General Strategies for RNA X-ray Crystallography
Abstract
1. Introduction
2. RNA Purification and Folding
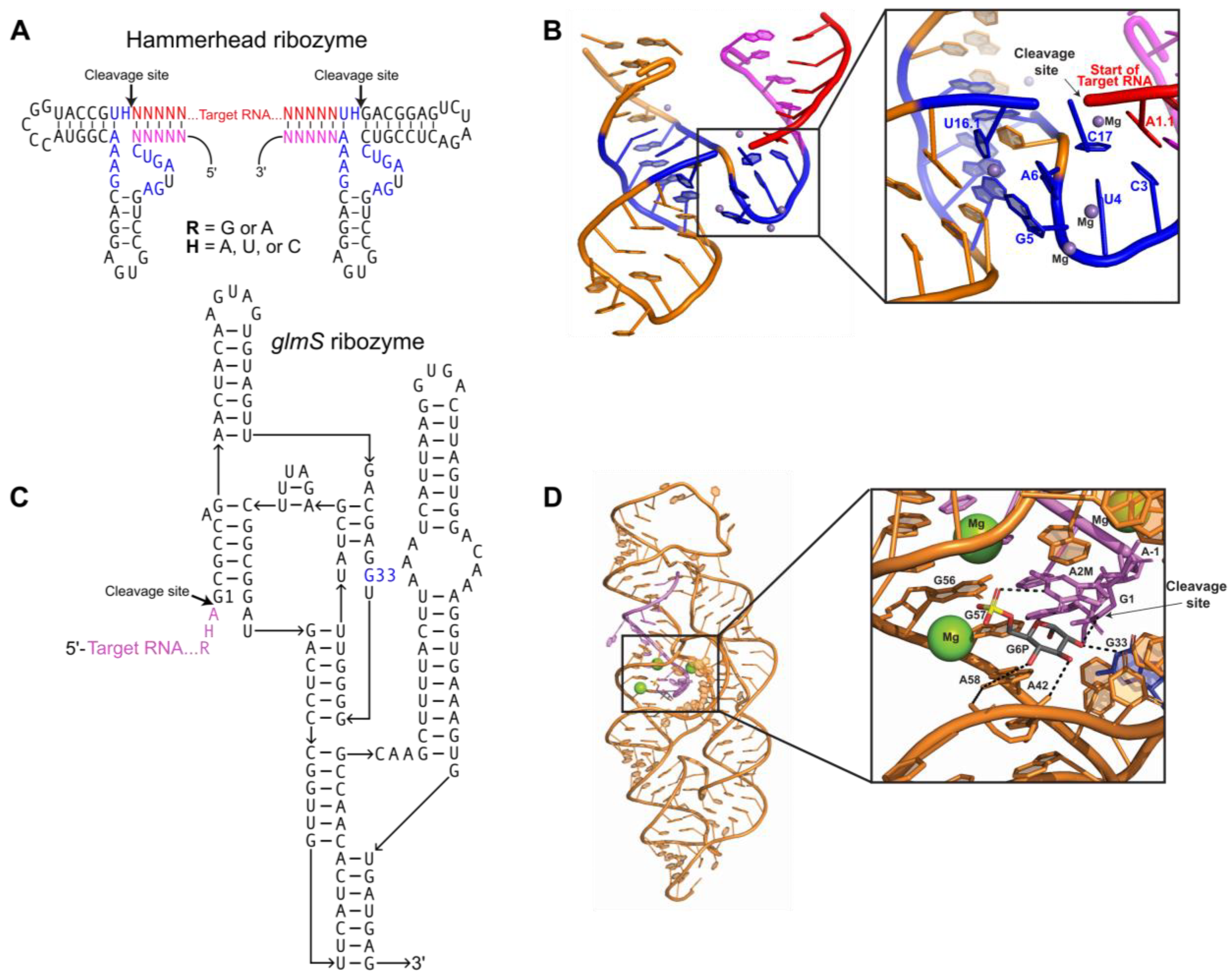
2.1. Producing Homogeneous Transcripts: Hammerhead Ribozyme
2.2. Producing Homogeneous Transcripts: glmS Ribozyme
2.3. Purification of Transcribed RNA
3. RNA-Driven Crystallization Modules
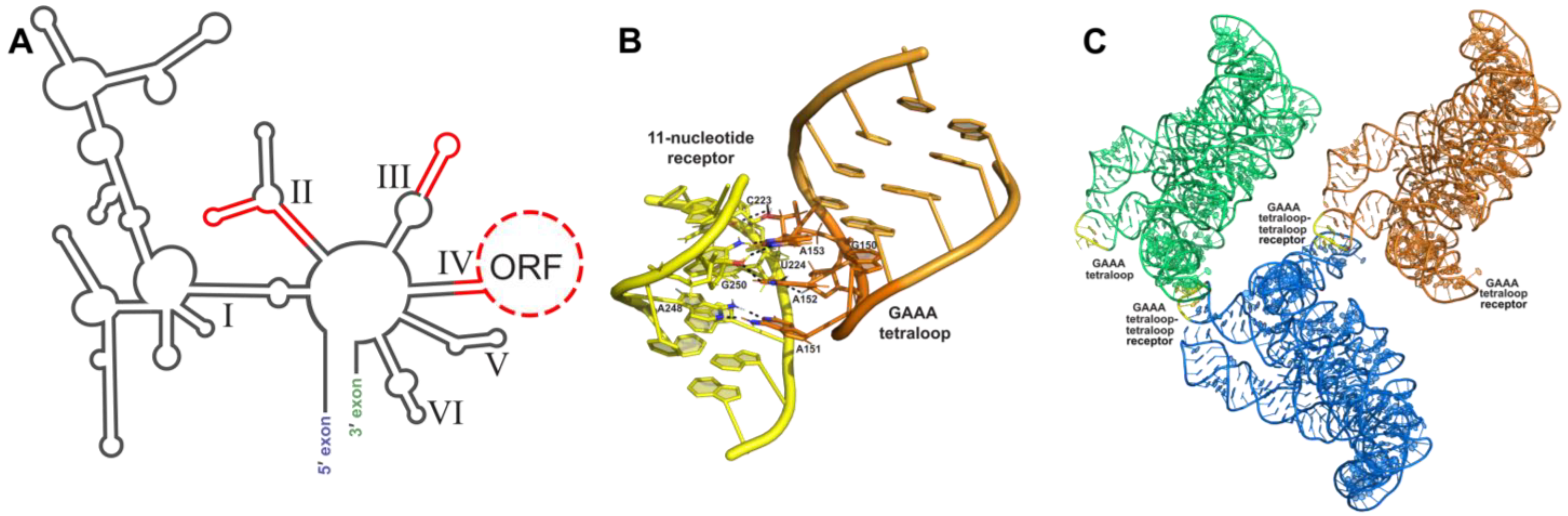
3.1. Tetraloop Interactions as RNA Crystallization Modules
3.2. Loop–Loop Interactions as RNA Crystallization Modules
4. Protein-Assisted RNA Crystallography
4.1. U1A Protein Module
4.2. Kink-Turn Module
4.3. Antibody Fragment Module
5. Solving the “Phase Problem” for RNA Crystals
5.1. Molecular Replacement Methods
5.2. Isomorphous Replacement and Anomalous Scattering
5.2.1. Isomorphous Replacement with Mg2+ Mimics
5.2.2. Engineering Heavy Metal Binding Sites
5.2.3. Incorporation of Selenium into Nucleic Acids
5.2.4. Soaking with Halogens
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Milligan, J.F.; Groebe, D.R.; Witherell, G.W.; Uhlenbeck, O.C. Oligoribonucleotide Synthesis Using T7 RNA Polymerase and Synthetic DNA Templates. Nucleic Acids Res. 1987, 15, 8783–8798. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.; Zheng, M.; Rüdisser, S. A Simple and Efficient Method to Reduce Nontemplated Nucleotide Addition at the 3 Terminus of RNAs Transcribed by T7 RNA Polymerase. RNA 1999, 5, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, L.E.; Zhou, Y.; McAllister, W.T. Termination and Slippage by Bacteriophage T7 RNA Polymerase. J. Mol. Biol. 1993, 232, 1030–1047. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, C.J.; Rathjen, P.D.; Forster, A.C.; Symons, R.H. Self-Cleavage of plus and Minus RNA Transcripts of Avocado Sunblotch Viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef] [PubMed]
- Buzayan, J.M.; Gerlach, W.L.; Bruening, G.; Keese, P.; Gould, A.R. Nucleotide Sequence of Satellite Tobacco Ringspot Virus RNA and Its Relationship to Multimeric Forms. Virology 1986, 151, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Symons, R.H. Plant Pathogenic RNAs and RNA Catalysis. Nucleic Acids Res. 1997, 25, 2683–2689. [Google Scholar] [CrossRef]
- Pata, J.D.; King, B.R.; Steitz, T.A. Assembly, Purification and Crystallization of an Active HIV-1 Reverse Transcriptase Initiation Complex. Nucleic Acids Res. 2002, 30, 4855–4863. [Google Scholar] [CrossRef]
- Nagai, K.; Oubridge, C.; Ito, N.; Jessen, T.H.; Avis, J.; Evans, P. Crystal Structure of the U1A Spliceosomal Protein Complexed with Its Cognate RNA Hairpin. Nucleic Acids Symp. Ser. 1995, 34, 1–2. [Google Scholar]
- Ruffner, D.E.; Stormo, G.D.; Uhlenbeck, O.C. Sequence Requirements of the Hammerhead RNA Self-Cleavage Reaction. Biochemistry 1990, 29, 10695–10702. [Google Scholar] [CrossRef]
- Stage-Zimmermann, T.K.; Uhlenbeck, O.C. Hammerhead Ribozyme Kinetics. RNA 1998, 4, 875–889. [Google Scholar] [CrossRef]
- Canny, M.D.; Jucker, F.M.; Kellogg, E.; Khvorova, A.; Jayasena, S.D.; Pardi, A. Fast Cleavage Kinetics of a Natural Hammerhead Ribozyme. J. Am. Chem. Soc. 2004, 126, 10848–10849. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, S.M.; Scott, W.G. Structural Simplicity and Mechanistic Complexity in the Hammerhead Ribozyme. Prog. Mol. Biol. Transl. Sci. 2018, 159, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Price, S.R.; Ito, N.; Oubridge, C.; Avis, J.M.; Nagai, K. Crystallization of RNA-Protein Complexes. I. Methods for the Large-Scale Preparation of RNA Suitable for Crystallographic Studies. J. Mol. Biol. 1995, 249, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Bassi, G.S.; Murchie, A.I.; Walter, F.; Clegg, R.M.; Lilley, D.M. Ion-Induced Folding of the Hammerhead Ribozyme: A Fluorescence Resonance Energy Transfer Study. EMBO J. 1997, 16, 7481–7489. [Google Scholar] [CrossRef]
