To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense
Abstract
1. Introduction
2. Results
2.1. Structure-Based Sequence Alignment of AmicoTA and Known DAATs
2.2. Substrate Specificity Profile of AmicoTA
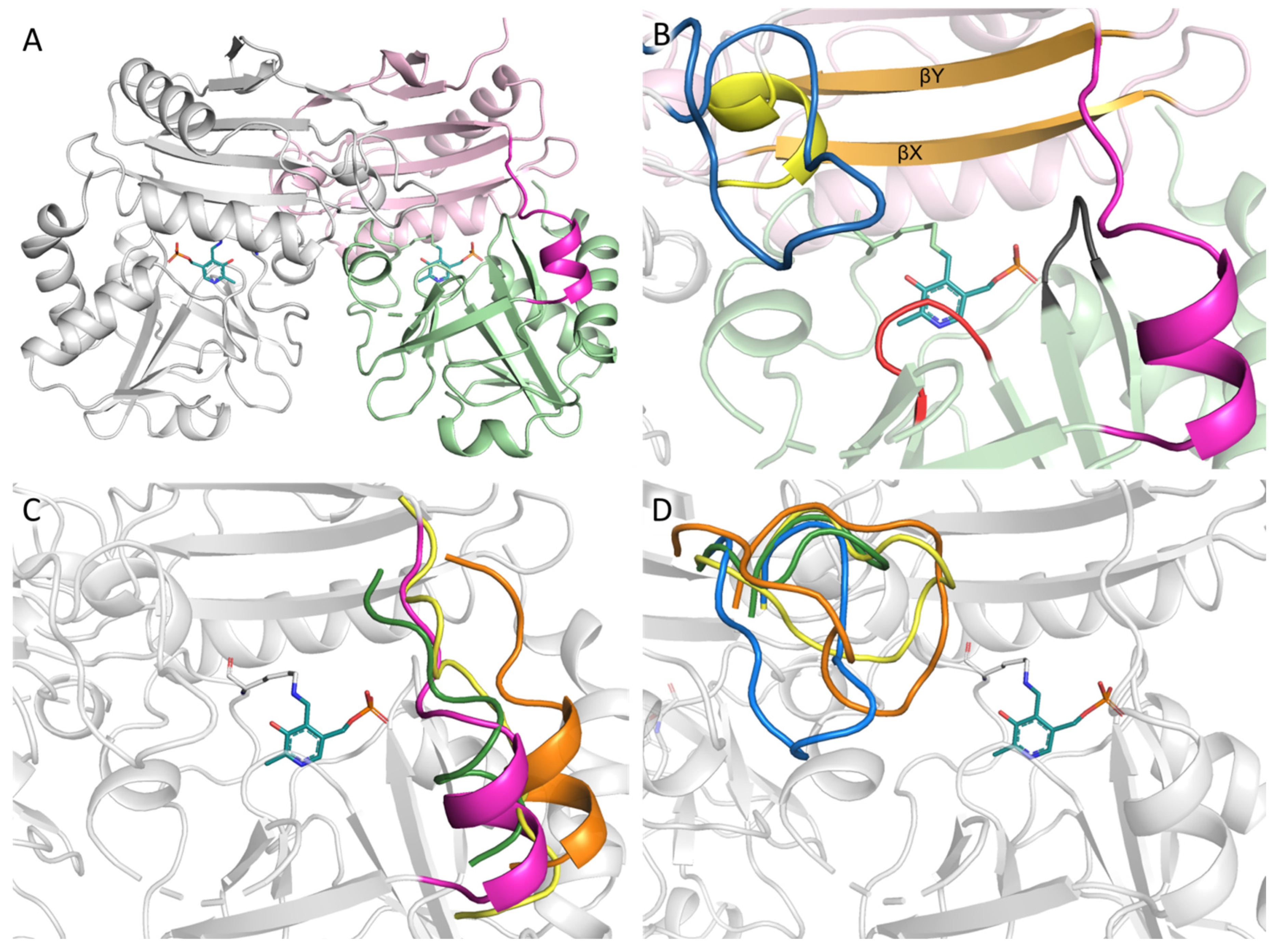
2.3. The Overall Structure of AmicoTA
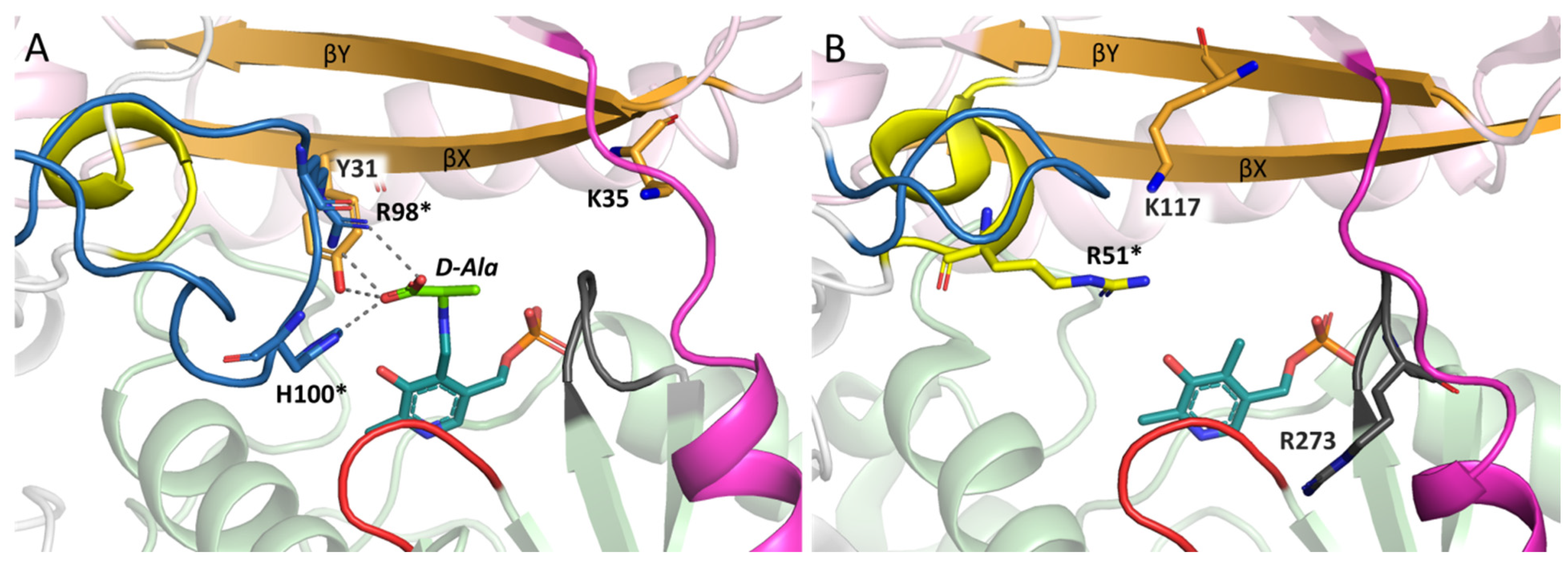
2.4. Active Site Organization in the AmicoTA Dimer
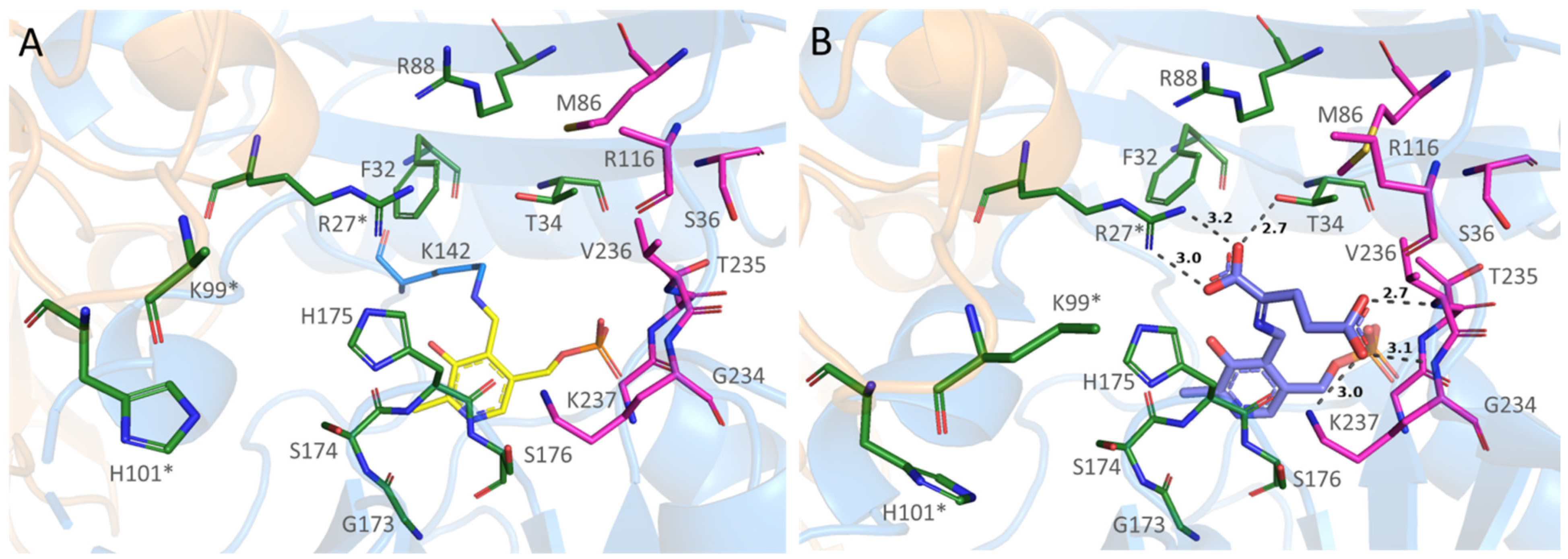
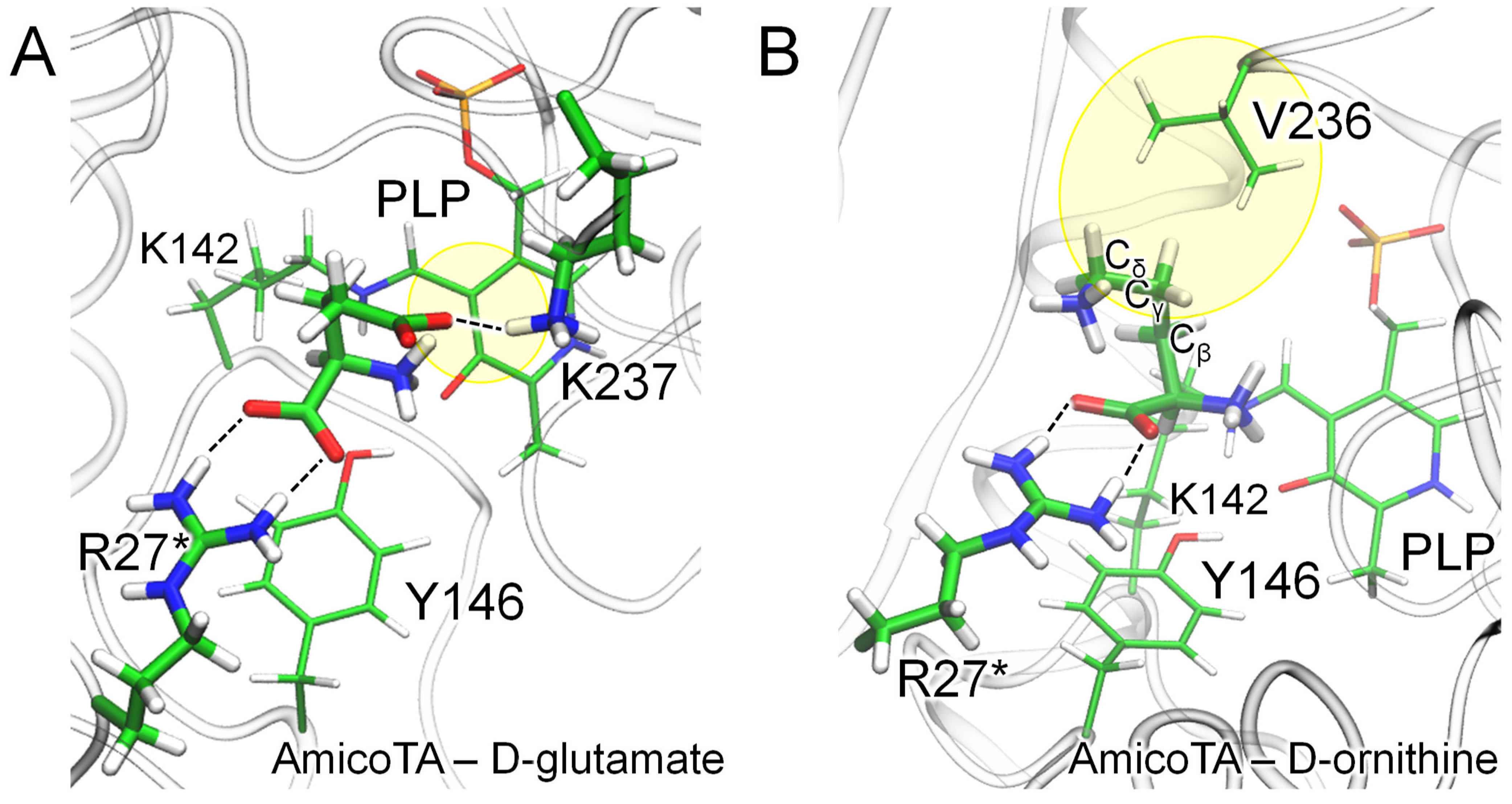
2.5. Structural Analysis of the Substrates Binding in the Complex of AmicoTA with D-Glutamate
2.6. Substrate Binding Modes Revealed by Molecular Dynamic Simulations
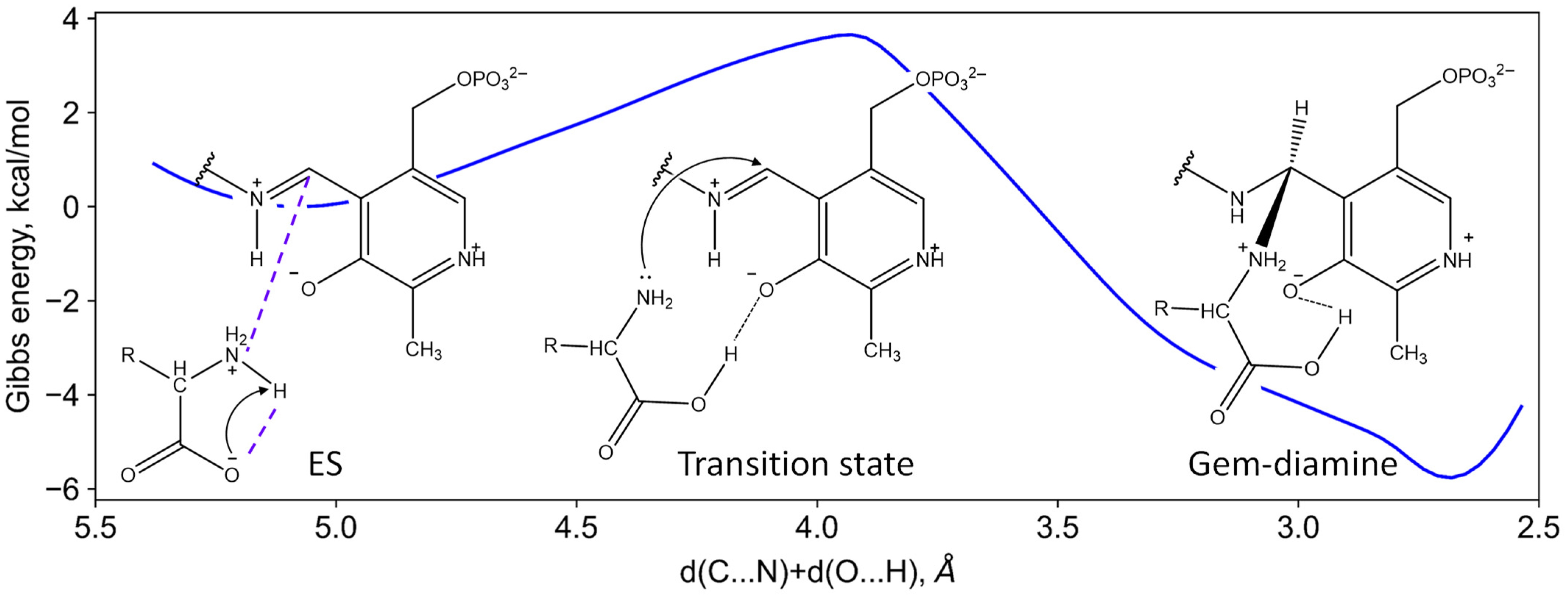
2.7. Michaelis Complex of AmicoTA with D-Glutamate and Substrate-Assisted Mechanism of Catalysis
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification of the Recombinant AmicoTA
4.2. Enzyme Activity Assay
4.3. Effect of pH and Temperature on the Overall Transamination Reaction
4.4. Analysis of Thermal Stability and Operational Stability
4.5. Half-Reaction Assay
4.6. Analysis of the Product Yield and Enantiomeric Excess in the Transamination Reaction
4.7. Crystallization and Data Collection
4.8. Structure Solution and Refinement
4.9. Structure Analysis and Validation
4.10. Molecular Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bell, E.L.; Finnigan, W.; France, S.P.; Green, A.P.; Hayes, M.A.; Hepworth, L.J.; Lovelock, S.L.; Niikura, H.; Osuna, S.; Romero, E.; et al. Biocatalysis. Nat. Rev. Methods Prim. 2021, 1, 46. [Google Scholar] [CrossRef]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef]
- Gu, X.; Zhao, J.; Chen, L.; Li, Y.; Yu, B.; Tian, X.; Min, Z.; Xu, S.; Gu, H.; Sun, J.; et al. Application of transition-metal catalysis, biocatalysis, and flow chemistry as state-of-the-art technologies in the synthesis of LCZ696. J. Org. Chem. 2020, 85, 6844–6853. [Google Scholar] [CrossRef] [PubMed]
- Slabu, I.; Galman, J.L.; Lloyd, R.C.; Turner, N.J. Discovery, Engineering, and Synthetic Application of Transaminase Biocatalysts. ACS Catal. 2017, 7, 8263–8284. [Google Scholar] [CrossRef]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal Phosphate Enzymes: Mechanistic, Structural, and Evolutionary Considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Toney, M.D. Reaction specificity in pyridoxal phosphate enzymes. Arch. Biochem. Biophys. 2005, 433, 279–287. [Google Scholar] [CrossRef]
- Braunstein, A.E. Amino Group Transfer. In The Enzymes; P.D., B., Ed.; Academic Press: London, UK, 1973; pp. 379–481. [Google Scholar]
