Structural Elucidation and Cytotoxic Activity of New Monoterpenoid Indoles from Gelsemium elegans
Abstract
1. Introduction
2. Results and Discussion
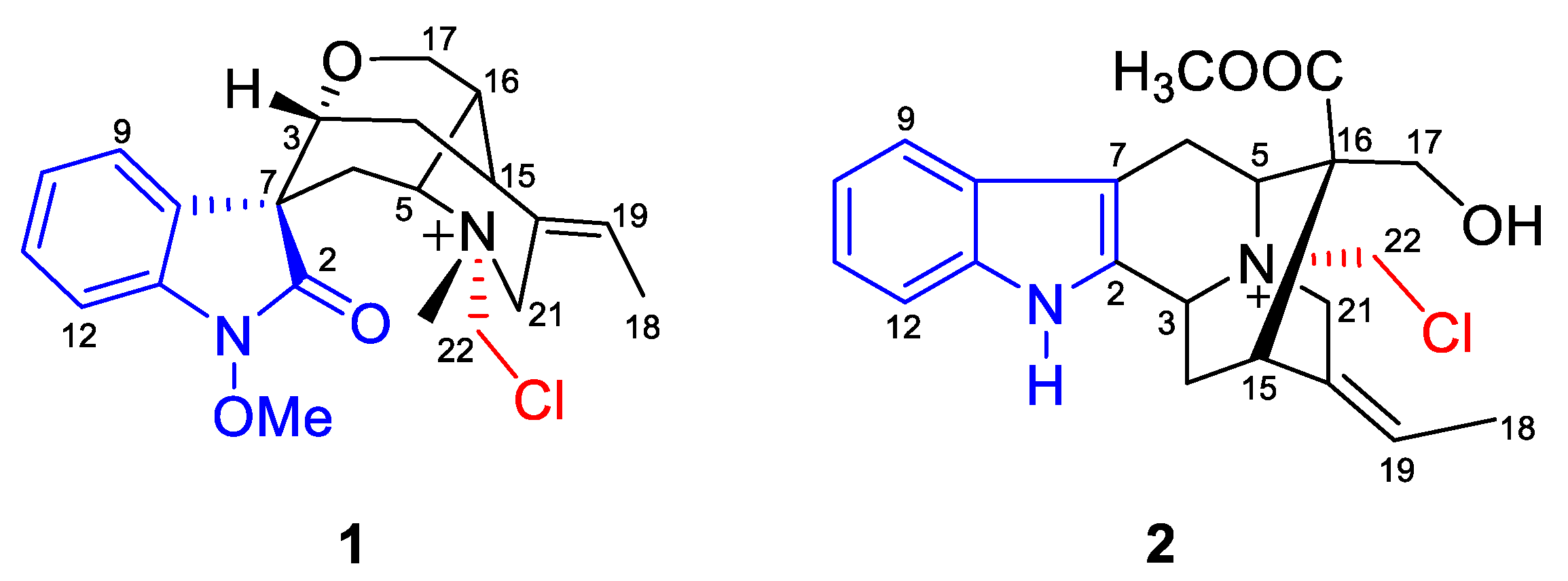
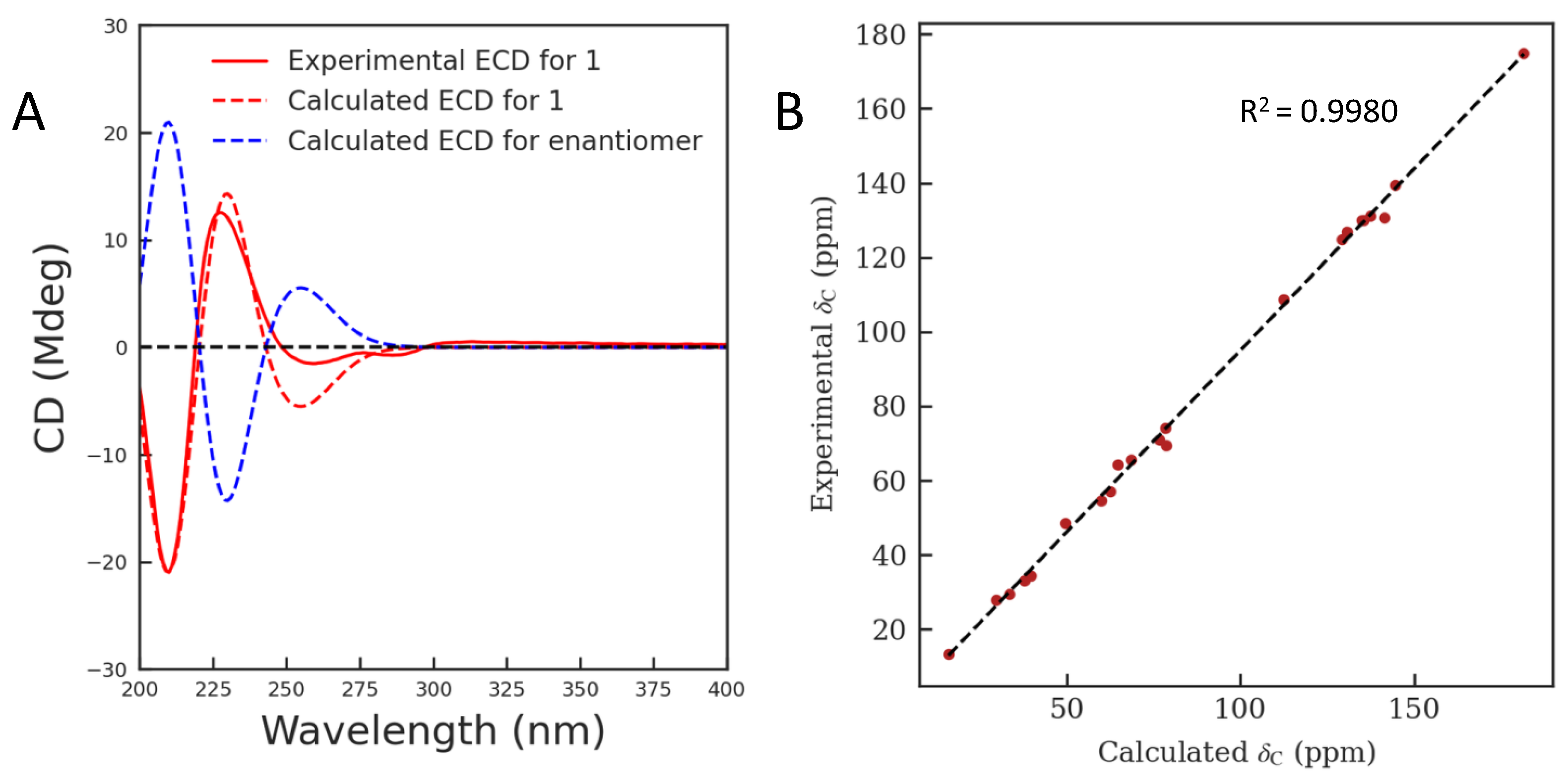
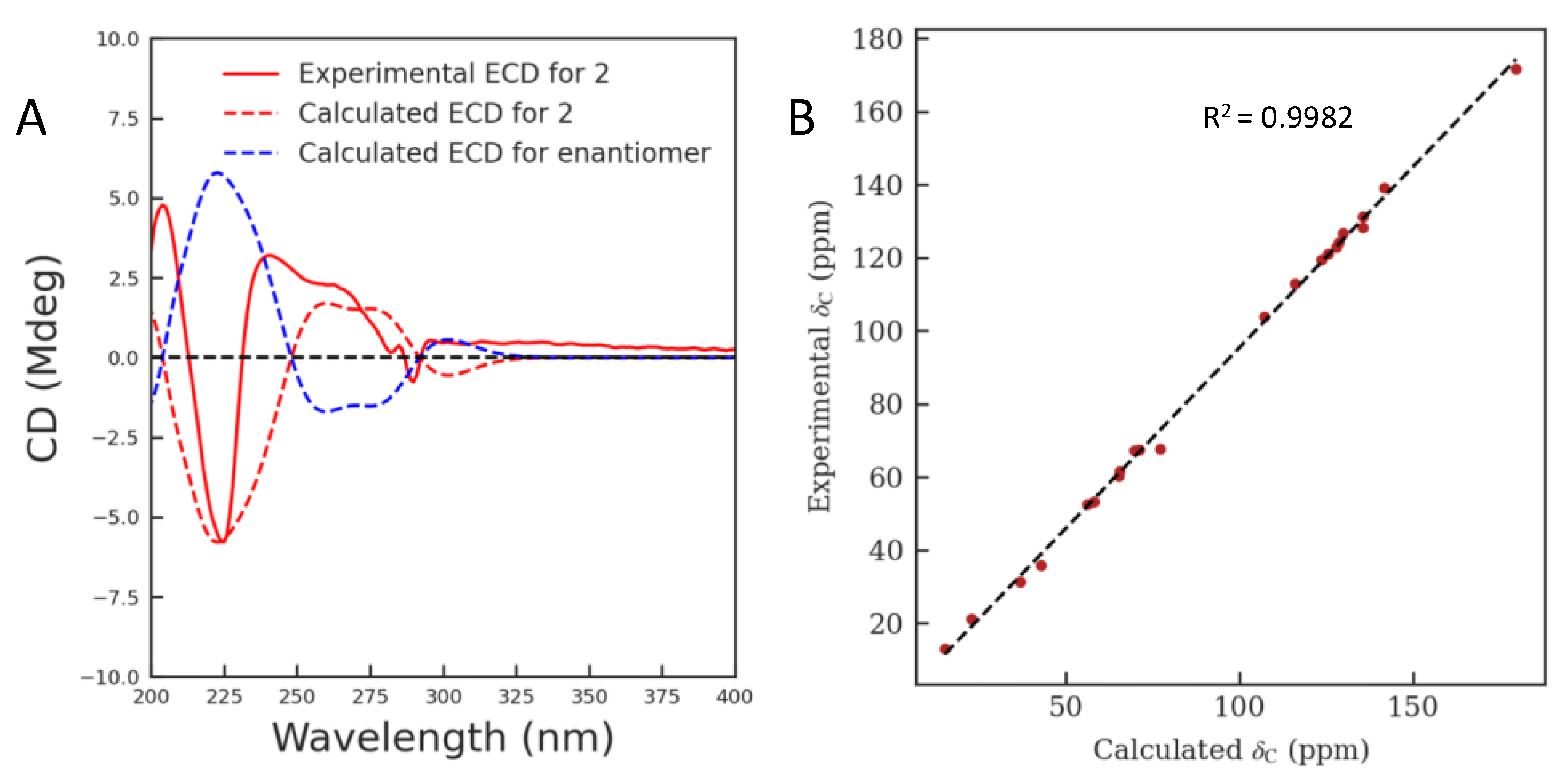
2.1. Structure Elucidation
2.2. The Cytotoxicity of Total Alkaloids and Compound 2
3. Experimental Section
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.3.1. Gelselegandine F (1)
3.3.2. Gelselegandine G (2)
3.4. ECD Calculation
3.5. 13C NMR Calculation
3.6. Cell Lines and Cell Culture
3.7. Cell Viability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhan, G.; Miao, R.; Zhang, F.; Hao, X.; Zheng, X.; Zhang, H.; Zhang, X.; Guo, Z. Monoterpene indole alkaloids with diverse skeletons from the stems of rauvolfia vomitoria and their acetylcholinesterase inhibitory activities. Phytochemistry 2020, 177, 112450. [Google Scholar] [CrossRef] [PubMed]
- Li, F.R.; Liu, L.; Liu, Y.P.; Wang, J.T.; Yang, M.L.; Khan, A.; Qin, X.J.; Wang, Y.D.; Cheng, G.G. HRESIMS-guided isolation of aspidosperma-scandine type bisindole alkaloids from Melodinus cochinchinensis and their anti-inflammatory and cytotoxic activities. Phytochemistry 2021, 184, 112–673. [Google Scholar] [CrossRef] [PubMed]