- Hammann, C.; Lilley, D.M.J. Folding and Activity of the Hammerhead Ribozyme. Chembiochem 2002, 3, 690–700. [Google Scholar] [CrossRef]
- Perriman, R.; Delves, A.; Gerlach, W.L. Extended Target-Site Specificity for a Hammerhead Ribozyme. Gene 1992, 113, 157–163. [Google Scholar] [CrossRef]
- Koizumi, M.; Hayase, Y.; Iwai, S.; Kamiya, H.; Inoue, H.; Ohtsuka, E. Design of RNA Enzymes Distinguishing a Single Base Mutation in RNA. Nucleic Acids Res. 1989, 17, 7059–7071. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Symons, R.H. Mutagenesis Analysis of a Self-Cleaving RNA. Nucleic Acids Res. 1989, 17, 5679–5685. [Google Scholar] [CrossRef]
- O’Rourke, S.M.; Estell, W.; Scott, W.G. Minimal Hammerhead Ribozymes with Uncompromised Catalytic Activity. J. Mol. Biol. 2015, 427, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.G. Ribozymes. Curr. Opin. Struct. Biol. 2007, 17, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.Y.; Fedor, M.J. The GlmS Riboswitch Integrates Signals from Activating and Inhibitory Metabolites in vivo. Nat. Struct. Mol. Biol. 2011, 18, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Ferré-D’Amaré, A.R.; Scott, W.G. Small Self-Cleaving Ribozymes. Cold Spring Harb. Perspect. Biol. 2010, 2, a003574. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.J.B.; Behera, V.; Walter, N.G. Nondenaturing Purification of Co-Transcriptionally Folded RNA Avoids Common Folding Heterogeneity. PLoS ONE 2010, 5, e12953. [Google Scholar] [CrossRef] [PubMed]
- Ke, A.; Zhou, K.; Ding, F.; Cate, J.H.D.; Doudna, J.A. A Conformational Switch Controls Hepatitis Delta Virus Ribozyme Catalysis. Nature 2004, 429, 201–205. [Google Scholar] [CrossRef]
- Ke, A.; Doudna, J.A. Crystallization of RNA and RNA-Protein Complexes. Methods 2004, 34, 408–414. [Google Scholar] [CrossRef]
- Batey, R.T.; Sagar, M.B.; Doudna, J.A. Structural and Energetic Analysis of RNA Recognition by a Universally Conserved Protein from the Signal Recognition Particle. J. Mol. Biol. 2001, 307, 229–246. [Google Scholar] [CrossRef]
- Pan, J.; Woodson, S.A. Folding Intermediates of a Self-Splicing RNA: Mispairing of the Catalytic Core. J. Mol. Biol. 1998, 280, 597–609. [Google Scholar] [CrossRef]
- Kladwang, W.; Hum, J.; Das, R. Ultraviolet Shadowing of RNA Can Cause Significant Chemical Damage in Seconds. Sci. Rep. 2012, 2, 517. [Google Scholar] [CrossRef]
- Turner, D.H.; Sugimoto, N.; Freier, S.M. RNA Structure Prediction. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 167–192. [Google Scholar] [CrossRef]
- Lukavsky, P.J.; Puglisi, J.D. Large-Scale Preparation and Purification of Polyacrylamide-Free RNA Oligonucleotides. RNA 2004, 10, 889–893. [Google Scholar] [CrossRef]
- Toor, N.; Keating, K.S.; Taylor, S.D.; Pyle, A.M. Crystal Structure of a Self-Spliced Group II Intron. Science 2008, 320, 77–82. [Google Scholar] [CrossRef]
- Chan, R.T.; Robart, A.R.; Rajashankar, K.R.; Pyle, A.M.; Toor, N. Crystal Structure of a Group II Intron in the Pre-Catalytic State. Nat. Struct. Mol. Biol. 2012, 19, 555–557. [Google Scholar] [CrossRef]
- Nachtergaele, S.; He, C. The Emerging Biology of RNA Post-Transcriptional Modifications. RNA Biol. 2016, 14, 156–163. [Google Scholar] [CrossRef]
- Kudrin, P.; Meierhofer, D.; Vågbø, C.B.; Ørom, U.A.V. Nuclear RNA-Acetylation Can Be Erased by the Deacetylase SIRT7. bioRxiv 2021, arXiv:2021.04.06.438707. [Google Scholar]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs Are Modified with N-Glycans and Displayed on the Surface of Living Cells. Cell 2021, 184, 3109–3124.e22. [Google Scholar] [CrossRef]