- Velick, S.F.; Vavra, J. A kinetic and equilibrium analysis of the glutamic oxaloacetate transaminase mechanism. J. Biol. Chem. 1962, 237, 2109–2122. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Popov, V.O.; Boyko, K.M. Structural insight into the substrate specificity of PLP fold type IV transaminases. Appl. Microbiol. Biotechnol. 2020, 104, 2343–2357. [Google Scholar] [CrossRef]
- Okada, K.; Hirotsu, K.; Hayashi, H.; Kagamiyama, H. Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: Induced fit and substrate recognition of the Enzyme. Biochemistry 2001, 40, 7453–7463. [Google Scholar] [CrossRef]
- Sugio, S.; Petsko, G.A.; Manning, J.M.; Soda, K.; Ringe, D. Crystal Structure of a D-Amino Acid Aminotransferase: How the Protein Controls Stereoselectivity. Biochemistry 1995, 34, 9661–9669. [Google Scholar] [CrossRef]
- Thomsen, M.; Skalden, L.; Palm, G.J.; Höhne, M.; Bornscheuer, U.T.; Hinrichs, W. Crystallographic characterization of the (R)-selective amine transaminase from Aspergillus fumigatus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Steffen-Munsberg, F.; Vickers, C.; Kohls, H.; Land, H.; Mallin, H.; Nobili, A.; Skalden, L.; van den Bergh, T.; Joosten, H.-J.; Berglund, P.; et al. Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv. 2015, 33, 566–604. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, J.F.; Eichele, G.; Ford, G.C.; Vincent, M.G.; Jansonius, J.N.; Gehring, H.; Christen, P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J. Mol. Biol. 1984, 174, 497–525. [Google Scholar] [CrossRef] [PubMed]
- Wybenga, G.G.; Crismaru, C.G.; Janssen, D.B.; Dijkstra, B.W. Structural Determinants of the β-Selectivity of a Bacterial Aminotransferase. J. Biol. Chem. 2012, 287, 28495–28502. [Google Scholar] [CrossRef]
- Grishin, N.V.; Phillips, M.A.; Goldsmith, E.J. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 1995, 4, 1291–1304. [Google Scholar] [CrossRef]
- Rudat, J.; Brucher, B.R.; Syldatk, C. Transaminases for the synthesis of enantiopure beta-amino acids. AMB Express 2012, 2, 11. [Google Scholar] [CrossRef]
- Funakoshi, M.; Sekine, M.; Katane, M.; Furuchi, T.; Yohda, M.; Yoshikawa, T.; Homma, H. Cloning and functional characterization of Arabidopsis thaliana D-amino acid aminotransferase - D-aspartate behavior during germination. FEBS J. 2008, 275, 1188–1200. [Google Scholar] [CrossRef]
