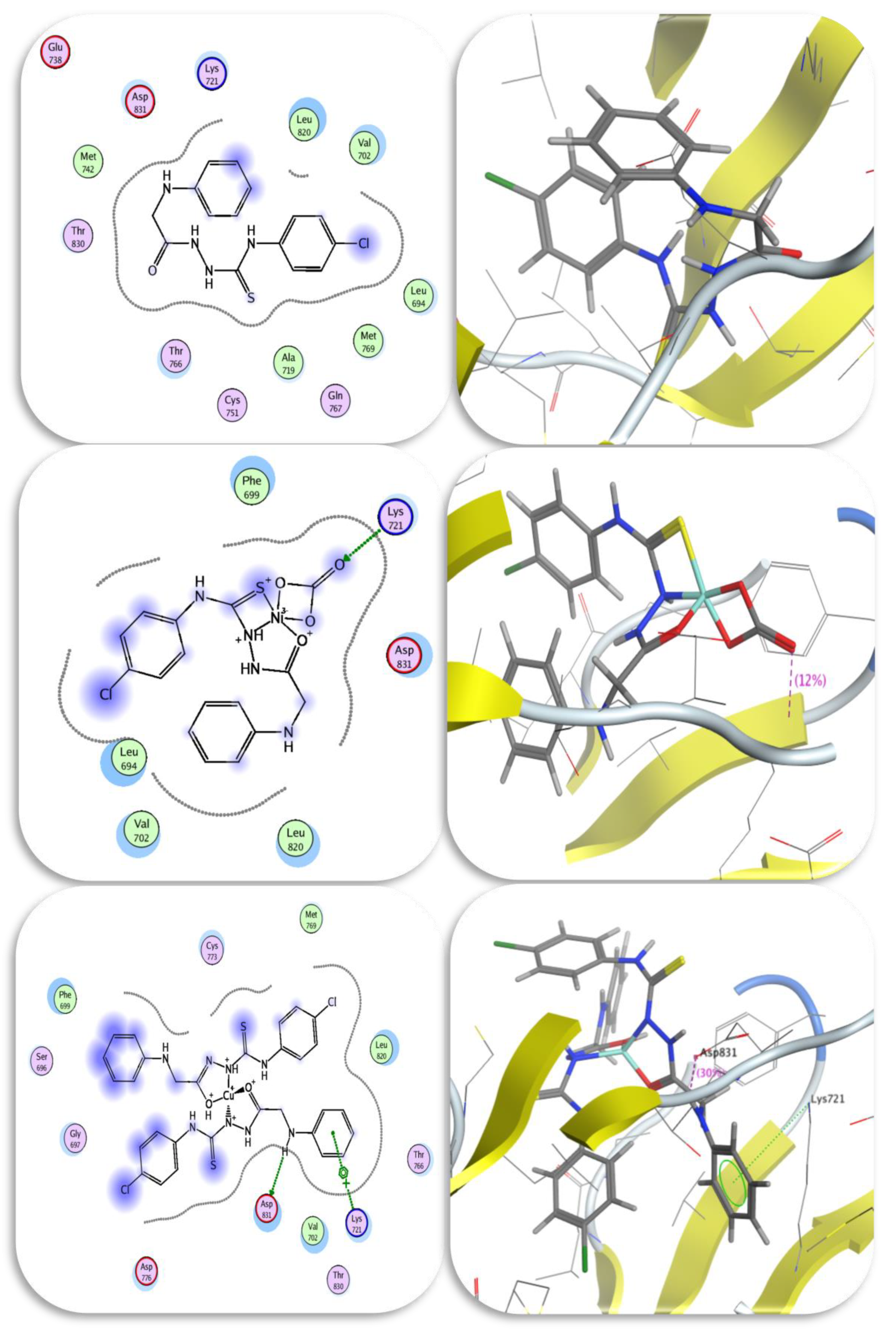
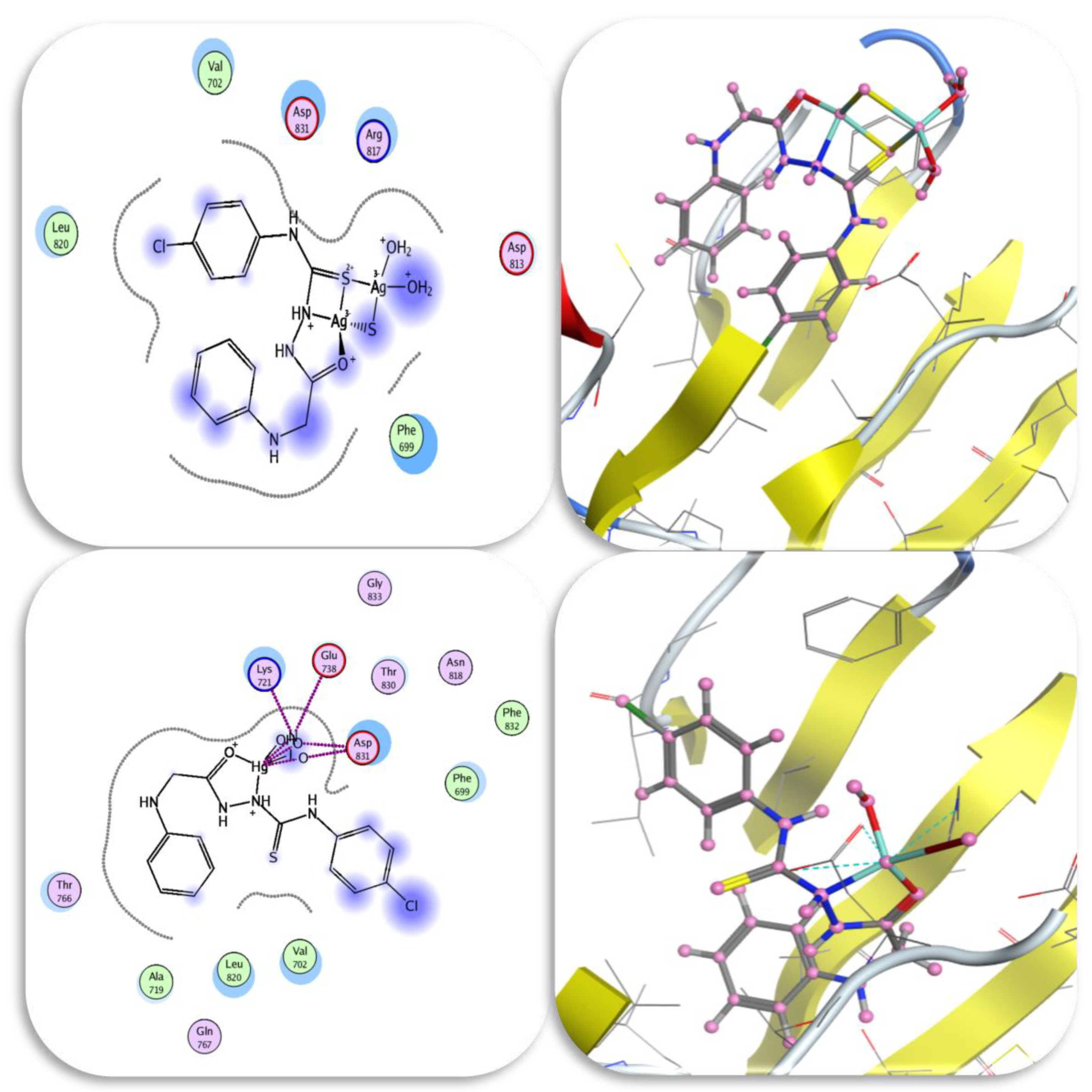

| No. | Compounds | Color Yield % | Mol. Wt | Found (cal.) % | Am | ||||
|---|---|---|---|---|---|---|---|---|---|
| C | H | N | Cl | M | |||||
| H2L | C15H15ClN4OS | Pale brown 75 | 334.82 | 53.43(53.81) | 4.46(4.52) | 16.47(16.73) | 10.83(10.59) | — | — |
| 1S | C16H17ClN4NiO5S | Buff 70 | 471.54 | 40.71(40.76) | 3.59(3.63) | 11.79(11.88) | 7.45(7.52) | 12.36(12.45) | 29 |
| 2S | C30H29Cl2CuN8O2S2 | DarkGreen 80 | 732.18 | 49.19(49.21) | 3.81(3.99) | 15.27(15.30) | 9.65(9.68) | 8.63(8.68) | 28 |
| 3S | C15H19Ag2ClN4O3S2 | Black 75 | 618.65 | 29.01(29.12) | 3.04(3.10) | 8.94(9.06) | 5.68(5.73) | 34.83(34.87) | 31 |
| 4S | C15H20ClHgIN4O4S | Grey 70 | 715.35 | 25.11(25.19) | 2.79(2.82) | 7.76(7.83) | 4.91(4.96) | 27.98(28.04) | 25 |
| No | ν(N4-H) | ν(N2-H) | ν(N1-H) | ν(C=O) | ν(C=S) | ν(M-O) | ν(M-N) | CO3/CO2 | (OH/H2O)coord. | (OH/H2O)hy. |
|---|---|---|---|---|---|---|---|---|---|---|
| H2L | 3335 | 3302 | 3100 | 1670 | 750 | — | — | — | — | — |
| 1S | 3296 | 3102 | 2926 | 1672–1634 | 755 | 549 | 448 | 1634–1595 | — | 3422 |
| 2S | 3519 | 3291 | 3101 | 1671 | 756 | 509 | 462 | — | — | — |