- Thamm, A.M.K.; Qu, Y.; De Luca, V. Discovery and metabolic engineering of ir-idoid/secoiridoid and monoterpenoid indole alkaloid biosynthesis. Phytochem. Rev. 2016, 15, 339–361. [Google Scholar] [CrossRef]
- Wei, X.; Dai, Z.; Yang, J.; Khan, A.; Yu, H.F.; Zhao, Y.L.; Wang, Y.F.; Liu, Y.P.; Yang, Z.F.; Huang, W.Y.; et al. Unprecedented sugar bridged bisindoles selective inhibiting glioma stem cells. Bioorg. Med. Chem. 2018, 76, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Zhao, Y.L.; Feng, T.; Cheng, G.G.; Zhang, B.H.; Li, Y.; Cai, X.H.; Luo, X.D. Melosuavines A-H, Cytotoxic Bisindole Alkaloid Derivatives from Melodinus suaveolens. J. Nat. Prod. 2013, 76, 2322–2329. [Google Scholar] [CrossRef] [PubMed]
- Beni, Z.; Hada, V.; Dubrovay, Z.; Szantay, C.J. Structure elucidation of indole-indoline type alkaloids: A retrospective account from the point of view of current NMR and MS technology. J. Pharm. Biomed. Anal. 2012, 69, 106–124. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, J.; Dai, Z.; Yu, H.F.; Ding, C.F.; Khan, A.; Zhao, Y.L.; Liu, Y.P.; Luo, X.D. Antitumor pyridine alkaloids hybrid with diverse units from Alangium chinense. Tetrahedron Lett. 2020, 61, 151502. [Google Scholar] [CrossRef]
- Li, Q.; Yang, S.; Teng, H.; Li, X.; Xie, W.; Wu, Z.; Yang, G.; Xu, J.; Chen, Y. Structural elucidation of two intricate polycyclic polyprenylated acylphloroglucinols using quantum chemical calculations and their hypoglycemic activities. Arab. J. Chem. 2022, 15, 104137. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef] [PubMed]