- Pucci, M.J.; Thanassi, J.A.; Ho, H.T.; Falk, P.J.; Dougherty, T.J. Staphylococcus haemolyticus contains two D-glutamic acid biosynthetic activities, a glutamate racemase and a D-amino acid transaminase. J. Bacteriol. 1995, 177, 336–342. [Google Scholar] [CrossRef]
- Radkov, A.D.; Moe, L.A. Bacterial synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 2014, 98, 5363–5374. [Google Scholar] [CrossRef]
- Gao, X.; Ma, Q.; Zhu, H. Distribution, industrial applications, and enzymatic synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 2015, 99, 3341–3349. [Google Scholar] [CrossRef]
- Tanizawa, K.; Masu, Y.; Asano, S.; Tanaka, H.; Soda, K. Thermostable D-amino acid aminotransferase from a thermophilic Bacillus species. J. Biol. Chem. 1989, 264, 2445–2449. [Google Scholar] [CrossRef] [PubMed]
- Barber, J.E.B.; Damry, A.M.; Calderini, G.F.; Walton, C.J.W.; Chica, R.A. Continuous colorimetric screening assay for detection of d-amino acid aminotransferase mutants displaying altered substrate specificity. Anal. Biochem. 2014, 463, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Yonaha, K.; Misono, H.; Yamamoto, T.; Soda, K. D-amino acid aminotransferase of Bacillus sphaericus. Enzymologic and spectrometric properties. J. Biol. Chem. 1975, 250, 6983–6989. [Google Scholar] [CrossRef]
- Lee, S.-G.; Hong, S.-P.; Song, J.J.; Kim, S.-J.; Kwak, M.-S.; Sung, M.-H. Functional and structural characterization of thermostable D-amino acid aminotransferases from Geobacillus spp. Appl. Environ. Microbiol. 2006, 72, 1588–1594. [Google Scholar] [CrossRef] [PubMed]
- Pavkov-Keller, T.; Strohmeier, G.A.; Diepold, M.; Peeters, W.; Smeets, N.; Schürmann, M.; Gruber, K.; Schwab, H.; Steiner, K. Discovery and structural characterisation of new fold type IV-transaminases exemplify the diversity of this enzyme fold. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Bakunova, A.K.; Nikolaeva, A.Y.; Rakitina, T.V.; Isaikina, T.Y.; Khrenova, M.G.; Boyko, K.M.; Popov, V.O.; Bezsudnova, E.Y. The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis: Biochemical and Structural Insights into the New Enzyme. Molecules 2021, 26, 5053. [Google Scholar] [CrossRef]
- Höhne, M.; Schätzle, S.; Jochens, H.; Robins, K.; Bornscheuer, U.T. Rational assignment of key motifs for function guides in silico enzyme identification. Nat. Chem. Biol. 2010, 6, 807–813. [Google Scholar] [CrossRef]
