A Natural Glucan from Black Bean Inhibits Cancer Cell Proliferation via PI3K-Akt and MAPK Pathway
Abstract
1. Introduction
2. Results
2.1. Purification and Component Identification of Black Bean Polysaccharide
2.1.1. Extraction and Purification of the Polysaccharide from Black Bean
2.1.2. Molecular Weight Determination
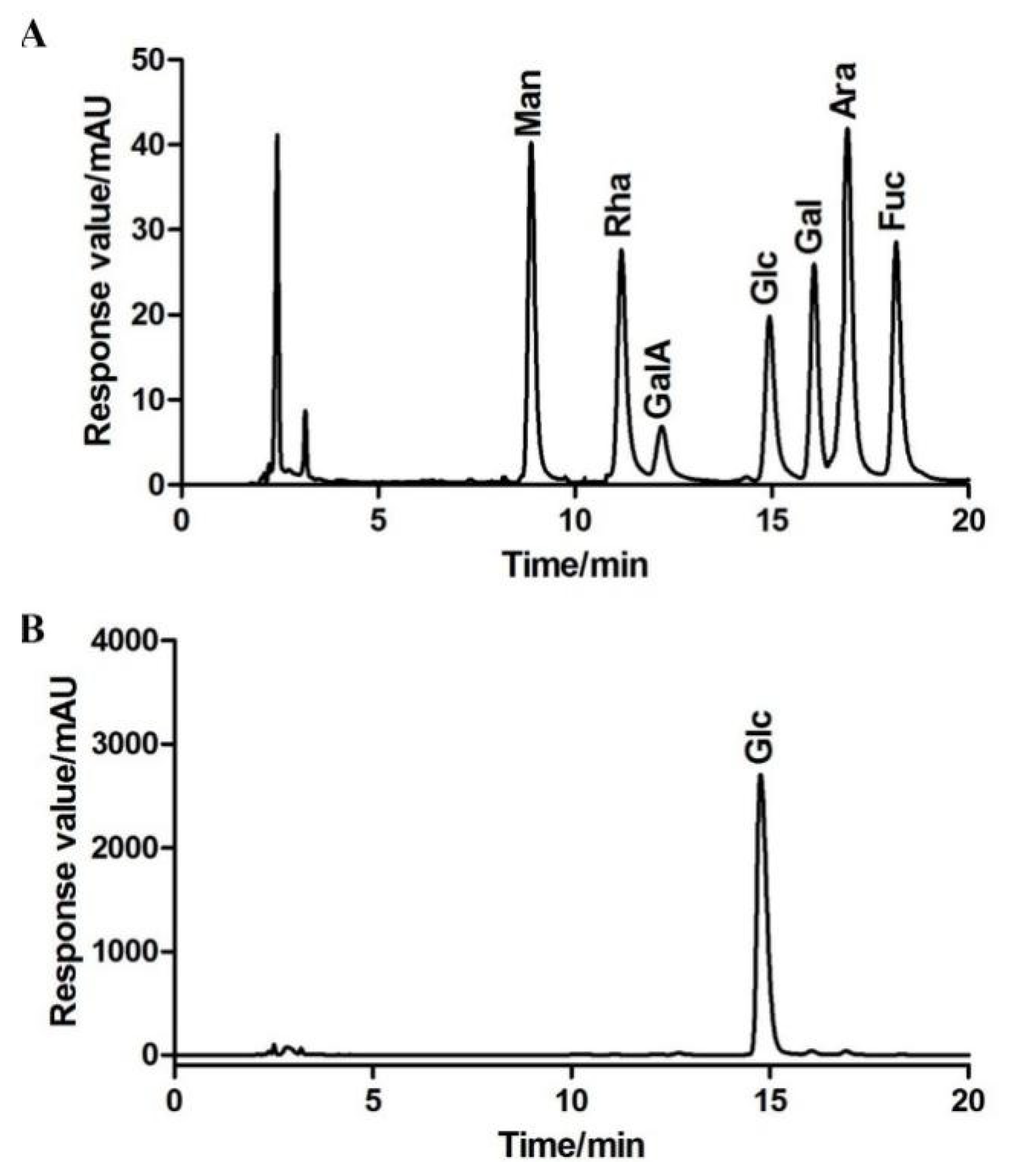
2.1.3. Monosaccharide Composition of BBWPW
2.2. Structural Characterization of Black Bean Polysaccharide
2.2.1. The Surface Morphology of BBWPW