- Tsui, K.Y.; Tombari, R.J.; Olson, D.E.; Tantillo, D.J. Reconsidering the structure of serlyticin-A. J. Nat. Prod. 2019, 82, 3464–3468. [Google Scholar] [CrossRef] [PubMed]
- Unzueta, P.A.; Greenwell, C.S.; Beran, G.J.O. Predicting density functional theory-quality nuclear magnetic resonance chemical shifts via Δ-machine learning. J. Chem. Theory Comput. 2021, 17, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.L.; Su, Y.P.; Liu, M.; Xu, Y.; Yang, J.; Liao, K.J.; Yu, C.X. Medicinal plants of the genus Gelsemium (Gelsemiaceae, Gentianales)—A review of their phytochemistry, pharmacology, toxicology and traditional use. J. Ethnopharmacol. 2014, 152, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.Y.; Wang, G.Y.; Li, N.P.; Chen, M.F.; Gu, J.H.; Zhang, D.M.; Wang, L.; Ye, W.C. Five new koumine-type alkaloids from the roots of Gelsemium elegans. Fitoterapia 2017, 118, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yang, J.; Ma, H.X.; Ding, C.F.; Yu, H.F.; Zhao, Y.L.; Liu, Y.P.; Khan, A.; Wang, Y.F.; Yang, Z.F.; et al. Antimicrobial indole alkaloids with adductive C9 aromatic unit from Gelsemium elegans. Tetrahedron Lett. 2018, 59, 2066–2070. [Google Scholar] [CrossRef]