- Kishimoto, K.; Yoshimura, T.; Esaki, N.; Sugio, S.; Manning, J.M.; Soda, K. Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 Å resolution: Implication of the structural role of Leu201 in transamination The leucine-to-alanine mutation at residue 201 of D-amino acid aminotransferase provides a unique enzym. Protein Eng. 1998, 11, 691–696. [Google Scholar]
- Kishimoto, K.; Yoshimura, T.; Soda, K.; Esaki, N. Mutation of Arginine 98, Which Serves as a Substrate-Recognition Site of D-Amino Acid Aminotransferase, Can Be Partly Compensated for by Mutation of Tyrosine 88 to an Arginyl Residue1. J. Biochem 1997, 122, 1182–1189. [Google Scholar] [CrossRef]
- Peisach, D.; Chipman, D.M.; Van Ophem, P.W.; Manning, J.M.; Ringe, D. Crystallographic study of steps along the reaction pathway of D -amino acid aminotransferase. Biochemistry 1998, 37, 4958–4967. [Google Scholar] [CrossRef]
- Ro, H.-S.; Hong, S.-P.; Seo, H.-J.; Yoshimura, T.; Esaki, N.; Soda, K.; Kim, H.-S.; Sung, M.-H. Site-directed mutagenesis of the amino acid residues in β-strand III [Val 30 -Val 36 ] of d -amino acid aminotransferase of Bacillus sp. YM-1. FEBS Lett. 1996, 398, 141–145. [Google Scholar] [CrossRef]
- Chertkov, O.; Sikorski, J.; Brambilla, E.; Lapidus, A.; Copeland, A.; del Rio, T.G.; Nolan, M.; Lucas, S.; Tice, H.; Cheng, J.F.; et al. Complete genome sequence of Aminobacterium colombiense type strain (ALA-1 T). Stand. Genomic Sci. 2010, 2, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Fuchikami, Y.; Yoshimura, T.; Gutierrez, A.; Soda, K.; Esaki, N. Construction and Properties of a Fragmentary D-Amino Acid Aminotransferase. J. Biochem. 1998, 124, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Metzler, D.E.; Ikawa, M.; Snell, E.E. A General Mechanism for Vitamin B 6 -catalyzed Reactions 1. J. Am. Chem. Soc. 1954, 76, 648–652. [Google Scholar] [CrossRef]
- Hirotsu, K.; Goto, M.; Okamoto, A.; Miyahara, I. Dual substrate recognition of aminotransferases. Chem. Rec. 2005, 5, 160–172. [Google Scholar] [CrossRef]
- Skalden, L.; Thomsen, M.; Höhne, M.; Bornscheuer, U.T.; Hinrichs, W. Structural and biochemical characterization of the dual substrate recognition of the ( R )-selective amine transaminase from Aspergillus fumigatus. FEBS J. 2015, 282, 407–415. [Google Scholar] [CrossRef]
- Isupov, M.N.; Boyko, K.M.; Sutter, J.-M.; James, P.; Sayer, C.; Schmidt, M.; Schönheit, P.; Nikolaeva, A.Y.; Stekhanova, T.N.; Mardanov, A.V.; et al. Thermostable branched-chain amino acid transaminases from thearchaea Geoglobus acetivorans and Archaeoglobus fulgidus: Biochemical and structural characterization. Front. Bioeng. Biotechnol. 2019, 7, 1–16. [Google Scholar] [CrossRef]
