Abstract
Background: Although X-ray fluorescence microscopy is becoming a widely used technique for single-cell analysis, sample preparation for this microscopy remains one of the main challenges in obtaining optimal conditions for the measurements in the X-ray regime. The information available to researchers on sample treatment is inadequate and unclear, sometimes leading to wasted time and jeopardizing the experiment’s success. Many cell fixation methods have been described, but none of them have been systematically tested and declared the most suitable for synchrotron X-ray microscopy. Methods: The HEC-1-A endometrial cells, human spermatozoa, and human embryonic kidney (HEK-293) cells were fixed with organic solvents and cross-linking methods: 70% ethanol, 3.7%, and 2% paraformaldehyde; in addition, HEK-293 cells were subjected to methanol/ C3H6O treatment and cryofixation. Fixation methods were compared by coupling low-energy X-ray fluorescence with scanning transmission X-ray microscopy and atomic force microscopy. Results: Organic solvents lead to greater dehydration of cells, which has the most significant effect on the distribution and depletion of diffusion elements. Paraformaldehyde provides robust and reproducible data. Finally, the cryofixed cells provide the best morphology and element content results. Conclusion: Although cryofixation seems to be the most appropriate method as it allows for keeping cells closer to physiological conditions, it has some technical limitations. Paraformaldehyde, when used at the average concentration of 3.7%, is also an excellent alternative for X-ray microscopy.
1. Introduction
X-ray fluorescence (XRF) microscopy is a valuable technique for studying biological samples [1] because it provides the distribution of elements in tissues and cells non-destructively, without the need for stains or fluorescent probes. Mapping the distribution and content of endogenous and exogenous elements and metals is pivotal, especially for those elements with great biological importance, named also as “life elements” [2]. In addition, the ability to map samples at the nanometre scale provides the opportunity to correlate elemental distribution with cell morphology and potentially investigate the physio-pathological state of the cell or explore relevant metabolic pathways [3,4,5].
Single-cell analysis is particularly attractive to researchers from life sciences, biology, and biomedicine, as it offers the opportunity to better understand cellular diversity and specificity. Indeed, multicellular organisms are composed of different tissues and cells, and conventional cell-based assays mainly represent the average response of a mixed population of cells [6,7,8]. As previously reported, elemental analysis on a single cell provides unique complementary information to large population analyses [7]. X-ray microscopy at the single-cell level allows for a detailed analysis of the representative elements of a particular cell state [6] or highlights morphometric features often related to cell function and activity [9]. In this scenario, proper sample preparation is the first and most important step to enable proper XRF analyses [10].
In the experimental setup for XRF Microscopy (XRFM), especially in vacuum conditions, for instance when dealing with soft X-rays, the biological constituents of samples must be fixed before being analysed. Membranes made of silicon nitride (Si3N4) are often preferred as sample carriers for cells, as they offer high transparency and minimal X-ray absorption for X-ray microscopy and are a very suitable surface for cell attachment.
Therefore, the crucial step to obtaining reliable and accurate X-ray data on concentrations and distributions of chemical elements in the samples lies in the correct handling and fixation methods [10]. The latter is chosen depending on the additional processing steps and final analyses planned before or after XRFM. The major challenge is to achieve a state as close as possible to the physiological condition of the cell.
Most commonly used cell fixation protocols have been adopted and adapted from immunocytochemistry; therefore, many fixation methods are described in the literature.
The most commonly used and readily available fixation methods use chemical-based fixatives that block autodegradation processes, increase stability, and preserve morphology [11]. Once applied to a biological sample, they form cross-links between amino acids, proteins, and lipids. Chemical fixation allows for better preservation of morphology than coagulating fixatives (e.g., solvents and acetic acid) generally used for light microscopy.
Aldehyde-based fixation with paraformaldehyde or formaldehyde preserves cell structure with less loss of biochemical information than fixation with organic solvents [12]; however, it is suboptimal for preserving most elements and immobilizing proteins for further analysis. Many small molecules (such as ions) or macromolecules (such as carbohydrates, lipids, and nucleic acids) are not efficiently cross-linked, resulting in their extraction, replacement, or loss [9]. Fixation with ethanol leads to dehydration and precipitation of proteins, resulting in alteration of cell membrane structure, cytoplasmic organelles, and nuclear content [12]. In addition, chemical fixation with organic solvents and cross-linking reagents can lead to denaturation of the protein antigens, which can be problematic for downstream antibody labelling techniques [13].
Some studies have attempted to assess the extent and impact of sample preparation for XRF, especially in the hard X-ray range, to map elements heavier than phosphorus [14]. Recently, a comprehensive study by Perrin et al. [15] showed a difference in the elemental distribution and concentration of several elements, up to Na and Mg, with different preparation protocols. They suggested frozen fixation as the optimal sample preparation for μPIXE (micro Proton Induced X-ray Emission) and μXRF (micro XRF), without an extensive investigation of the cells’ morphology. Similarly, Jin et al. [16] conducted a systematic study comparing elemental maps of frozen cells with chemically fixed cells, also considering different wash buffers for rinsing the cells. They claim that plunge-frozen cells followed by freeze-drying and plunge-frozen cells measured in cryo-conditions are preferable for quantitative elemental evaluations, as they found significant loss of K and Cl in chemically fixed cells, along with increased content and altered distribution of Ca, possible loss of P, and possible redistribution of Zn [16]. Although cryofixation is considered by many the ideal currently available method for cell imaging [10], this preparation is limited by the need for a cryosystem in the preparation facility and by a potential rapid degradation of fixative-free samples. In cryofixation, the sample must be frozen fast enough for the water to change from its normal liquid state to its solid state (vitrification) without an ice crystal phase in between. Ice has a larger volume than water and can therefore cause deformation of the biological ultrastructure. While a variety of samples are suitable for rapid freezing, only a small volume of the sample will produce amorphous (ice-crystal free) ice. Viruses, proteins, organelles, and cells can be quickly iced; however, their volume makes tissues and multicellular organisms much more challenging to be properly cryofixated. Some experimental set-ups enable investigating frozen cells in a frozen hydrated state, under cooling [16]. Even if this condition allows for better sample preservation and reduces radiation damage effects, it requires a complex cryo-stage and cryo-transfer of the samples, which are not available in all facilities, making the experiments much more difficult starting from the sample preparation step till the final analysis [17].
Finally, sample preparation seems to influence the extent of radiation damage caused by soft X-rays during analyses [18,19,20]; therefore, this effect should also be taken into consideration during the selection of the ideal sample preparation.
In summary, the information available to researchers on sample treatment is still insufficient and sometimes ambiguous, leading to the uncertain success of the experiment.
In this study, we combined XRFM with atomic force microscopy (AFM) to evaluate how different fixatives can alter and influence the content and distribution of light chemical elements, diffusible ions, and cell morphology. The comparison will provide helpful information for synchrotron scientists to determine the most appropriate protocol for a particular study or cell model.