- Song, D.; Hu, X.Y.; Liang, J.J.; Liu, X.; Pu, X.; Zhang, L.Y.; Zhou, Y.; Wei, X. Chemical constituents with osteoclasts inhibitory activity from Gelsemium elegans. Chem. Nat. Compd. 2022, 58, 962–966. [Google Scholar] [CrossRef]
- Ouyang, S.; Wang, L.; Zhang, Q.W.; Wang, G.C.; Wang, Y.; Huang, X.J.; Zhang, X.Q.; Jiang, R.W.; Yao, X.S.; Che, C.T.; et al. Six new monoterpenoid indole alkaloids from the aerial part of Gelsemium elegans. Tetrahedron 2011, 67, 4807–4813. [Google Scholar] [CrossRef]
- Wei, X.; Guo, R.; Wang, X.; Liang, J.J.; Yu, H.F.; Ding, C.F.; Feng, T.T.; Zhang, L.Y.; Liu, X.; Hu, X.Y.; et al. New monoterpenoid indoles with osteoclast activities from Gelsemium elegans. Molecules 2021, 26, 7457. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.L.; Zhu, J.P.; Hesse, M. Indole alkaloids from Aistonia angustjfolia. Planta Med. 1989, 55, 463–466. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Veremeersch, T.; Flynn, C.J.; Maguire, A.R.; Hutchison, G.R. Confab—Systematic generation of diverse low-energy conformers. J. Cheminformatics 2011, 3, 3–8. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09 Revision D.01; Gaussian Inc.: Wallingford, UK, 2009. [Google Scholar]
 ), 1H-1H COSY (
), 1H-1H COSY ( ) and ROESY (
) and ROESY ( ) correlations of compound 1.