- Telzerow, A.; Paris, J.; Håkansson, M.; González-Sabín, J.; Ríos-Lombardía, N.; Schürmann, M.; Gröger, H.; Morís, F.; Kourist, R.; Schwab, H.; et al. Amine Transaminase from Exophiala Xenobiotica —Crystal Structure and Engineering of a Fold IV Transaminase that Naturally Converts Biaryl Ketones. ACS Catal. 2019, 9, 1140–1148. [Google Scholar] [CrossRef]
- Fasella, P.; Giartosio, A.; Hammes, G.G. The Interaction of Aspartate Aminotransferase with α-Methylaspartic Acid *. Biochemistry 1966, 5, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Mizuno, T.; Kagamiyama, H. A hydrogen-bonding network modulating enzyme function: Asparagine-194 and tyrosine-225 of Escherichia coli aspartate aminotransferase. Biochemistry 1993, 32, 1810–1815. [Google Scholar] [CrossRef] [PubMed]
- Hammes, G.G. A golden era for understanding enzyme mechanisms. Protein Sci. 1998, 7, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Sayer, C.; Martinez-Torres, R.J.; Richter, N.; Isupov, M.N.; Hailes, H.C.; Littlechild, J.A.; Ward, J.M. The substrate specificity, enantioselectivity and structure of the ( R )-selective amine: Pyruvate transaminase from Nectria haematococca. FEBS J. 2014, 281, 2240–2253. [Google Scholar] [CrossRef]
- Boyko, K.M.; Stekhanova, T.N.; Nikolaeva, A.Y.; Mardanov, A.V.; Rakitin, A.L.; Ravin, N.V.; Bezsudnova, E.Y.; Popov, V.O. First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for l-amino acids and R-amines. Extremophiles 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.; Skalden, L.; Palm, G.J.; Höhne, M.; Bornscheuer, U.T.; Hinrichs, W. Crystallization and preliminary X-ray diffraction studies of the ( R )-selective amine transaminase from Aspergillus fumigatus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1415–1417. [Google Scholar] [CrossRef]
- Karpeisky, M.Y.; Ivanov, V.I. A Molecular Mechanism for Enzymatic Transamination. Nature 1966, 210, 493–496. [Google Scholar] [CrossRef]
- Inoue, K.; Kuramitsu, S.; Aki, K.; Watanabe, Y.; Takagi, T.; Nishigai, M.; Ikai, A.; Kagamiyama, H. Branched-Chain Amino Acid Aminotransferase of Escherichia coli: Overproduction and Properties1. J. Biochem. 1988, 104, 777–784. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Boyko, K.M.; Popov, V.O. Properties of bacterial and archaeal branched-chain amino acid aminotransferases. Biochem. 2017, 82, 1572–1591. [Google Scholar] [CrossRef]