- Peabody, D.S. The RNA Binding Site of Bacteriophage MS2 Coat Protein. EMBO J. 1993, 12, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Srikantan, S.; Gorospe, M. MS2-TRAP (MS2-Tagged RNA Affinity Purification): Tagging RNA to Identify Associated MiRNAs. Methods 2012, 58, 81–87. [Google Scholar] [CrossRef]
- Lim, F.; Peabody, D.S. RNA Recognition Site of PP7 Coat Protein. Nucleic Acids Res. 2002, 30, 4138–4144. [Google Scholar] [CrossRef] [PubMed]
- Fritz, S.E.; Haque, N.; Hogg, J.R. Highly Efficient in Vitro Translation of Authentic Affinity-Purified Messenger Ribonucleoprotein Complexes. RNA 2018, 24, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Youngman, E.M.; Green, R. Affinity Purification of in Vivo-Assembled Ribosomes for in Vitro Biochemical Analysis. Methods 2005, 36, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Ferré-D’Amaré, A.R. Use of the Spliceosomal Protein U1A to Facilitate Crystallization and Structure Determination of Complex RNAs. Methods 2010, 52, 159–167. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Moxon, S.; Marshall, M.; Khanna, A.; Eddy, S.R.; Bateman, A. Rfam: Annotating Non-Coding RNAs in Complete Genomes. Nucleic Acids Res. 2005, 33, D121–D124. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.L.; Garst, A.D.; Batey, R.T. Determining Structures of RNA Aptamers and Riboswitches by X-Ray Crystallography. Methods Mol. Biol. 2009, 535, 135–163. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Winker, S.; Gutell, R.R. Architecture of Ribosomal RNA: Constraints on the Sequence of “Tetra-Loops”. Proc. Natl. Acad. Sci. USA 1990, 87, 8467–8471. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. Stitching Together RNA Tertiary Architectures. J. Mol. Biol. 1999, 294, 829–849. [Google Scholar] [CrossRef]
- Richardson, K.E.; Adams, M.S.; Kirkpatrick, C.C.; Gohara, D.W.; Znosko, B.M. Identification and Characterization of New RNA Tetraloop Sequence Families. Biochemistry 2019, 58, 4809–4820. [Google Scholar] [CrossRef] [PubMed]
- Ferré-D’Amaré, A.R.; Zhou, K.; Doudna, J.A. A General Module for RNA Crystallization. J. Mol. Biol. 1998, 279, 621–631. [Google Scholar] [CrossRef]
- Coonrod, L.A.; Lohman, J.R.; Berglund, J.A. Utilizing the GAAA Tetraloop/Receptor to Facilitate Crystal Packing and Determination of the Structure of a CUG RNA Helix. Biochemistry 2012, 51, 8330–8337. [Google Scholar] [CrossRef]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragón, A. Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef]
- Robart, A.R.; Chan, R.T.; Peters, J.K.; Rajashankar, K.R.; Toor, N. Crystal Structure of a Eukaryotic Group II Intron Lariat. Nature 2014, 514, 193–197. [Google Scholar] [CrossRef]
- Toor, N.; Rajashankar, K.; Keating, K.S.; Pyle, A.M. Structural Basis for Exon Recognition by a Group II Intron. Nat. Struct. Mol. Biol. 2008, 15, 1221–1222. [Google Scholar] [CrossRef]
- Montange, R.K.; Batey, R.T. Structure of the S-Adenosylmethionine Riboswitch Regulatory mRNA Element. Nature 2006, 441, 1172–1175. [Google Scholar] [CrossRef]
- Grundy, F.J.; Henkin, T.M. The S Box Regulon: A New Global Transcription Termination Control System for Methionine and Cysteine Biosynthesis Genes in Gram-Positive Bacteria. Mol. Microbiol. 1998, 30, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Nahvi, A.; Sudarsan, N.; Barrick, J.E.; Breaker, R.R. An mRNA Structure That Controls Gene Expression by Binding S-Adenosylmethionine. Nat. Struct. Biol. 2003, 10, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pley, H.W.; Flaherty, K.M.; McKay, D.B. Model for an RNA Tertiary Interaction from the Structure of an Intermolecular Complex between a GAAA Tetraloop and an RNA Helix. Nature 1994, 372, 111–113. [Google Scholar] [CrossRef]