) correlations of compound 1.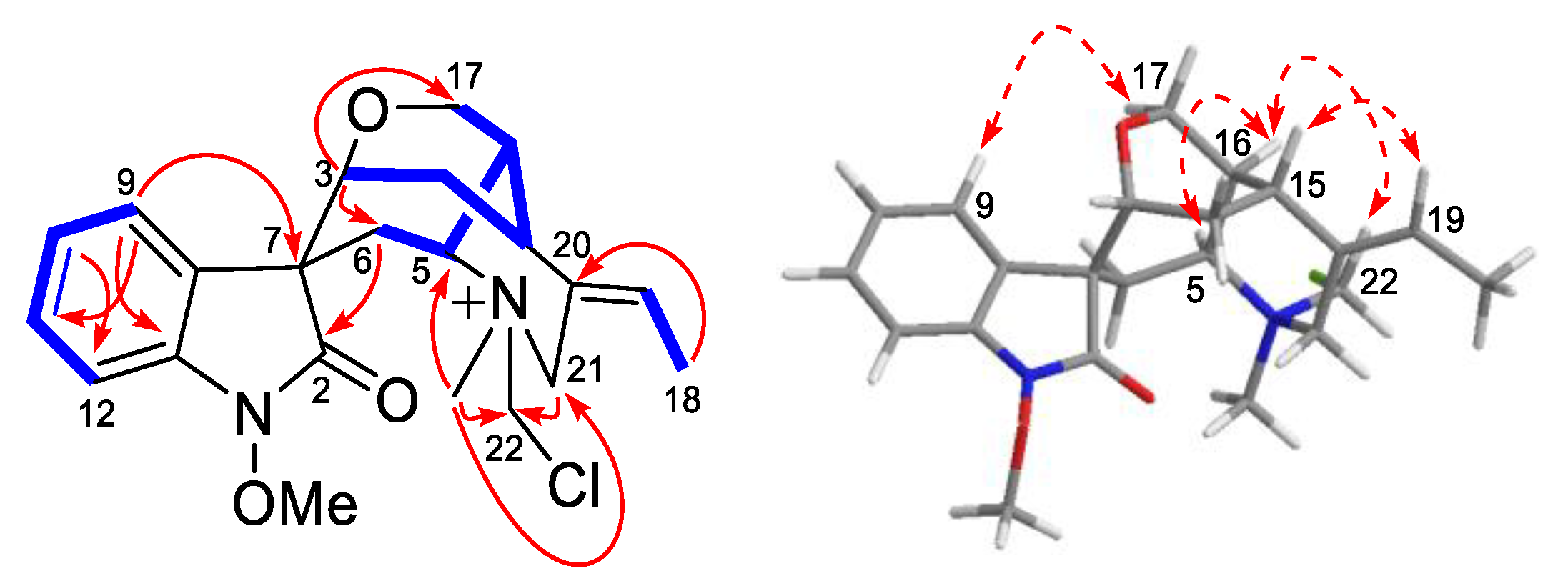
 ), 1H-1H COSY (
), 1H-1H COSY ( ) and ROESY (
) and ROESY ( ) correlations of compound 2.
) correlations of compound 2.