| 3S | 3641 | 3294 | 3018 | 1627 | 756 | 501 | 462 | — | 956 | — |
| 4S | 3448 | 3294 | 2924 | 1674 | 756 | 509 | 416 | — | 941 | 3471 |
| No | Compounds | λmax (DMF, nm) | Meff BM. |
|---|---|---|---|
| H2L | C15H15ClN4OS | 260, 300 | — |
| 1S | C16H17ClN4NiO5S | 286, 374 | 2.84 |
| 2S | C30H29Cl2CuN8O2S2 | 281, 371 | Dia |
| 3S | C15H19Ag2ClN4O3S2 | 281 | — |
| 4S | C15H20ClHgIN4O4S | 299, 378 | — |
| No | Compound | Angle 2θ | d-Value nm | FWHM | Grain Size nm |
|---|---|---|---|---|---|
| H2L | C15H15ClN4OS | 15.852 | 0.560592 | 0.215 | 41.50 |
| 20.645 | 0.430118 | 0.256 | 35.12 | ||
| 23.817 | 0.372922 | 0.220 | 41.02 | ||
| 3S | C15H19Ag2ClN4O3S2 | 20.717 | 0.429305 | 0.165 | 54.35 |
| 31.492 | 0.283903 | 0.176 | 52.03 | ||
| 34.373 | 0.259788 | 0.345 | 26.75 | ||
| 4S | C15H20ClHgIN4O4S | 20.677 | 0.429818 | 0.172 | 52.28 |
| 21.547 | 0.413320 | 0.179 | 50.11 | ||
| 24.882 | 0.358438 | 0.141 | 64.12 |
| No | Compound | TAG(A)/oC | Wt. Loss Calc. (Found) % | Leaving Species |
|---|---|---|---|---|
| H2L | At 190 | - | Melting | |
| C15H15ClN4OS | 190–633 | 99.9 | Gradual decomp. | |
| 1S | C16H17ClN4NiO5S | 41–178 | 3.82 (3.86) | H2O |
| 178–288 | 63.27 (63.21) | C15H14N4OS | ||
| 288–391 | 17.06 (17.11) | HCl + CO2 | ||