- Pley, H.W.; Flaherty, K.M.; McKay, D.B. Three-Dimensional Structure of a Hammerhead Ribozyme. Nature 1994, 372, 68–74. [Google Scholar] [CrossRef]
- Costa, M.; Michel, F. Frequent Use of the Same Tertiary Motif by Self-Folding RNAs. EMBO J. 1995, 14, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Michel, F. Rules for RNA Recognition of GNRA Tetraloops Deduced by in Vitro Selection: Comparison with in Vivo Evolution. EMBO J. 1997, 16, 3289–3302. [Google Scholar] [CrossRef] [PubMed]
- Abramovitz, D.L.; Pyle, A.M. Remarkable Morphological Variability of a Common RNA Folding Motif: The GNRA Tetraloop-Receptor Interaction. J. Mol. Biol. 1997, 266, 493–506. [Google Scholar] [CrossRef]
- Murphy, F.L.; Cech, T.R. GAAA Tetraloop and Conserved Bulge Stabilize Tertiary Structure of a Group I Intron Domain. J. Mol. Biol. 1994, 236, 49–63. [Google Scholar] [CrossRef]
- Cate, J.H.; Gooding, A.R.; Podell, E.; Zhou, K.; Golden, B.L.; Kundrot, C.E.; Cech, T.R.; Doudna, J.A. Crystal Structure of a Group I Ribozyme Domain: Principles of RNA Packing. Science 1996, 273, 1678–1685. [Google Scholar] [CrossRef]
- Szewczak, A.A.; Podell, E.R.; Bevilacqua, P.C.; Cech, T.R. Thermodynamic Stability of the P4-P6 Domain RNA Tertiary Structure Measured by Temperature Gradient Gel Electrophoresis. Biochemistry 1998, 37, 11162–11170. [Google Scholar] [CrossRef]
- Baird, N.J.; Westhof, E.; Qin, H.; Pan, T.; Sosnick, T.R. Structure of a Folding Intermediate Reveals the Interplay between Core and Peripheral Elements in RNA Folding. J. Mol. Biol. 2005, 352, 712–722. [Google Scholar] [CrossRef]
- Chauhan, S.; Woodson, S.A. Tertiary Interactions Determine the Accuracy of RNA Folding. J. Am. Chem. Soc. 2008, 130, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Sosnick, T.R.; Pan, T. Modular Construction of a Tertiary RNA Structure: The Specificity Domain of the Bacillus Subtilis RNase P RNA. Biochemistry 2001, 40, 11202–11210. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, I.; Brenowitz, M. Perturbation of the Hierarchical Folding of a Large RNA by the Destabilization of Its Scaffold’s Tertiary Structure. J. Mol. Biol. 2005, 354, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Treiber, D.K.; Williamson, J.R. Concerted Kinetic Folding of a Multidomain Ribozyme with a Disrupted Loop-Receptor Interaction. J. Mol. Biol. 2001, 305, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz Krummel, D.A.; Oubridge, C.; Leung, A.K.W.; Li, J.; Nagai, K. Crystal Structure of Human Spliceosomal U1 SnRNP at 5.5 Å Resolution. Nature 2009, 458, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Quigley, G.J.; Rich, A. Structural Domains of Transfer RNA Molecules. Science 1976, 194, 796–806. [Google Scholar] [CrossRef]
- Lehnert, V.; Jaeger, L.; Michel, F.; Westhof, E. New Loop-Loop Tertiary Interactions in Self-Splicing Introns of Subgroup IC and ID: A Complete 3D Model of the Tetrahymena Thermophila Ribozyme. Chem. Biol. 1996, 3, 993–1009. [Google Scholar] [CrossRef]
- Batey, R.T.; Rambo, R.P.; Doudna, J.A. Tertiary Motifs in RNA Structure and Folding. Angew. Chem. Int. Ed. Engl. 1999, 38, 2326–2343. [Google Scholar] [CrossRef]
- Gregorian, R.S.; Crothers, D.M. Determinants of RNA Hairpin Loop-Loop Complex Stability. J. Mol. Biol. 1995, 248, 968–984. [Google Scholar] [CrossRef]
- Eisinger, J. Complex Formation between Transfer RNA’S with Complementary Anticodons. Biochem. Biophys. Res. Commun. 1971, 43, 854–861. [Google Scholar] [CrossRef]