As cell models, we have chosen (i) the human HEC-1-A adenocarcinoma endometrial cell line as a model of an adherent immortalised tumour cell line, (ii) human spermatozoa as an example of primary suspension cells, and (iii) the immortalised human embryonic 293 cell line (HEK-293), which are genetically identical cells, thus ensuring repeatability of the experiments. As fixation methods, we have selected (i) 70% ethanol (EtOH), (ii) paraformaldehyde (PFA) 2% for 2′, and (iii) PFA 3.7% for 20′ for all the three cell lines. Additionally, for the HEK-293 cell line, two additional fixation methods were added: methanol/C3H6O (Acetone) and cryofixation.
The advantages and limitations of each preparation method were investigated using low-energy X-ray fluorescence (LEXRF), in conjunction with scanning transmission X-ray microscopy (STXM) and AFM. The advantage of AFM lies in its ability to non-destructively study the morphology of biological samples at the cellular and subcellular level with a spatial resolution in the nanometre range. Therefore, it is an ideal approach to assessing volume and thickness differences in biological samples. In this work, it was used prior to X-ray analyses.
2. Results
As shown in Figure 1, three different fixation procedures were applied to all samples selected for this study; HEC-1-A endometrial cells, spermatozoa, and the HEK-293 cell line were prepared with three different protocols, i.e., 70% ethanol (EtOH), paraformaldehyde (PFA) 2% for 2′, and PFA 3.7% for 20′. For the last cell line, i.e., HEK-293, two additional fixation methods were tested: methanol/C3H6O and cryofixation. AFM measurements with a spatial resolution of 200 nm were first performed on the different selected cells, which were then analysed by XRFM and STXM. For XRFM, the cells were analysed at 1.5 keV to determine the intracellular distribution and XRF intensities of essential life elements such as Na, Mg, and O; at the same time, absorption and differential phase contrast images were acquired in STXM mode. While XRFM allows for elemental distribution inspection, STXM imaging provides morphological information; absorption images are sensitive to different sample thickness and/or density, and phase contrast images highlight borders or structures.
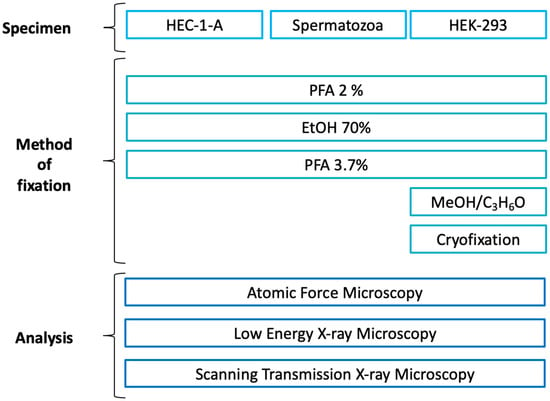
Figure 1.
Experimental plan of the experiment indicating the fixation protocols used for each type of sample. Paraformaldehyde (PFA); Ethanol (EtOH); Methanol (MeOH); Acetone (C3H6O).
The XRF results were compared with and complemented by the AFM images.
2.1. HEC-1-A Cells
2.1.1. XRFM Analysis
The XRF elemental maps of selected HEC-1-A endometrial cells fixed with the three different protocols, i.e., 70% ethanol (EtOH), paraformaldehyde (PFA) 2% for 2′, and PFA 3.7% for 20′, are shown in Figure 2. Absorption and phase contrast images (STXM) show no significant change in cell morphology with PFA 3.7% for 20′ and PFA 2% for 2′ protocols. At the same time, a less defined membrane edge is observed with ethanol fixation. Indeed, in the differential phase contrast image (PhC) of Figure 2a, the cells’ borders appear more blurred or less sharp for EtOH treatment than in the corresponding PFA 3.7% or PFA 2% fixations (PhC in Figure 2b,c). In Figure 2d, we show the average Na, O, and Mg values extracted from all the XRF maps collected for the three protocols. Compared to the standard PFA of 3.7% for 20′, fixation with PFA of 2% for 2′ can result in lower light element content, especially Na and, to a lesser extent, Mg. Fixation with 70% ethanol retains the highest Na levels but results in a deficiency of Mg (Figure 2d). It is worth noting that even if we did not quantify the elements’ amount, average XRF counts of oxygen can be selected to compare the different fixation methods in terms of precision index, as shown in Figure 2e. The coefficient of variation was determined to assess the extent of variability relative to the population mean, as it is widely used to express the precision and repeatability of an assay; a higher coefficient of variation (CV) reflects a large dispersion around the mean. Although the mean fluorescence counts (MFC) are not statistically different when considering the uncertainties, PFA 2% has the highest CV (18.37%) compared to EtOH and PFA 3.7%, for which CV was 5.95% and 9.20%, respectively (see Table S1. HEC-1-A descriptive analysis). All in all, fixation with PFA 3.7% for 20′ seems to preserve the internal cellular structure and thereby the Na and Mg content in the cells more efficiently (Figure 2d).
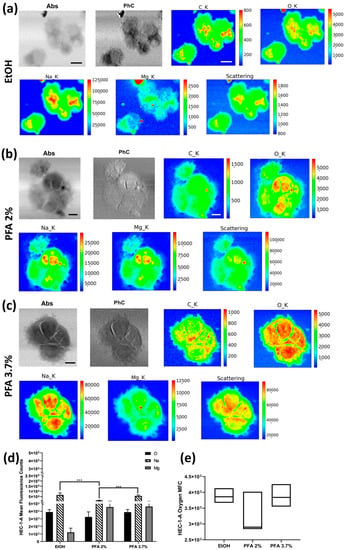
Figure 2.
STXM and XRF analysis of the HEC-1-A cell line. Absorption (Abs) and differential phase contrast (PhC) images, alongside with O, Na, Mg, and Scattering maps of HEC-1A endometrial cells fixed with three different methods: (a) EtOH 70% (mapped area 60 × 52 µm2), (b) PFA 2% (mapped area 68 × 68 µm2), and (c) PFA 3.7% (mapped area 52 × 52 µm2). Scale bars are 10 µm. (d) Normalised average XRF counts of O, Na, and Mg in HEC-1-A adenocarcinoma endometrial cells fixed with EtOH 70%, PFA 2%, and 3.7% evaluated on at least 6 cells/fixation method. (e) Min-to-max box plot comparing oxygen MFC as obtained by descriptive statistical analysis used to determine the coefficient of variation. A two-way ANOVA with multiple comparisons test was performed to test the statistical difference among the fixation methods. *** p < 0.001. The STXM images (Abs and PhC) and XRF images were acquired at 1500 eV excitation energy with a step size of 200 nm and 800 nm, respectively, and an acquisition time of 10 ms/pixel and 6 s/pixel, respectively.