- Amorim Franco, T.M.; Hegde, S.; Blanchard, J.S. Chemical mechanism of the branched-chain aminotransferase IlvE from Mycobacterium tuberculosis. Biochemistry 2016, 55, 6295–6303. [Google Scholar] [CrossRef]
- Goto, M.; Miyahara, I.; Hirotsu, K.; Conway, M.; Yennawar, N.; Islam, M.M.; Hutson, S.M. Structural Determinants for Branched-chain Aminotransferase Isozyme-specific Inhibition by the Anticonvulsant Drug Gabapentin. J. Biol. Chem. 2005, 280, 37246–37256. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, N.M.F.S.A.; Fernandes, P.A.; Ramos, M.J. Computational Mechanistic Studies Addressed to the Transimination Reaction Present in All Pyridoxal 5′-Phosphate-Requiring Enzymes. J. Chem. Theory Comput. 2011, 7, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Boyko, K.; Gorbacheva, M.; Rakitina, T.; Korzhenevskiy, D.; Vanyushkina, A.; Kamashev, D.; Lipkin, A.; Popov, V. Expression, purification, crystallization and preliminary X-ray crystallographic analysis of the histone-like HU protein from Spiroplasma melliferum KC3. Acta Crystallogr. Sect. FStructural Biol. Commun. 2015, 71, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Nurizzo, D.; Mairs, T.; Guijarro, M.; Rey, V.; Meyer, J.; Fajardo, P.; Chavanne, J.; Biasci, J.C.; McSweeney, S.; Mitchell, E. The ID23-1 structural biology beamline at the ESRF. J. Synchrotron Radiat. 2006, 13, 227–238. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Vagin, A.A.; Isupov, M.N. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001, 57, 1451–1456. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ 1 and χ 2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2009, 31, 671–690. [Google Scholar] [CrossRef]
- Seritan, S.; Bannwarth, C.; Fales, B.S.; Hohenstein, E.G.; Isborn, C.M.; Kokkila-Schumacher, S.I.L.; Li, X.; Liu, F.; Luehr, N.; Snyder, J.W.; et al. TeraChem: A graphical processing unit -accelerated electronic structure package for large-scale ab initio molecular dynamics. WIREs Comput. Mol. Sci. 2021, 11. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Bernardi, R.C.; Rudack, T.; Scheurer, M.; Riplinger, C.; Phillips, J.C.; Maia, J.D.C.; Rocha, G.B.; Ribeiro, J.V.; Stone, J.E.; et al. NAMD goes quantum: An integrative suite for hybrid simulations. Nat. Methods 2018, 15, 351–354. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
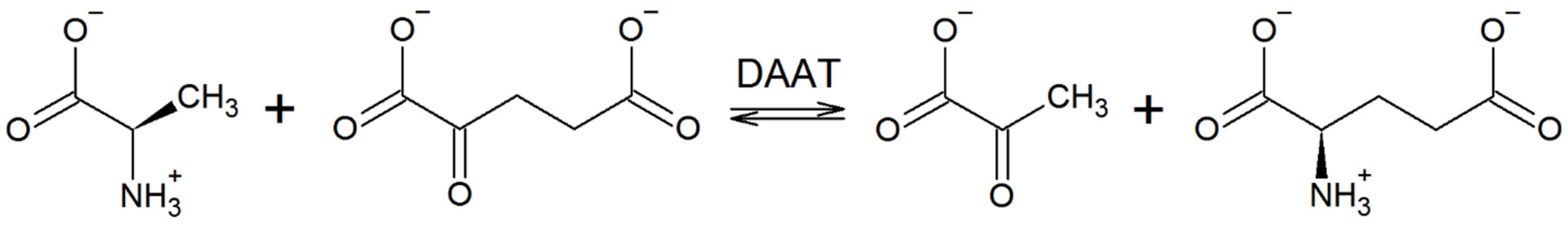


| DAAT from | Identity, % | O-Pocket α-Helix | βX-Strand | βY-Strand | O-Pocket Loop | Interdomain Loop | β-turn1 | β-turn2 |