- Eisinger, J.; Gross, N. The Anticodon-Anticodon Complex. J. Mol. Biol. 1974, 88, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Söll, D.G.; Crothers, D.M. Studies of the Complex between Transfer RNAs with Complementary Anticodons. I. Origins of Enhanced Affinity between Complementary Triplets. J. Mol. Biol. 1976, 103, 499–519. [Google Scholar] [CrossRef] [PubMed]
- Labuda, D.; Grosjean, H.; Striker, G.; Pörschke, D. Codon:Anticodon and Anticodon:Anticodon Interaction: Evaluation of Equilibrium and Kinetic Parameters of Complexes Involving a G:U Wobble. Biochim. Biophys. Acta 1982, 698, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Houssier, C.; Grosjean, H. Temperature Jump Relaxation Studies on the Interactions between Transfer RNAs with Complementary Anticodons. The Effect of Modified Bases Adjacent to the Anticodon Triplet. J. Biomol. Struct. Dyn. 1985, 3, 387–408. [Google Scholar] [CrossRef]
- Romby, P.; Giegé, R.; Houssier, C.; Grosjean, H. Anticodon-Anticodon Interactions in Solution. Studies of the Self-Association of Yeast or Escherichia Coli tRNAAsp and of Their Interactions with Escherichia Coli tRNAVal. J. Mol. Biol. 1985, 184, 107–118. [Google Scholar] [CrossRef]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the Primary Site of the Human Immunodeficiency Virus Type 1 RNA Dimerization in Vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef]
- Paillart, J.C.; Marquet, R.; Skripkin, E.; Ehresmann, B.; Ehresmann, C. Mutational Analysis of the Bipartite Dimer Linkage Structure of Human Immunodeficiency Virus Type 1 Genomic RNA. J. Biol. Chem. 1994, 269, 27486–27493. [Google Scholar] [CrossRef]
- Brunel, C.; Marquet, R.; Romby, P.; Ehresmann, C. RNA Loop-Loop Interactions as Dynamic Functional Motifs. Biochimie 2002, 84, 925–944. [Google Scholar] [CrossRef]
- Oubridge, C.; Ito, N.; Evans, P.R.; Teo, C.H.; Nagai, K. Crystal Structure at 1.92 A Resolution of the RNA-Binding Domain of the U1A Spliceosomal Protein Complexed with an RNA Hairpin. Nature 1994, 372, 432–438. [Google Scholar] [CrossRef]
- Rupert, P.B.; Ferré-D’Amaré, A.R. Crystal Structure of a Hairpin Ribozyme-Inhibitor Complex with Implications for Catalysis. Nature 2001, 410, 780–786. [Google Scholar] [CrossRef]
- Cochrane, J.C.; Lipchock, S.V.; Strobel, S.A. Structural Investigation of the GlmS Ribozyme Bound to Its Catalytic Cofactor. Chem. Biol. 2007, 14, 97–105. [Google Scholar] [CrossRef]
- Adams, P.L.; Stahley, M.R.; Kosek, A.B.; Wang, J.; Strobel, S.A. Crystal Structure of a Self-Splicing Group I Intron with Both Exons. Nature 2004, 430, 45–50. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R.; Zhou, K.; Doudna, J.A. Crystal Structure of a Hepatitis Delta Virus Ribozyme. Nature 1998, 395, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Edwards, T.E.; Ferré-D’Amaré, A.R. Structural Basis for Specific, High-Affinity Tetracycline Binding by an in Vitro Evolved Aptamer and Artificial Riboswitch. Chem. Biol. 2008, 15, 1125–1137. [Google Scholar] [CrossRef]
- Kulshina, N.; Baird, N.J.; Ferré-D’Amaré, A.R. Recognition of the Bacterial Second Messenger Cyclic Diguanylate by Its Cognate Riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.D.; Lipchock, S.V.; Ames, T.D.; Wang, J.; Breaker, R.R.; Strobel, S.A. Structural Basis of Ligand Binding by a C-Di-GMP Riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1218–1223. [Google Scholar] [CrossRef] [PubMed]