2.1.2. AFM
AFM microscopy was performed prior to XRF measurements on all cell lines to investigate the effects of the fixation procedure on cell morphology and volume changes. The AFM images of HEC-1-A endometrial cells fixed by the different methods and grown on Si3N4 membranes are shown in Figure 3. The three representative cell groups selected in Figure 2 are shown in the first panels of Figure 3a (EtOH), Figure 3b (PFA 2%), and Figure 3c (PFA 3.7%), together with other cells prepared with the same fixation method not shown in Figure 2 for spatial constraints. The AFM images confirm that there is no significant difference in cell morphology between the PFA 2% and PFA 3.7% protocols. Cells fixed with ethanol (Figure 3a) appear emptier and less compact, with the membrane edges tending to become less sharp and less defined. This is in agreement with what has been previously highlighted by comparing the differential phase contrast images of Figure 2.
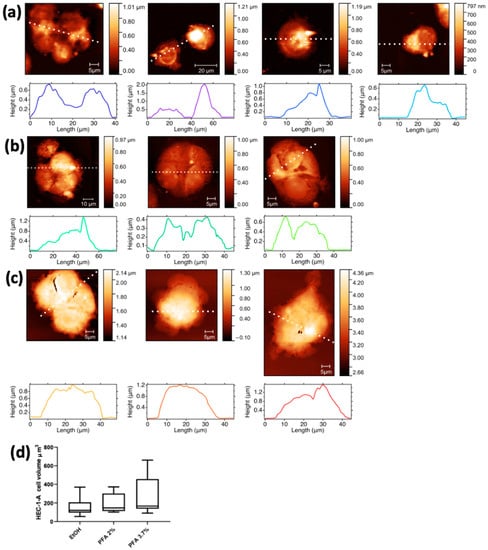
Figure 3.
AFM images with corresponding surface profiles of the HEC-1A endometrial cells. The first column shows the same cells depicted in Figure 2, fixed with different methods: (a) EtOH, (b) PFA 2%, and (c) PFA 3.7%. The other pictures of panels a-c show additional cells prepared with the same three fixation methods. (d) Min-to-max box plot of HEC-1-A cell volume (µm3).
Cells in PFA 3.7% have a higher mean volume (264.1 ± 212.3) overall compared to PFA 2% (196.0 ± 104.8) and EtOH 70% (163.3 ± 104.8); although the three values are comparable within their uncertainties, there is a progressive reduction of the cell volume from PFA 3.7% to EtOH.
2.2. Spermatozoa
2.2.1. XRFM Analysis
Semen was collected from three healthy volunteer donors. Fixation with PFA 2% (Figure 4b) and 3.7% (Figure 4c) does not alter the morphological phenotype of the spermatozoa, which is characterised by a typically oval head and regular shape of the neck, midpiece, and tail. In contrast, when fixed with ethanol 70%, they appear dehydrated, and the tails are strongly curved (Figure 4a). Figure 4d shows the average counts of O, Na, and Mg on the heads of the spermatozoa for each of the preparations (values are normalised for the number of analysed spermatozoa). Fixation with PFA 3.7% can maintain a higher amount of Na and Mg (Figure 4c) compared to ethanol (Figure 4a) and PFA 2% (Figure 4b) fixations. Due to the large variability in the sample under study, ethanol and PFA 2% do not really show statistically significant differences compared with each other. Figure 4e shows the distribution of sperm oxygen levels; in this case, the coefficient of variation for PFA 3.7% (17.14%) is higher than for EtOH 70% (10.51%) and PFA 2% (7.1%), respectively. Spermatozoa XRF data show a trend which can be considered in agreement with that shown for HEC-1-A cells concerning PFA 3.7% preparation. However, a lower reproducibility level is observed, with high values of uncertainties that can be attributed to the peculiar feature of this cell type; indeed, we found a high degree of variability between different donors and sometimes between cells from the same donor (see Table S2. Spermatozoa descriptive analysis).
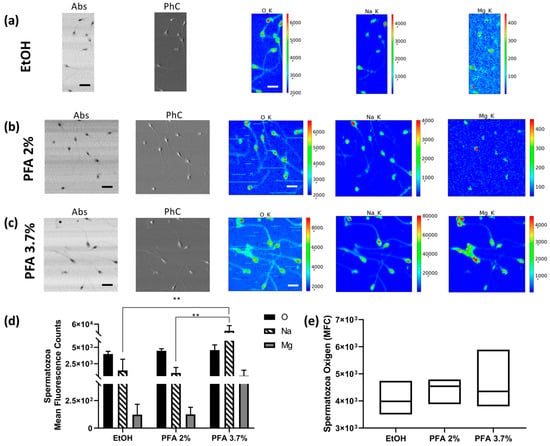
Figure 4.
STXM and XRF analysis of human spermatozoa. Absorption (Abs) and phase contrast (PhC) images, together with O, Na and Mg maps of spermatozoa fixed with different methods: (a) EtOH 70% (mapped area 30 × 75 µm2), (b) PFA 2% (mapped area 70 × 70 µm2), and (c) PFA 3.7% (mapped area 70 × 70 µm2). Scale bars are 10 µm. (d) Normalised average XRF counts of O, Na, and Mg in spermatozoa fixed with EtOH 70%, PFA 2%, and 3.7% evaluated on at least 6–10 cells/patient/fixation method. (e) Min-to-max box plot comparing oxygen MFC as obtained by descriptive statistical analysis used to determine the coefficient of variation. A two-way ANOVA with multiple comparisons test was performed to test the statistical difference among the fixation methods. ** p < 0.005. The STXM images (Abs and PhC) and XRF images were acquired at 1500 eV excitation energy with a step size of 400 nm and 1 μm, respectively, and an acquisition time of 5 ms/pixel and 10 s/pixel, respectively.
2.2.2. AFM
Figure 5 shows the AFM results for the same spermatozoa groups shown in Figure 4. The mean values of cell volume are comparable for the PFA 3.7% and PFA 2% fixatives, while a significantly lower volume is calculated for the cells fixed with ethanol (Figure 5d).
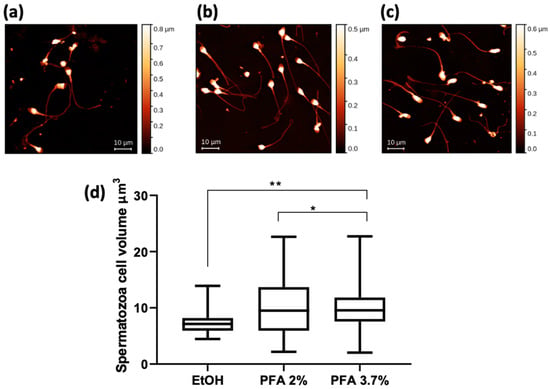
Figure 5.