| Residue | >391 | 15.85 (15.81) | NiO | |
| 2S | C30H29Cl2CuN8O2S2 | 163–363 | 91.32 (91.28) | Decomp. |
| Residue | >800 | 8.68 (8.71) | Cu | |
| 3S | C15H19Ag2ClN4O3S2 | 244–361 | 56.72 (56.76) | Decomp. |
| Residue | >650 | 43.28 (43.22) | 2AgO + 3C | |
| 4S | C15H20ClHgIN4O4S | 105–385 | 59.65 (59.60) | Decomp. |
| Residue | >800 | 40.35 (40.39) | HgO + 6C |
| Parameter | H2L | 1S | 2S | 3S | 4S |
|---|---|---|---|---|---|
| ET, Hartree | −1733.21933 | −1332.83777 | −1994.48187 | −1354.25992 | −1029.79482 |
| EHOMO, Ev | −5.94 | −6.17 | −5.14 | −3.71 | −4.86 |
| ELUMO, Ev | −5.43 | −2.99 | −1.65 | −2.76 | −4.21 |
| ΔE, eV | 0.51 | 3.17 | 3.48 | 0.95 | 0.64 |
| I = −E HOMO, eV | 5.94 | 6.17 | 5.14 | 3.71 | 4.86 |
| A = −E LUMO, eV | 5.43 | 2.99 | 1.65 | 2.76 | 4.21 |
| χ, eV | 22.27 | 2.88 | 1.95 | 6.81 | 13.96 |
| η, eV | 0.255 | 1.58 | 1.74 | 0.47 | 0.32 |
| S, eV−1 | 1.96 | 0.31 | 0.28 | 1.05 | 1.53 |
| µ, eV | −5.68 | −4.58 | −3.39 | −3.24 | −4.53 |
| Dipole moment (debye) | 2.4081 | 17.30 | 4.28 | 11.40 | 4.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshater, H.; Al-Sulami, A.I.; Aly, S.A.; Abdalla, E.M.; Sakr, M.A.; Hassan, S.S. Antitumor and Antibacterial Activity of