- Shechner, D.M.; Grant, R.A.; Bagby, S.C.; Koldobskaya, Y.; Piccirilli, J.A.; Bartel, D.P. Crystal Structure of the Catalytic Core of an RNA-Polymerase Ribozyme. Science 2009, 326, 1271–1275. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Schmeing, T.M.; Moore, P.B.; Steitz, T.A. The Kink-Turn: A New RNA Secondary Structure Motif. EMBO J. 2001, 20, 4214–4221. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lilley, D.M.J. The Molecular Recognition of Kink-Turn Structure by the L7Ae Class of Proteins. RNA 2013, 19, 1703–1710. [Google Scholar] [CrossRef]
- Lilley, D.M.J. The L7Ae Proteins Mediate a Widespread and Highly Functional Protein–RNA Interaction. Biochemist 2019, 41, 40–44. [Google Scholar] [CrossRef]
- Zhang, J.; Ferré-D’Amaré, A.R. Cocrystal Structure of a T-Box Riboswitch Stem I Domain in Complex with Its Cognate tRNA. Nature 2013, 500, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferré-D’Amaré, A.R. New Molecular Engineering Approaches for Crystallographic Studies of Large RNAs. Curr. Opin. Struct. Biol. 2014, 26, 9–15. [Google Scholar] [CrossRef]
- Koide, S. Engineering of Recombinant Crystallization Chaperones. Curr. Opin. Struct. Biol. 2009, 19, 449–457. [Google Scholar] [CrossRef]
- Dutzler, R.; Campbell, E.B.; Cadene, M.; Chait, B.T.; MacKinnon, R. X-Ray Structure of a ClC Chloride Channel at 3.0 A Reveals the Molecular Basis of Anion Selectivity. Nature 2002, 415, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Tereshko, V.; Uysal, S.; Koide, A.; Margalef, K.; Koide, S.; Kossiakoff, A.A. Toward Chaperone-Assisted Crystallography: Protein Engineering Enhancement of Crystal Packing and X-Ray Phasing Capabilities of a Camelid Single-Domain Antibody (VHH) Scaffold. Protein Sci. 2008, 17, 1175–1187. [Google Scholar] [CrossRef]
- Iwata, S.; Ostermeier, C.; Ludwig, B.; Michel, H. Structure at 2.8 A Resolution of Cytochrome c Oxidase from Paracoccus Denitrificans. Nature 1995, 376, 660–669. [Google Scholar] [CrossRef]
- Lieberman, R.L.; Culver, J.A.; Entzminger, K.C.; Pai, J.C.; Maynard, J.A. Crystallization Chaperone Strategies for Membrane Proteins. Methods 2011, 55, 293–302. [Google Scholar] [CrossRef]
- Piccirilli, J.A.; Koldobskaya, Y. Crystal Structure of an RNA Polymerase Ribozyme in Complex with an Antibody Fragment. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Fellouse, F.A.; Esaki, K.; Birtalan, S.; Raptis, D.; Cancasci, V.J.; Koide, A.; Jhurani, P.; Vasser, M.; Wiesmann, C.; Kossiakoff, A.A.; et al. High-Throughput Generation of Synthetic Antibodies from Highly Functional Minimalist Phage-Displayed Libraries. J. Mol. Biol. 2007, 373, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Gilbreth, R.N.; Esaki, K.; Tereshko, V.; Koide, S. High-Affinity Single-Domain Binding Proteins with a Binary-Code Interface. Proc. Natl. Acad. Sci. USA 2007, 104, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.-D.; Tereshko, V.; Frederiksen, J.K.; Koide, A.; Fellouse, F.A.; Sidhu, S.S.; Koide, S.; Kossiakoff, A.A.; Piccirilli, J.A. Synthetic Antibodies for Specific Recognition and Crystallization of Structured RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, G. Crystallography Made Crystal Clear: A Guide for Users of Macromolecular Models; Elsevier Science & Technology: Burlington, VT, USA, 2006; ISBN 978-0-08-045554-9. [Google Scholar]
- Taylor, G.L. Introduction to Phasing. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 325–338. [Google Scholar] [CrossRef]
- Evans, P.; McCoy, A. An Introduction to Molecular Replacement. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 1–10. [Google Scholar] [CrossRef]