AFM images of human spermatozoa fixed with different methods: (a) EtOH, (b) PFA 2%, and (c) PFA 3.7%. Scale bar is 10 µm. (d) Spermatozoa cell volume data expressed as a min-to-max box plot. A one-way ANOVA with multiple comparisons test was performed to test the statistical difference among the mean volumes obtained after fixations. ** p < 0.005 * p < 0.05.
2.3. HEK-293
2.3.1. XRFM Analysis
The HEK-293 cell line was also fixed with PFA 2%, 3.7%, and EtOH. Moreover, additional fixation methods were tested on the HEK-293 cell line, as shown in Figure 1 and described in Section 4. All XRF data are summarised in Figure 6. The PFA 3.7% protocol again proves to be a valid preparation method that appropriately preserves all metals and “living elements”. Although the ethanol method does not seem effective in preserving the Mg content in the cells, the Na XRF intensity is well-preserved (Figure 6a). Interestingly, the methanol/C3H6O and cryofixation methods show the highest Mg XRF counts in the cells, followed by PFA 3.7%, while PFA 2% and ethanol favour the loss of Mg XRF intensity. As for Na, methanol/C3H6O has the highest level, followed by PFA 3.7%, while cryofixation, EtOH, and PFA 2% show acceptable levels, although lower (Figure 6f). The washing processes increase the depletion of Na and Mg (see Figure S1). It is worth noting that the overall average O XRF counts are less affected by the preparation compared to Na and Mg and can be considered as a reference for comparing the possible Na and Mg depletions among the different fixation methods. From the absorption images and elemental distribution maps, it is possible to appreciate how fixation with ethanol and 3.7% PFA give similar results, while the absorption image of the cell fixed with 2% PFA is blurred, indicating partial degradation of the sample. Although the Na and Mg XRF intensity seems to be less affected by methanol/C3H6O and cryofixation, Figure 6d shows how methanol/ C3H6O fixation increases the precipitation of Na aggregates on the cells, which is visible in both the absorption and phase contrast images and even more so in the Na-XRF map; this is probably due to a reaction between the solvent and the PBS buffer used to wash the cells. This artefact accounts for the highest value of Na for methanol/C3H6O fixed cells. Cells that have been fixed with methanol/ C3H6O and then washed do not show the same Na clusters (Figure S1), but the cells appear to be “depleted”, and the total amounts of O, Na, and Mg decrease significantly. Similar considerations apply when comparing ethanol fixation with the same fixation followed by subsequent washing (see Supplementary Materials). The PFA 2% protocol achieves similar results to the ethanol protocol concerning the O, Na, and Mg XRF counts, but the absorption and phase contrast images clearly show impaired cell morphology. The coefficient of variation of oxygen, used as the dispersion parameter of the XRF measurement, changes between the different methods, with very high values for the cryofixed cells and the ethanol-treated cells. At the same time, the other protocols give relatively low values of CV (see Table S3. HEK-293 descriptive analysis).
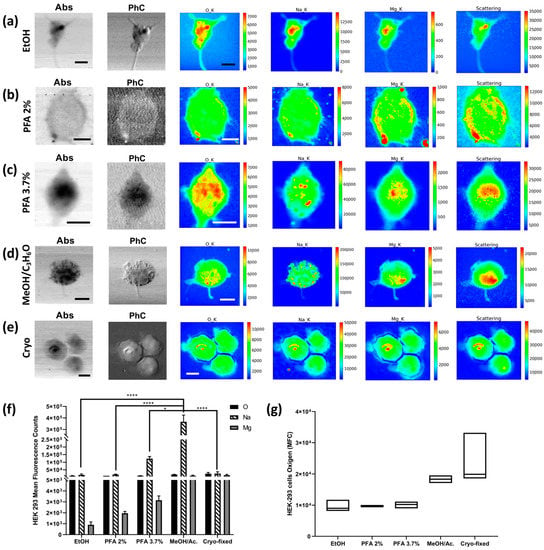
Figure 6.
STXM and XRF analysis of HEK-293 cells. Absorption (Abs) and phase contrast (PhC) images, together with O, Na, Mg, and Scattering maps of HEK-293 cells fixed with different methods: (a) EtOH 70% (mapped area 45 × 45 µm2), (b) PFA 2% (mapped area 34 × 34 µm2), (c) PFA 3.7% (mapped area 25 × 30 µm2), (d) 1:1 MeOH/C3H6O (mapped area 40 × 40 µm2), and (e) cryofixed (mapped area 50 × 50 µm2). Scale bars are 10 µm. (f) Normalised average X-ray fluorescence counts of O, Na, and Mg in HEK-293. A two-way ANOVA with multiple comparisons test was performed on at least on 4 cells/fixation method to test the statistical difference among the fixation methods. * p < 0.05, **** p < 0.0005. (g) Min-to-max box plot comparing oxygen MFC as obtained by descriptive statistical analysis used to determine the coefficient of variation. The STXM images (Abs and PhC) and XRF images were acquired at 1500 eV excitation energy with a step size of 500 nm and an acquisition time of 33 ms/pixel and 6 s/pixel, respectively.
2.3.2. AFM
Interestingly, AFM images of methanol/C3H6O preparation (Figure 7d) show that the morphology of HEK-293 cells is significantly altered, with submicrometric features appearing like clumps on their surface. As mentioned earlier, according to the XRF maps, these features appear to be closely associated with Na nanoclusters. When prepared with ethanol (Figure 7a), the morphology is better preserved, although the cells appear somewhat depleted. This is even more pronounced in the cells fixed with PFA 2% (Figure 7b), as their overall height appears reduced compared to methanol and ethanol fixation; moreover, the cells prepared with PFA 2% have visible nanometric holes on their surface, clearly indicating some stress. In contrast, cells prepared with PFA 3.7% (Figure 7c) appear more filled, as evidenced by their height, and have a smoother surface; overall, they appear healthier and with a better-preserved morphology. AFM images with corresponding surface profiles collected on a selection of (a) 1:1 MeOH/C3H6O washed cells and of (b) 70% ethanol washed cells are reported in Supporting Materials in Figure S2.
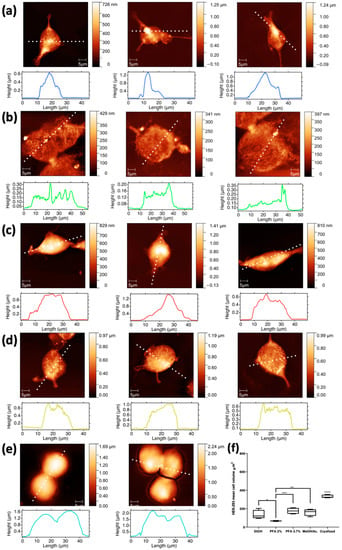
Figure 7.