| NO. | Compound 1 a | Compound 2 a | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 2 | 174.9 | 131.2 | ||
| 3 | 3.65 (d, J = 7.0, 1H) | 74.1 | 5.29 (d, J = 10.3, 1H) | 61.6 |
| 5 | 4.56 (m, 1H) | 71.0 | 3.93 (m, 1H) | 67.4 |
| 6 | 2.78 (dd, J = 17.3, 4.3, 1H) 2.64 (dd, J = 17.3, 8.2, 1H) | 29.4 | 3.26 (dd, J = 18.3, 4.8, 1H) 3.91 (d, J = 4.6, 1H) | 21.1 |
| 7 | 57.0 | 103.9 | ||
| 8 | 131.2 | 126.7 | ||
| 9 | 7.54 (d, J = 7.5, 1H) | 126.8 | 7.51 (d, J = 7.9, 1H) | 119.5 |
| 10 | 7.20 (td, J = 7.6, 0.9, 1H) | 124.8 | 7.11 (m, 1H) | 121.2 |
| 11 | 7.41 (td, J = 7.7, 0.9, 1H) | 130.0 | 7.21 (m, 1H) | 124.3 |
| 12 | 7.11 (d, J = 7.7, 1H) | 108.7 | 7.40 (d, J = 8.2, 1H) | 112.9 |
| 13 | 139.5 | 139.1 | ||
| 14 | 2.41 (ddd, J = 15.7, 11.5, 7.3, 1H) 2.28 (dd, J = 15.7, 5.5, 1H) | 27.8 | 2.53 (m, 1H) 3.07 (dd, J = 13.3, 4.4, 1H) | 31.3 |
| 15 | 2.92 (m, 1H) | 34.5 | 3.13 (d, J = 4.1, 1H) | 35.9 |
| 16 | 2.86 (dd, J = 10.1, 5.1, 1H) | 33.1 | 53.3 | |
| 17 | 4.33 (d, J = 11.5, 1H) 4.19 (dd, J = 11.5, 4.8, 1H) | 65.6 | 3.85 (m, 2H) | 67.5 |
| 18 | 1.86 (d, J = 6.9, 3H) | 13.4 | 1.73 (d, J = 7.0, 3H) | 13.0 |
| 19 | 6.03 (q, J = 6.9, 1H) | 130.8 | 5.73 (qd, J = 7.0,4.5, 1H) | 122.9 |
| 20 | 130.0 | 128.4 | ||
| 21 | 5.48 (d, J = 11.1, 1H) 4.40 (d, J = 14.5, 1H) | 54.7 | 4.77 (d, J = 16.4, 1H), 4.60 (d, J = 16.2, 1H) | 60.2 |
| 22 | 5.46 (s, 2H) | 69.4 | 5.52 (d, J = 10.4, 1H) 5.40 (d, J = 10.4, 1H) | 67.8 |
| N1-OCH3 | 4.03 (s, 3H) | 64.3 | ||
| N4-CH3 | 3.27 (s, 3H) | 48.6 | ||
| COOCH3 | 3.01 (s, 3H) | 52.7 | ||
| COOCH3 | 171.7 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.; Liang, J.-J.; Pu, S.-B.; Zhang, P.-P.; Peng, Y.-L.; Liu, X.; Feng, T.-T.; Pu, X.; Zhou, Y.; Liu, X.-W.; et al. Structural Elucidation and Cytotoxic Activity of New Monoterpenoid Indoles from Gelsemium elegans. Molecules 2023, 28, 2531. https://doi.org/10.3390/molecules28062531
Song D, Liang J-J, Pu S-B, Zhang P-P, Peng Y-L, Liu X, Feng T-T, Pu X, Zhou Y, Liu X-W, et al. Structural Elucidation and Cytotoxic Activity of New Monoterpenoid Indoles from Gelsemium elegans. Molecules. 2023; 28(6):2531. https://doi.org/10.3390/molecules28062531
Chicago/Turabian StyleSong, Da, Jia-Jun Liang, Shi-Biao Pu, Pan-Pan Zhang, Yun-Lin Peng, Xia Liu, Ting-Ting Feng, Xiang Pu, Ying Zhou, Xiong-Wei Liu, and et al. 2023. "Structural Elucidation and Cytotoxic Activity of New Monoterpenoid Indoles from Gelsemium elegans" Molecules 28, no. 6: 2531. https://doi.org/10.3390/molecules28062531
APA StyleSong, D., Liang, J.-J., Pu, S.-B., Zhang, P.-P., Peng, Y.-L., Liu, X., Feng, T.-T., Pu, X., Zhou, Y., Liu, X.-W., & Wei, X. (2023). Structural Elucidation and Cytotoxic Activity of New Monoterpenoid Indoles from Gelsemium elegans. Molecules, 28(6), 2531. https://doi.org/10.3390/molecules28062531