- Marcia, M.; Humphris-Narayanan, E.; Keating, K.S.; Somarowthu, S.; Rajashankar, K.; Pyle, A.M. Solving Nucleic Acid Structures by Molecular Replacement: Examples from Group II Intron Studies. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2174–2185. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Template Convolution to Enhance or Detect Structural Features in Macromolecular Electron-Density Maps. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 179–185. [Google Scholar] [CrossRef]
- Giorgetti, A.; Raimondo, D.; Miele, A.E.; Tramontano, A. Evaluating the Usefulness of Protein Structure Models for Molecular Replacement. Bioinformatics 2005, 21 (Suppl. 2), ii72–ii76. [Google Scholar] [CrossRef]
- Thompson, J.; Baker, D. Incorporation of Evolutionary Information into Rosetta Comparative Modeling. Proteins 2011, 79, 2380–2388. [Google Scholar] [CrossRef]
- McCoy, A.J.; Sammito, M.D.; Read, R.J. Implications of AlphaFold2 for Crystallographic Phasing by Molecular Replacement. Acta Crystallogr. D Struct. Biol. 2022, 78, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; McHugh, R.; Anishchenko, I.; Baker, D.; DiMaio, F. Accurate Prediction of Nucleic Acid and Protein-Nucleic Acid Complexes Using RoseTTAFoldNA. bioRxiv 2022, arXiv:2022.09.09.507333. [Google Scholar]
- Watkins, A.M.; Rangan, R.; Das, R. FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure 2020, 28, 963–976.e6. [Google Scholar] [CrossRef] [PubMed]
- Grigg, J.C.; Ke, A. Structural Determinants for Geometry and Information Decoding of TRNA by T Box Leader RNA. Structure 2013, 21, 2025–2032. [Google Scholar] [CrossRef]
- Grigg, J.C.; Price, I.R.; Ke, A. TRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-TRNA Sensing Mechanisms by the T-Box Riboswitch. Crystals 2022, 12, 694. [Google Scholar] [CrossRef]
- Xiao, H.; Murakami, H.; Suga, H.; Ferré-D’Amaré, A.R. Structural Basis of Specific tRNA Aminoacylation by a Small in Vitro Selected Ribozyme. Nature 2008, 454, 358–361. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R.; Doudna, J.A. Methods to Crystallize RNA. In Current Protocols in Nucleic Acid Chemistry; Wiley & Sons: New York, NY, USA, 2001; Chapter 7, Unit 7.6. [Google Scholar] [CrossRef]
- Rupert, P.B.; Ferré-D’Amaré, A.R. Crystallization of the Hairpin Ribozyme: Illustrative Protocols. Methods Mol. Biol. 2004, 252, 303–311. [Google Scholar] [CrossRef]
- Pyle, A. Metal Ions in the Structure and Function of RNA. J. Biol. Inorg. Chem. 2002, 7, 679–690. [Google Scholar] [CrossRef]
- Wedekind, J.E. Metal Ion Binding and Function in Natural and Artificial Small RNA Enzymes from a Structural Perspective. Met. Ions Life Sci. 2011, 9, 299–345. [Google Scholar]
- Jenkins, J.L.; Wedekind, J.E. The Quick and the Dead: A Guide to Fast Phasing of Small Ribozyme and Riboswitch Crystal Structures. Methods Mol. Biol. 2016, 1490, 265–280. [Google Scholar] [CrossRef]
- Batey, R.T.; Kieft, J.S. Soaking Hexammine Cations into RNA Crystals to Obtain Derivatives for Phasing Diffraction Data. Methods Mol. Biol. 2016, 1320, 219–232. [Google Scholar] [CrossRef]
- Keel, A.Y.; Rambo, R.P.; Batey, R.T.; Kieft, J.S. A General Strategy to Solve the Phase Problem in RNA Crystallography. Structure 2007, 15, 761–772. [Google Scholar] [CrossRef]
- Jou, R.; Cowan, J.A. Ribonuclease H Activation by Inert Transition-Metal Complexes. Mechanistic Probes for Metallocofactors: Insights on the Metallobiochemistry of Divalent Magnesium Ion. J. Am. Chem. Soc. 1991, 113, 6685–6686. [Google Scholar] [CrossRef]
- Cate, J.H.; Yusupov, M.M.; Yusupova, G.Z.; Earnest, T.N.; Noller, H.F. X-Ray Crystal Structures of 70S Ribosome Functional Complexes. Science 1999, 285, 2095–2104. [Google Scholar] [CrossRef]