AFM images with corresponding surface profiles collected on a selection of HEK-293 cells fixed with the different methods: (a) EtOH 70%, (b) PFA 2%, (c) PFA 3.7%, (d) 1:1 MeOH/C3H6O, and (e) cryofixed. Scale bar is 5 µm. Cell volume calculated from the AFM images collected on a selection of HEK-293 cells fixed with ethanol and PFA 3.7% and compared with the rest of the applied fixation methods (f). The second column shows the same cells depicted in Figure 6 (where a, b and e are rotated of 90 degrees left, 90 degrees right and 180 degrees respectively compared to Figure 6), while the other columns depict additional cells prepared with the same five fixation methods. * p < 0.05, ** p < 0.005, *** p < 0.001, **** p < 0.0005.
3. Discussion
XRF analysis is a powerful, non-destructive, analytical technique with wide application in biomedical science, allowing the chemical characterisation, with good detection limit and high spatial resolution, of trace elements that may play an essential role in cellular biochemical processes. We have recently presented some important results on cells derived from reproductive tissues, observing Na, Cu, and Mg distribution in spermatozoa [21] and ovarian tissues [22]. In another work, we also demonstrated that it is possible to merge compositional and morphological information to quantitatively derive the element concentration by combining XRFM with AFM and STXM analyses [23]. The faithfulness of XRF measurements is highly dependent on the proper preparation of the samples, which is indeed a critical step in obtaining the appropriate information; it requires careful attention in order to preserve the cellular morphology and the elemental localisation inside the cells.
The present study illustrates the effect of several cell fixation methods in single-cell analyses on the localisation and intracellular levels of light elements by synchrotron radiation XRF. As already reported, the most affected elements are the most diffusible ones, namely the light ones, such as Na, Mg, K, Cl, and Ca [15,16]. However, in the available literature, there is a lack of a comprehensive evaluation of the morphological changes correlated to elemental preservation or possible losses after fixation.
We focused on analysing the impact of preparation techniques on the determination of “life elements” O, Na, and Mg inside the cells, using the TwinMic soft X-ray microscope, which also allows for morphological investigations through high-resolution (sub-micrometric length scale) absorption and phase contrast images [23]. Even if the concentration of intracellular elements inside the cells cannot be represented by XRF counts nor MFC parameter alone, the present study aims at illustrating the effect of different fixation methods on cell morphology and elemental distribution. Oxygen XRF counts can be used as a reference to compare Na and Mg changes, while AFM microscopy performed prior to X-ray analyses indeed enabled us to examine at nanometric resolution the changes in cell volumes and morphology induced by the different fixation methods.
In order to provide a more systematic and comprehensive understanding, we decided to compare different fixation protocols on three cell models. Cell fixation methods were chosen accordingly to the following motives: (i) cross-linker, (ii) organic solvent, and (iii) frozen methods. Usually, chemical fixation preserves structures in a state (both chemically and structurally) as close to living tissue as possible by stabilising the proteins, nucleic acids, and other macromolecules. For instance, PFA 3.7% fixation is among the most used for X-ray microscopy and immune- and histochemistry analyses, having the ability to permeabilise cells, in addition to a quick fixation period. Different chemical fixatives are characterised by different permeabilisation grades, reflecting anomalous elemental diffusion from the cells. For this reason, we tested a few examples of chemical fixatives: PFA 3.7%, PFA 2%, ethanol 70%, and 1:1 methanol/ C3H6O. While the first three methods were applied to all cell models, methanol/ C3H6O and cryofixation were used only for the HEK-293 cells, which are highly reproducible, thus allowing for a more comprehensive analysis.
Our results confirm that the PFA 3.7% protocol is an excellent compromise to preserve both morphological features, including cell volume, and light element content to a reasonable extent. The findings align with Perrin et al. [15], confirming that Na XRF intensity is well-preserved in PFA 3.7% treated cells and even better for the MeOH/C3H6O protocol. However, when investigating at a submicrometric length scale, our results demonstrate that the high Na levels after methanol fixation are due to nanometric Na agglomerates or precipitates onto the cells (Figure S1), thus excluding this fixative since it introduces evident artefacts. Moreover, our results evidence that the fixations with organic solvents greatly affect the morphology or the volume, probably due to their dehydrating effect.
While we still observe significant Mg levels for both PFA 3.7% and MeOH/C3H6O fixations, for Perrin et al. [15] its elemental content seems to be reduced below the detection limits in both cases. PFA 2% does not show any advantage compared to PFA 3.7%, as the cells treated with this method usually present a lower cell volume and a lower XRF counts of Mg. Furthermore, we compared those results with ethanol fixation, which is a frequently used protocol, highlighting good preservation of Na levels, but not of Mg and cellular volume. All the observations mentioned above concerning PFA 3.7%, PFA 2%, and ethanol effects on elemental XRF intensities, and thus on elemental content, are substantially confirmed in all cell models used in this study (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7), providing a good consistency to our results. However, AFM analyses revealed that the way PFA 2% affects the three cell models differs in terms of cell volume, with HEC-1-A endometrial cells the least affected (Figure 3). Spermatozoa cells show the highest variability in cell volume modifications when fixed with PFA 2%, suggesting that the cell dimension, which in this case is highly variable, may affect the final deformation. The maximal effect for the HEK-293 embryonic line is likely due to their undifferentiated phenotype, as seen in Figure 7b, where the PFA 2% fixed cells seem to collapse and cover a wider surface. In addition, they present irregular cellular profiles. Hence, too mild of a treatment with PFA is insufficient to cross-link the structural cell component, thus resulting in rapid degradation.
Our data with HEK-293 cells suggest cryofixation as the best method, as it ensures the highest cell volume, very likely the closest to the physiological one. Moreover, in agreement with Perrin et al. [15], it maintains a high level of Mg while inducing a moderate loss of Na (Figure 6e). However, cryofixation with freeze-drying is a sensitive step because biological samples usually contain a high percentage of water. If not properly performed, it can lead to elemental redistribution, loss of elements, shrinking of the specimens, and severe morphology distortion [24].
Cryofixation provides the most truthful representation of the cellular biological structure; however, a limit is the lower repeatability compared to other protocols, as shown by the measurement of the oxygen coefficient of variation (Figure 6g). This is probably due to an intrinsic technical difficulty of operating in a standardised way [25]. To summarise, PFA fixation at 3.7% is the recommended protocol in the absence of cryofixation, even though our results and other ones from literature [15,16,26] suggest that all chemical fixatives affect some aspects of the measurement.
Finally, attention must be paid to water washing after fixation since our results demonstrate that it has some influence on elemental XRF counts, as organic fixatives are known to perforate cell membranes, inducing a release of elemental content outside the cell.
The main analytical constraints of the methods under study are summarised in Table 1 according to our study and the literature [15,16,26].

Table 1.
Summary of the main analytical constraints of the fixation methods under study for synchrotron X-ray microscopy.