|---|---|---|---|---|---|---|---|---|
| Bacillus sp. YM-1 | 100 | 21DRGYGFG27 | 29GVYEVVKVY37 | 85GHIYFQVT92 | 93RGTSPRAHQFPENTVKP109 | 117NPRPLENLEKG128 | 178GSSS181 | 240STTS243 |
| B. sphaericus | 68.0 | 22DRGYQFG28 | 30GIYEVIKVY38 | 86GHVYFQIT93 | 94RGTTSRNHIFPDASVPA110 | 119GERSIEQFEKG129 | 179CSSA182 | 241SVSS244 |
| B. thailandensis | 37.5 | 32DRGFIFG38 | 40GVYEVVPIY48 | 05AIVYIQVT112 | 113RGVAKRGHAFPANAVP128 | 137LALPTDAQRAQG147 | 198GSSS201 | 260SATK263 |
| G. toebii SK1 | 48.0 | 25ERGLQFG31 | 33GVYEVARIY41 | 90AILYLQVT97 | 98RGSFPRNHAFPAENRP113 | 122MPRKIREIEQG132 | 182GSSS185 | 244STTS247 |
| L. salivarius | 35.9 | 23DRALYFG29 | 31GCYDATTFK39 | 87GILYWQTS94 | 95RGSGLRNHIFPEDSQP110 | 118PYGLVPFDTE127 | 177CAHS180 | 240SSAC242 |
| C. pusillum | 23.1 | 46DLGITRG52 | 54GVFETIAVI62 | 114LFAKLILT121 | 122RGIEGEGRP130 | 139GEDFSQQRLG148 | 208GPTS211 | 270SSVR273 |
| H. hydrossis | 25.9 | 23DLSILRG29 | 31GIFDYFLAR39 | 87AGIRLVLT94 | 95GGYSPDGYTVNP107 | 115DLPASAWEFSAQG127 | 177SARS180 | 238STIK241 |
| M. tuberculosis | 23.6 | 21DLAAVRG27 | 29GVFETLLVR37 | 7GALRLIYS94 | 95RGREGGSAP103 | 112VPARVIGARRDG123 | 183GPRS186 | 252SSMT255 |
| A. thaliana | 23.8 | 99DHMVHRG105 | 107GVFDTALII115 | 163GSLRYWLS170 | 171AGPGDFLLSPSQCLLP186 | 195TNFAINPIG203 | 256GPNM259 | 324GSGI327 |
| AmicoTA | 28.7 | 22DLIIQRG28 | 30GVFETISTH38 | 85TMVRPYIT92 | 93GDSFGKDHLFSSSRYFV110 | 115IRKPDPILYEKG126 | 173GSHS176 | 234GTVK237 |
| Group | Sequence Motif 1 | Sequence Motif 2 | ||||
|---|---|---|---|---|---|---|
| 26 | 31 | 33 | 35 | x88xx | 98xx | |
| Canonical DAATs | F | Y | [VA] | [KRPT] | xYxQ | RxH |
| Non-canonical DAATs | R | F | [TY] | [LA] | x[RK]x[IVW] | Non-conservative |
| Amino Donor | 103 × kmax, s−1 | KD, mM | kmax/KD, M−1 s−1 |
|---|---|---|---|
| D-Glutamate | 1080 ± 40 | 2.1 ± 0.2 | 515 ± 70 |
| D-Alanine | 280 ± 10 | 50 ± 6 | 5.6 ± 0.8 |
| D-Aspartate | 118 ± 6 | 120 ± 15 | 1.0 ± 0.2 |
| D-Leucine | 33 ± 2 | 100 ± 10 | 0.33 ± 0.05 |
| D-Ornithine | 2.1 ± 0.1 | 10 ± 1 | 0.21 ± 0.02 |
| D-Phenylalanine | 2.9 ± 0.2 | 19 ± 3 | 0.15 ± 0.03 |
| L-Alanine | ND | ||
| L-Leucine | ND | ||
| (R)-(+)-1-phenylethylamine | ND | ||
| (S)-(−)-1-phenylethylamine | ND | ||
| Substrate | Co-Substrate | kcat, s−1 | Km, mM | kcat/Km, M−1 s−1 |
|---|---|---|---|---|
| D-alanine | 5 mM α-ketoglutarate | 9.9 ± 1.3 | 465 ± 60 | 22 ± 6 |
| α-ketoglutarate | 500 mM D-alanine | 9.9 ± 1.3 | 7.2 ± 0.8 | 1375 ± 330 |
| D-glutamate | 500 mM pyruvate | 1.40 ± 0.06 | 5.2 ± 0.6 | 270 ± 40 |