Ni(II), Cu(II), Ag(I), and Hg(II) Complexes with Ligand Derived from Thiosemicarbazones: Characterization and Theoretical Studies. Molecules 2023, 28, 2590. https://doi.org/10.3390/molecules28062590
Alshater H, Al-Sulami AI, Aly SA, Abdalla EM, Sakr MA, Hassan SS. Antitumor and Antibacterial Activity of Ni(II), Cu(II), Ag(I), and Hg(II) Complexes with Ligand Derived from Thiosemicarbazones: Characterization and Theoretical Studies. Molecules. 2023; 28(6):2590. https://doi.org/10.3390/molecules28062590
Chicago/Turabian StyleAlshater, Heba, Ahlam I. Al-Sulami, Samar A. Aly, Ehab M. Abdalla, Mohamed A. Sakr, and Safaa S. Hassan. 2023. "Antitumor and Antibacterial Activity of Ni(II), Cu(II), Ag(I), and Hg(II) Complexes with Ligand Derived from Thiosemicarbazones: Characterization and Theoretical Studies" Molecules 28, no. 6: 2590. https://doi.org/10.3390/molecules28062590
APA StyleAlshater, H., Al-Sulami, A. I., Aly, S. A., Abdalla, E. M., Sakr, M. A., & Hassan, S. S. (2023). Antitumor and Antibacterial Activity of Ni(II), Cu(II), Ag(I), and Hg(II) Complexes with Ligand Derived from Thiosemicarbazones: Characterization and Theoretical Studies. Molecules, 28(6), 2590. https://doi.org/10.3390/molecules28062590