To note, the study was performed on a limited population of cells due to two main factors: (i) access to a synchrotron facility is granted only for scientific merit, assessed by a peer review process carried out every 6 months; (ii) low-energy XRF is not an efficient phenomenon, as below 2 keV the Auger effect is the dominant process, which results in a low florescence yield and thus long measurement times. Despite these intrinsic limitations, the reported results still provide very useful information to the X-ray microscopy and XRF community, and further complement and confirm the results available in literature, which suffer as well from similar constraints [15,16].
4. Materials and Methods
Silicon nitride membranes Si3N4 (100 nm thick; Silson, Southam, UK) were used as support for both XRFM and AFM analyses for all the specimens under study. Si3N4 were previously sterilised under a laminar flow hood with EtOH 70%, which was left to evaporate overnight in a petri dish.
4.1. Cellular Models
4.1.1. HEC-1-A Endometrial Cells
HEC-1-A endometrial cells from human adenocarcinoma (ATCC HTB-112) were cultured in McCoy’s 5a Medium supplemented with 10% foetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin (Sigma Aldrich, Milan, Italy). For the experiments 50.000 cells were seeded in 12 multiwell plates containing silicon nitride membranes (100 nm thick; Silson, Southam, UK). After 24 h, the cells were fixed with three different protocols:
- Paraformaldehyde (PFA) 3.7% in phosphate buffer saline (PBS) for 20 min at room temperature
- PFA 2% for 2 min at room temperature
- 70% ice-cold ethanol for 3 min at −20 °C
Then the membranes were washed twice in PBS and twice in distilled water and air-dried.
4.1.2. Spermatozoa
Three fresh semen samples were collected from European Caucasian male volunteer donors enrolled at the assisted procreation unit (Institute for Maternal and Child Health IRCCS Burlo Garofolo, Trieste, Italy). All participants signed an informed consent. All experimental procedures were conducted according to the ethical standards of the Declaration of Helsinki (7th version 2013); the study was approved by the regional (FVG) Ethics Committee (Comitato Etico Unico Regionale FVG) (CEUR-2019-Os-191). The specimens were collected via masturbation after 2–7 days of sexual abstinence. After liquefaction, the samples were observed under an optic microscope and they presented normal parameters in terms of concentration and motility (according to the World Health Organization guidelines) [27].
The semen from each donor were divided in 3 groups and fixed with:
- PFA 3.7% for 20 min at room temperature
- PFA 2% for 2 min at room temperature
- 70% ice-cold ethanol for 3 min at −20 °C
Then the samples were washed twice in physiological solution (0.9% NaCl in water), and after centrifugation at 300× g for 10 min they were resuspended in physiological solution, deposited on 100 nm thick Si3N4 membranes, and air-dried. The membranes were briefly dipped twice in distilled water and air-dried.
4.1.3. HEK-293 Cell Line
Human Embryonic Kidney 293 cells (HEK-293) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Biochrom, Berlin, Germany) supplemented with 10% foetal bovine serum (FBS; Biochrom, Berlin, Germany) on silicon nitride membranes (100 nm thick) at 37 °C and 5% CO2 atmosphere. Cells were left to attach overnight and washed with phosphate buffer saline (PBS; Euroclone, Milan, Italy). Subsequently, cells were subjected to the following fixation methods:
- PFA 3.7% for 20 min at room temperature
- PFA 2% for 2 min at room temperature
- 70% ice-cold ethanol for 3 min at −20 °C
- 1:1 MeOH/C3H6O for 3 min at −20 °C
Then all membranes were washed twice in PBS and twice in distilled water and air-dried.
- 5.
- Cryofixation: cells were washed with ammonium acetate buffer solution 100 mM pH 7.4 prepared with high purity water (Fisher Scientific™ Accu100 Ultrapure Water System), the excess washing buffer was removed, and then Si3N4 membranes were rinsed in ethane (−160 °C) by using a home-made machine; cells were subsequently freeze-dried by leaving the specimens overnight in a controlled pressure system, allowing the ice to sublimate and slowly reach atmospheric pressure and temperature.
All reagents used for fixation are highly pure ones bought from Sigma Aldrich—Merck (Milano, Italy) (PFA 158127, Ethanol 32221, PBS P4417). The PFA was also filtered with a 0.2 micron filter prior to use to eliminate aggregate samples.
4.2. AFM Microscopy
The AFM measurements were performed at the NanoInnovation Laboratory located at Elettra—Sincrotrone Trieste (Trieste, Italy). Suitable cells, prepared as described in Section 4.1, were selected through visible light microscopy and then mapped with AFM microscopy prior to XRF analysis. This was done in order to avoid any possible artefacts caused by radiation damage induced by soft X-rays. AFM micrographs were acquired in contact mode in air with an XE100 (Park Instruments) or a MFP3D (Asylume Research/Oxford Instruments) instrument. For the acquisition of images, we used soft cantilevers (Mikromasch CSC38, radius of curvature <10 nm, spring constant 0.006 N/m) at a 0.1–0.2 Hz scanning rate and a 80–160 nm pixel resolution. Images were analysed with Gwyddion software [28]; volume extraction was carried out by flattening the image to remove background, selecting the cells by a height-threshold based mask, and measuring the zero-basis volume of the single cells. Graphs of AFM trace profiles were obtained using the Igor Pro software (Wavemetrics, Lake Oswego, OR, USA).
4.3. XRF Microscopy
The XRF measurements were performed at the TwinMic beamline of Elettra—Sincrotrone Trieste (Trieste, Italy) [29]. The TwinMic microscope was operated in scanning transmission mode (STXM), where the specimen is raster-scanned across a microprobe delivered by a zone plate diffractive optic. The transmitted photons are collected through an X-ray-visible light converting system by a fast readout CCD camera providing absorption and differential phase contrast images [30]. Simultaneously, the XRF emitted by the sample is collected by eight SDDs located symmetrically in front of the sample [31,32]. The sample plane is located perpendicularly to the beam axis and the measurements are conducted at room temperature and in high vacuum condition. For the present experiment, an incident energy of 1.5 keV was chosen for optimal excitation of Na and Mg, allowing the detection of O as well. The zone plate optic (600 µm in diameter with an outermost zone width of 50 nm and a central stop of 140 µm) was focusing the incident X-ray photons on the sample on a spot size of 200 nm to 1 µm in diameter, according to the analysed sample and the incident beam intensity available during the experiments. The collected XRF spectra were deconvolved with the PyMCA software package [33]. For endometrial cells, at least three areas were mapped per fixation type, for a total of six analysed cells/fixation method, while for the HEK-293 cell line, at least four cells were mapped per fixation protocols. As far as for the sperm cells, for each of the three donors, a range of six to ten were analysed for each type of fixation.
The presented XRF counts and the relative MFC were calculated as average values on the whole cell.