| pyruvate | 30 mM D-glutamate | 1.40 ± 0.06 | 160 ± 20 | 9 ± 1 |
| Holo form of AmicoTA | The Complex of AmicoTA with D-Glutamate | |
|---|---|---|
| Diffraction source | ESRF (ID23-1 beamline) | Institute of Organic Chemistry RAS (Rigaku OD XtaLAB Synergy-S) |
| Wavelength (Å) | 0.98 | 1.54 |
| Temperature (K) | 100 | |
| Detector | PILATUS 6M | HyPix-6000HE |
| Crystal-to-detector distance (mm) | 250 | 33 |
| Rotation range per image (°) | 0.05 | 0.3 |
| Total rotation range (°) | 120 | 300 |
| Space group | P212121 | P212121 |
| a, b, c (Å) | 61.19, 80.97, 98.96 | 61.55, 90.01, 100.86 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Average mosaicity (°) | 0.273 | 1.04 |
| Resolution range (Å) | 43.78–1.90 (1.94–1.90) | 21.48–1.90 (1.94–1.90) |
| Completeness (%) | 93.0 (92.5) | 98.8 (97.5) |
| Average redundancy | 4.5 (4.4) | 10.8 (11.3) |
| 〈I/σ(I)〉 | 10.1 (1.5) | 19.3 (2.6) |
| Rmeas (%) | 8.9 (100.4) | 11.1 (116.8) |
| CC1/2 | 99.8 (55.8) | 99.9 (83.1) |
| Rfact (%) | 18.9 | 19.5 |
| Rfree. (%) | 24.5 | 21.1 |
| Bonds (Å) | 0.02 | 0.01 |
| Angles (°) | 1.98 | 1.66 |
| Most favored (%) | 98.5 | 98.7 |
| Allowed (%) | 1.5 | 1.3 |
| PDB entry code | 8AHR | 8AYK |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shilova, S.A.; Khrenova, M.G.; Matyuta, I.O.; Nikolaeva, A.Y.; Rakitina, T.V.; Klyachko, N.L.; Minyaev, M.E.; Boyko, K.M.; Popov, V.O.; Bezsudnova, E.Y. To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense. Molecules 2023, 28, 2109. https://doi.org/10.3390/molecules28052109
Shilova SA, Khrenova MG, Matyuta IO, Nikolaeva AY, Rakitina TV, Klyachko NL, Minyaev ME, Boyko KM, Popov VO, Bezsudnova EY. To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense. Molecules. 2023; 28(5):2109. https://doi.org/10.3390/molecules28052109
Chicago/Turabian StyleShilova, Sofia A., Maria G. Khrenova, Ilya O. Matyuta, Alena Y. Nikolaeva, Tatiana V. Rakitina, Natalia L. Klyachko, Mikhail E. Minyaev, Konstantin M. Boyko, Vladimir O. Popov, and Ekaterina Yu. Bezsudnova. 2023. "To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense" Molecules 28, no. 5: 2109. https://doi.org/10.3390/molecules28052109
APA StyleShilova, S. A., Khrenova, M. G., Matyuta, I. O., Nikolaeva, A. Y., Rakitina, T. V., Klyachko, N. L., Minyaev, M. E., Boyko, K. M., Popov, V. O., & Bezsudnova, E. Y. (2023). To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense. Molecules, 28(5), 2109. https://doi.org/10.3390/molecules28052109