- Cate, J.H.; Doudna, J.A. Metal-Binding Sites in the Major Groove of a Large Ribozyme Domain. Structure 1996, 4, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sussman, J.L.; Wang, A.H.; Seeman, N.C.; Rich, A. Three-Dimensional Tertiary Structure of Yeast Phenylalanine Transfer RNA. Science 1974, 185, 435–440. [Google Scholar] [CrossRef]
- Robertus, J.D.; Ladner, J.E.; Finch, J.T.; Rhodes, D.; Brown, R.S.; Clark, B.F.; Klug, A. Structure of Yeast Phenylalanine tRNA at 3 A Resolution. Nature 1974, 250, 546–551. [Google Scholar] [CrossRef]
- Golden, B.L. Heavy Atom Derivatives of RNA. In Methods in Enzymology; RNA—Ligand Interactions, Part A; Academic Press: Cambridge, MA, USA, 2000; Volume 317, pp. 124–132. [Google Scholar]
- Golden, B.L.; Gooding, A.R.; Podell, E.R.; Cech, T.R. X-Ray Crystallography of Large RNAs: Heavy-Atom Derivatives by RNA Engineering. RNA 1996, 2, 1295–1305. [Google Scholar]
- Wedekind, J.E.; McKay, D.B. Purification, Crystallization, and X-Ray Diffraction Analysis of Small Ribozymes. Meth. Enzymol. 2000, 317, 149–168. [Google Scholar] [CrossRef]
- Masquida, B.; Westhof, E. On the Wobble GoU and Related Pairs. RNA 2000, 6, 9–15. [Google Scholar] [CrossRef]
- Varani, G.; McClain, W.H. The G·U Wobble Base Pair. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Colmenarejo, G.; Tinoco, I. Structure and Thermodynamics of Metal Binding in the P5 Helix of a Group I Intron Ribozyme. J. Mol. Biol. 1999, 290, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Stefan, L.R.; Zhang, R.; Levitan, A.G.; Hendrix, D.K.; Brenner, S.E.; Holbrook, S.R. MeRNA: A Database of Metal Ion Binding Sites in RNA Structures. Nucleic Acids Res. 2006, 34, D131–D134. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.D.; Rambo, R.P.; Van Tyne, D.; Batey, R.T. Structure of the SAM-II Riboswitch Bound to S-Adenosylmethionine. Nat. Struct. Mol. Biol. 2008, 15, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Huang, Z. Selenium Derivatization of Nucleic Acids for X-Ray Crystal-Structure and Function Studies. Chem. Biodivers. 2010, 7, 753–785. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Sheng, J.; Carrasco, N.; Huang, Z. Selenium Derivatization of Nucleic Acids for Crystallography. Nucleic Acids Res. 2007, 35, 477–485. [Google Scholar] [CrossRef]
- Höbartner, C.; Rieder, R.; Kreutz, C.; Puffer, B.; Lang, K.; Polonskaia, A.; Serganov, A.; Micura, R. Syntheses of RNAs with up to 100 Nucleotides Containing Site-Specific 2′-Methylseleno Labels for Use in X-Ray Crystallography. J. Am. Chem. Soc. 2005, 127, 12035–12045. [Google Scholar] [CrossRef]
- Höbartner, C.; Micura, R. Chemical Synthesis of Selenium-Modified Oligoribonucleotides and Their Enzymatic Ligation Leading to an U6 SnRNA Stem-Loop Segment. J. Am. Chem. Soc. 2004, 126, 1141–1149. [Google Scholar] [CrossRef]
- Dauter, M.; Dauter, Z. Many Ways to Derivatize Macromolecules and Their Crystals for Phasing. Methods Mol. Biol. 2017, 1607, 349–356. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, R.W.; Smathers, C.M.; Robart, A.R. General Strategies for RNA X-ray Crystallography. Molecules 2023, 28, 2111. https://doi.org/10.3390/molecules28052111
Jackson RW, Smathers CM, Robart AR. General Strategies for RNA X-ray Crystallography. Molecules. 2023; 28(5):2111. https://doi.org/10.3390/molecules28052111
Chicago/Turabian StyleJackson, Ryland W., Claire M. Smathers, and Aaron R. Robart. 2023. "General Strategies for RNA X-ray Crystallography" Molecules 28, no. 5: 2111. https://doi.org/10.3390/molecules28052111
APA StyleJackson, R. W., Smathers, C. M., & Robart, A. R. (2023). General Strategies for RNA X-ray Crystallography. Molecules, 28(5), 2111. https://doi.org/10.3390/molecules28052111