A comparison about the microscopies used in the manuscript and a summary of a few experimental details are given in Table S5. Please note that in the table we consider generic applications of those microscopies, but we acknowledge that special ones exist as well which may have quite different specifications (i.e., HS-AFM, high energy XRF, fly scan STXM, no-contact AFM, etc.).
4.4. Statistical Analysis
Data are expressed as means ± standard deviation or min-to-max box plots. One- or two-way ANOVA with multiple comparisons tests were carried out for statistical comparisons among the different fixation methods. Descriptive analysis was performed to calculate coefficients of variation (CV). Differences with a p < 0.05 were considered statistically significant. Data analyses were developed on GraphPad Prism 8.4.2 (GraphPad Software Inc., La Jolla, CA, USA).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/molecules28041992/s1: Figure S1: XRF Analysis of washed HEK-293 cells; Figure S2: AFM Analysis of washed HEK-293 cells; Table S1: HEC-1-A descriptive analysis; Table S2: Spermatozoa descriptive analysis; Table S3: HEK-293 descriptive analysis; Table S4: HEK-293 Student’s t-test analysis; Table S5: Comparison between AFM, STXM and LEXRF.
Author Contributions
Conceptualisation, L.M., L.P., L.Z. and A.G.; methodology, A.G., G.K.; software, G.K.; validation, S.K., E.M. and S.I.; formal analysis, G.K., P.P. and D.E.B.; investigation, L.M., P.P., V.B., G.G., S.K. and D.E.B.; resources, A.G.; data curation, G.K.; writing—original draft preparation, L.M., L.P., L.Z. and A.G.; writing—review and editing, L.M., L.P., L.Z., V.B., E.M., S.I. and A.G.; visualization, G.K., V.B. and G.G.; supervision, L.P. and A.G.; funding acquisition, L.P., S.C., G.R. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the Italian Ministry of Health, through the contribution given to the Institute for Maternal and Child Health IRCCS Burlo Garofolo Trieste, Italy (5mille15D1; 5xmille SD03/22). Part of this work has been performed in the frame of the INTEGRA CERIC-ERIC research project.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Institute for Maternal and Child Health IRCCS Burlo Garofolo and by the regional Ethical Committee (Comitato Etico Unico Regionale FVG CEUR-2019-Os-191 22 October 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and/or analysed during the current study are published as FAIR and open on a suitable data repository in Elettra Sincrotrone Trieste [34]. For any additional information, you may contact the corresponding author.
Acknowledgments
We acknowledge Elettra Sincrotrone Trieste (Trieste, Italy) for granting access to its synchrotron radiation facility and the TwinMic beamline (Proposals n° 20160109, 20200410 and 20205077). The authors acknowledge the kind assistance of Stefania Luppi and Monica Martinelli in the preparation of clinical samples.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples are available from the authors.
References
- Vogt, S. Applications of Synchrotron Radiation: Micro Beams in Cell Micro Biology and Medicine. Phys. Today 2008, 61, 53–54. [Google Scholar] [CrossRef]
- Merolle, L.; Ragazzi, M.; Gianoncelli, A.; Altissimo, M.; Ciarrocchi, A.; Bedolla, D.E.; Marraccini, C.; Baricchi, R.; Pertinhez, T.A. Mapping Fundamental Life Elements in Papillary Thyroid Carcinoma Tissue. J. Inst. 2018, 13, C05018. [Google Scholar] [CrossRef]
- Poitry-Yamate, C.; Gianoncelli, A.; Kaulich, B.; Kourousias, G.; Magill, A.W.; Lepore, M.; Gajdosik, V.; Gruetter, R. Feasibility of Direct Mapping of Cerebral Fluorodeoxy-D-Glucose Metabolism in Situ at Subcellular Resolution Using Soft X-ray Fluorescence. J. Neurosci. Res. 2013, 91, 1050–1058. [Google Scholar] [CrossRef]
- Paunesku, T.; Vogt, S.; Maser, J.; Lai, B.; Woloschak, G. X-ray Fluorescence Microprobe Imaging in Biology and Medicine. J. Cell. Biochem. 2006, 99, 1489–1502. [Google Scholar] [CrossRef] [PubMed]
- Paunesku, T.; Wanzer, M.B.; Kirillova, E.N.; Muksinova, K.N.; Revina, V.S.; Romanov, S.A.; Lyubchansky, E.R.; Grosche, B.; Birschwilks, M.; Vogt, S.; et al. X-ray Fluorescence Microscopy for Investigation of Archival Tissues. Health Phys 2012, 103, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Merolle, L.; Sponder, G.; Sargenti, A.; Mastrototaro, L.; Cappadone, C.; Farruggia, G.; Procopio, A.; Malucelli, E.; Parisse, P.; Gianoncelli, A.; et al. Overexpression of the Mitochondrial Mg Channel MRS2 Increases Total Cellular Mg Concentration and Influences Sensitivity to Apoptosis. Metallomics 2018, 10, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Malucelli, E.; Procopio, A.; Fratini, M.; Gianoncelli, A.; Notargiacomo, A.; Merolle, L.; Sargenti, A.; Castiglioni, S.; Cappadone, C.; Farruggia, G.; et al. Single Cell versus Large Population Analysis: Cell Variability in Elemental Intracellular Concentration and Distribution. Anal. Bioanal. Chem. 2018, 410, 337–348. [Google Scholar] [CrossRef]
- Zeb, Q.; Wang, C.; Shafiq, S.; Liu, L. Chapter 6—An Overview of Single-Cell Isolation Techniques. In Single-Cell Omics; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 101–135. ISBN 978-0-12-814919-5. [Google Scholar]
- Weinhardt, V.; Chen, J.-H.; Ekman, A.; McDermott, G.; Le Gros, M.A.; Larabell, C. Imaging Cell Morphology and Physiology Using X-rays. Biochem Soc Trans 2019, 47, 489–508. [Google Scholar] [CrossRef]
- Marguí, E.; Queralt, I.; Van Grieken, R. Sample Preparation for X-ray Fluorescence Analysis. In Encyclopedia of Analytical Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 1–25. ISBN 978-0-470-02731-8. [Google Scholar]
- Histotechnology: A Self-Instructional Text 5th Edition. Available online: https://store.ascp.org/productlisting/productdetail?productId=97915824 (accessed on 23 June 2022).
- Hobro, A.J.; Smith, N.I. An Evaluation of Fixation Methods: Spatial and Compositional Cellular Changes Observed by Raman Imaging. Vib. Spectrosc. 2017, 91, 31–45. [Google Scholar] [CrossRef]
- Rodig, S.J. Fixing Attached Cells for Staining. Cold Spring Harb. Protoc. 2020, 2020, 099689. [Google Scholar] [CrossRef]
- Amporndanai, K. Biological Specimens Preparation for X-ray Microscopy. 2014. Available online: https://www.desy.de/f/students/2014/reports/amporndanai_kangsa.pdf (accessed on 27 October 2022).
- Perrin, L.; Carmona, A.; Roudeau, S.; Ortega, R. Evaluation of Sample Preparation Methods for Single Cell Quantitative Elemental Imaging Using Proton or Synchrotron Radiation Focused Beams. J. Anal. At. Spectrom. 2015, 30, 2525–2532. [Google Scholar] [CrossRef]
- Jin, Q.; Paunesku, T.; Lai, B.; Gleber, S.-C.; Chen, S.I.; Finney, L.; Vine, D.; Vogt, S.; Woloschak, G.; Jacobsen, C. Preserving Elemental Content in Adherent Mammalian Cells for Analysis by Synchrotron-Based x-Ray Fluorescence Microscopy. J. Microsc. 2017, 265, 81–93. [Google Scholar] [CrossRef]
- Osanna, A.; Jacobsen, C.; Kalinovsky, A.; Kirz, J.; Maser, J.; Wang, S. X-ray Microscopy: Preparations for Studies of Frozen Hydrated Specimens. Scanning Microsc. Suppl. 1996, 10, 349–356, discussion 356–358. [Google Scholar]
- Gianoncelli, A.; Vaccari, L.; Kourousias, G.; Cassese, D.; Bedolla, D.E.; Kenig, S.; Storici, P.; Lazzarino, M.; Kiskinova, M. Soft X-ray Microscopy Radiation Damage On Fixed Cells Investigated With Synchrotron Radiation FTIR Microscopy. Sci. Rep. 2015, 5, 10250. [Google Scholar] [CrossRef]
- Bedolla, D.E.; Mantuano, A.; Pickler, A.; Mota, C.L.; Braz, D.; Salata, C.; Almeida, C.E.; Birarda, G.; Vaccari, L.; Barroso, R.C.; et al. Effects of Soft X-ray Radiation Damage on Paraffin-Embedded Rat Tissues Supported on Ultralene: A Chemical Perspective. J. Synchrotron Radiat. 2018, 25, 848–856. [Google Scholar] [CrossRef]
- Surowka, A.D.; Gianoncelli, A.; Birarda, G.; Sala, S.; Cefarin, N.; Matruglio, A.; Szczerbowska-Boruchowska, M.; Ziomber-Lisiak, A.; Vaccari, L. Soft X-ray Induced Radiation Damage in Thin Freeze-Dried Brain Samples Studied by FTIR Microscopy. J. Synchrotron Radiat. 2020, 27, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Zupin, L.; Pascolo, L.; Gianoncelli, A.; Gariani, G.; Luppi, S.; Giolo, E.; Ottaviani, G.; Crovella, S.; Ricci, G. Synchrotron Radiation Soft X-ray Microscopy and Low Energy X-ray Fluorescence to Reveal Elemental Changes in Spermatozoa Treated with Photobiomodulation Therapy. Anal. Methods 2020, 12, 3691–3696. [Google Scholar] [CrossRef]
- Pascolo, L.; Venturin, I.; Gianoncelli, A.; Bortul, R.; Zito, G.; Giolo, E.; Salomé, M.; Bedolla, D.E.; Altissimo, M.; Zweyer, M.; et al. Light Element Distribution in Fresh and Frozen–Thawed Human Ovarian Tissues: A Preliminary Study. Reprod. BioMed. Online 2018, 37, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Gianoncelli, A.; Bonanni, V.; Gariani, G.; Guzzi, F.; Pascolo, L.; Borghes, R.; Billè, F.; Kourousias, G. Soft X-ray Microscopy Techniques for Medical and Biological Imaging at TwinMic—Elettra. Appl. Sci. 2021, 11, 7216. [Google Scholar] [CrossRef]
- Castillo-Michel, H.A.; Larue, C.; del Real, A.P.; Cotte, M.; Sarret, G. Practical review on the use of synchrotron based micro- and nano- X-ray fluorescence mapping and X-ray absorption spectroscopy to investigate the interactions between plants and engineered nanomaterials. Plant Physiol. Biochem. 2017, 110, 13–32. [Google Scholar] [CrossRef]
- Pushie, M.J.; Pickering, I.J.; Korbas, M.; Hackett, M.J.; George, G.N. Elemental and Chemically Specific X-ray Fluorescence Imaging of Biological Systems. Chem. Rev. 2014, 114, 8499–8541. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Shimura, M.; Fujii, M.; Maeshima, K.; Yumoto, H.; Mimura, H.; Sano, Y.; Yabashi, M.; Nishino, Y.; Tamasaku, K.; et al. Elemental mapping of frozen-hydrated cells with cryo-scanning X-ray fluorescence microscopy. X-ray Spectrom. 2010, 39, 260–266. [Google Scholar] [CrossRef]
- WHO|WHO Manual for the Standardized Investigation, Diagnosis and Management of the Infertile Male. Available online: https://www.who.int/reproductivehealth/publications/infertility/0521774748/en/ (accessed on 11 May 2020).
- Nečas, D.; Klapetek, P. Gwyddion: An Open-Source Software for SPM Data Analysis. Open Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Kourousias, G.; Merolle, L.; Altissimo, M.; Bianco, A. Current Status of the TwinMic Beamline at Elettra: A Soft X-ray Transmission and Emission Microscopy Station. J. Synchrotron Radiat. 2016, 23, 1526–1537. [Google Scholar] [CrossRef] [PubMed]
- Gianoncelli, A.; Morrison, G.R.; Kaulich, B.; Bacescu, D.; Kovac, J. Scanning Transmission X-ray Microscopy with a Configurable Detector. Appl. Phys. Lett. 2006, 89, 251117. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Kaulich, B.; Alberti, R.; Klatka, T.; Longoni, A.; De Marco, A.; Marcello, A.; Kiskinova, M. Simultaneous Soft X-ray Transmission and Emission Microscopy. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2009, 608, 195–198. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Kourousias, G.; Stolfa, A.; Kaulich, B. Recent Developments at the TwinMic Beamline at ELETTRA: An 8 SDD Detector Setup for Low Energy X-ray Fluorescence. J. Phys. Conf. Ser. 2013, 425, 182001. [Google Scholar] [CrossRef]
- Sole, A.; Papillon, E.; Cotte, M.; Walter, P.; Susini, J. A Multiplatform Code for the Analysis of Energy-Dispersive X-ray Fluorescence Spectra. Spectrochim. Acta Part B At. Spectrosc. 2007, 62, 63–68. [Google Scholar] [CrossRef]
- Kourousias, G.; Bedolla Orozco, D.E.; Bonanni, V.; Gariani, G.; Gianoncelli, A.; Iotti, S.; Kenig, S.; Malucelli, E.; Merolle, L.; Parisse, P.; et al. Manuscript Dataset Open and FAIR 2022; Elettra-Sincrotrone Trieste: Trieste, Italy, 2022. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).