2.2.2. Infrared Spectra Analysis
2.2.3. NMR Analysis
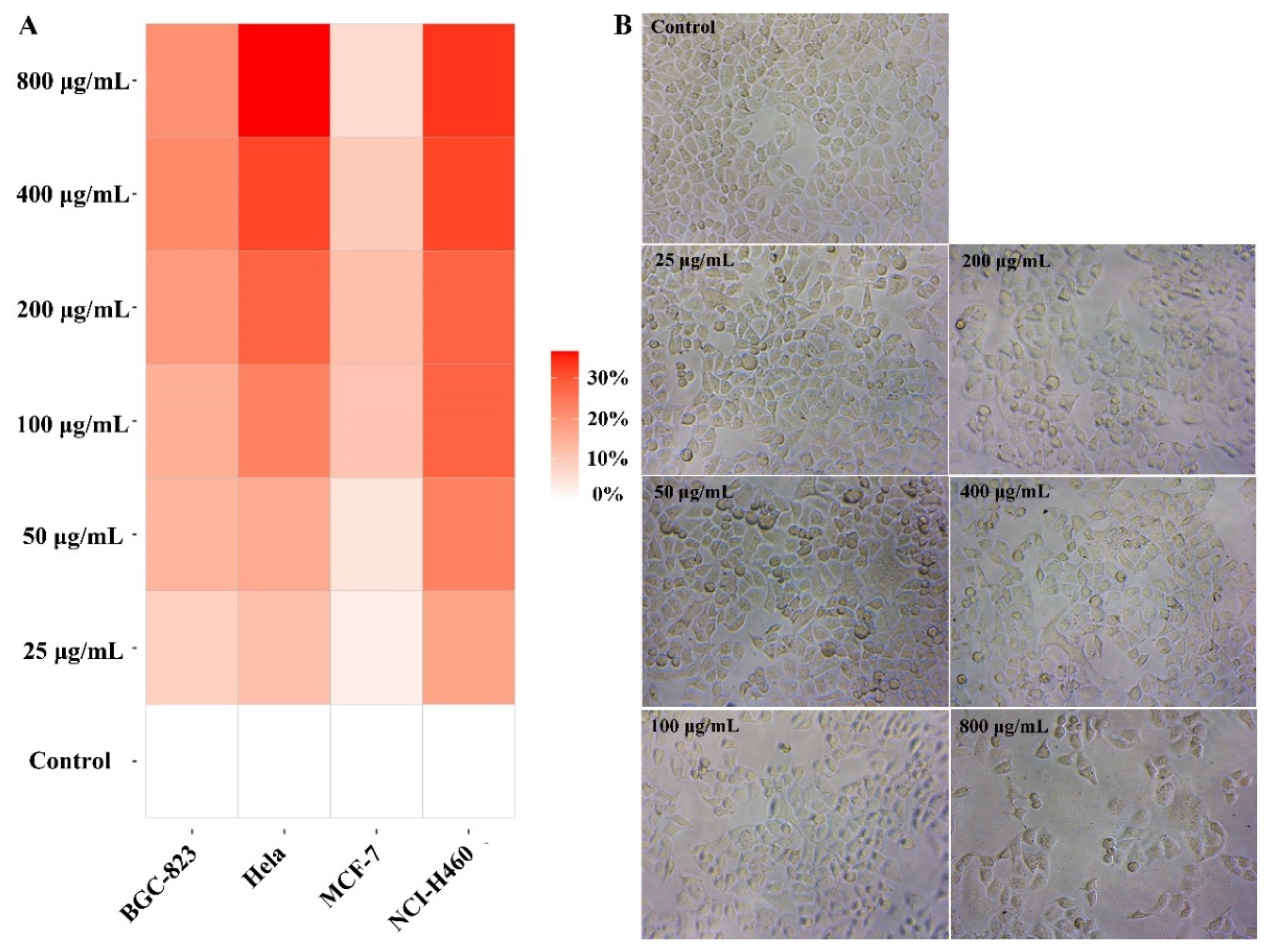
2.3. BBWPW Inhibits the Proliferation of Cancer Cells
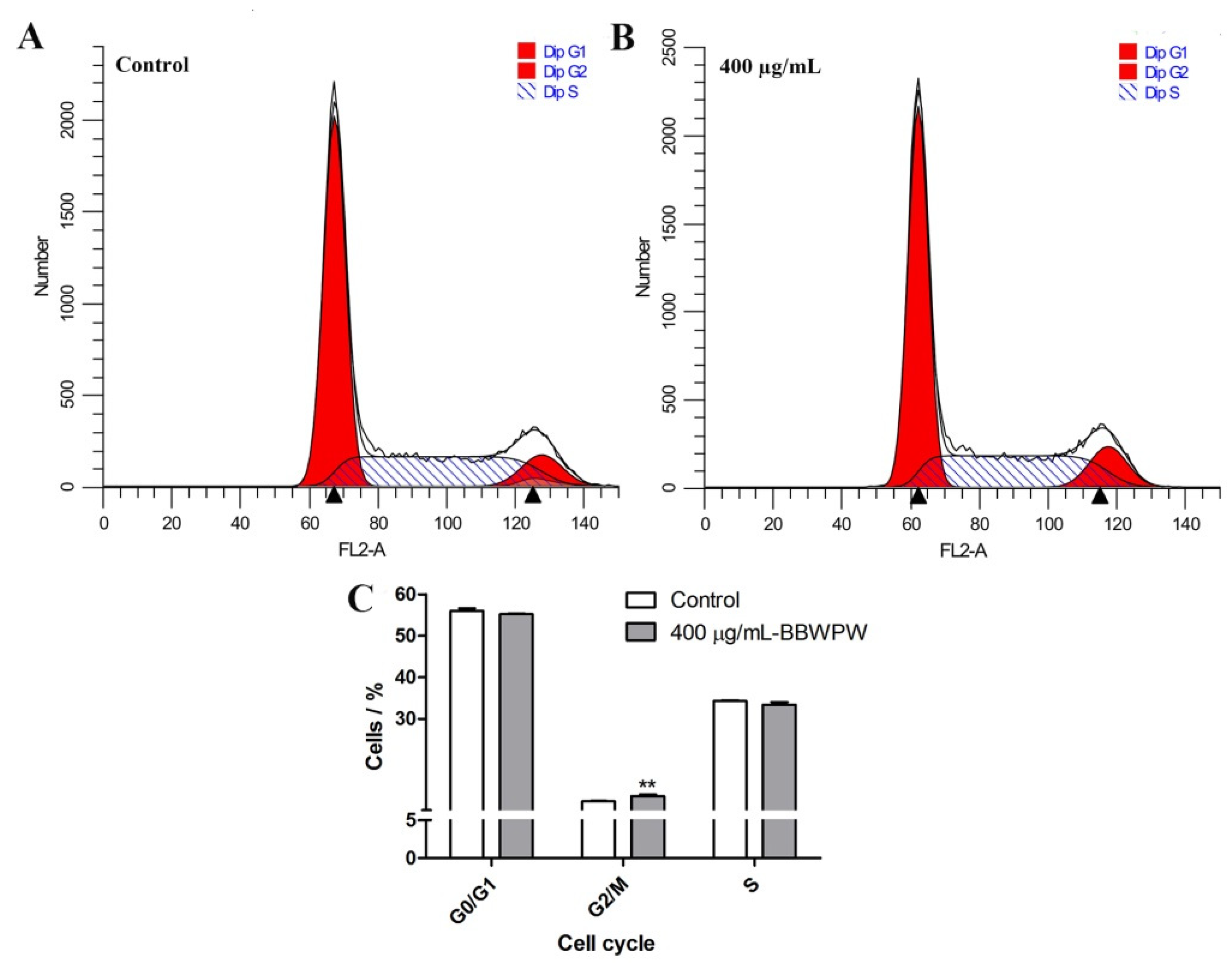
2.4. BBWPW Induces HeLa Cell Arrest in the G2/M Phase
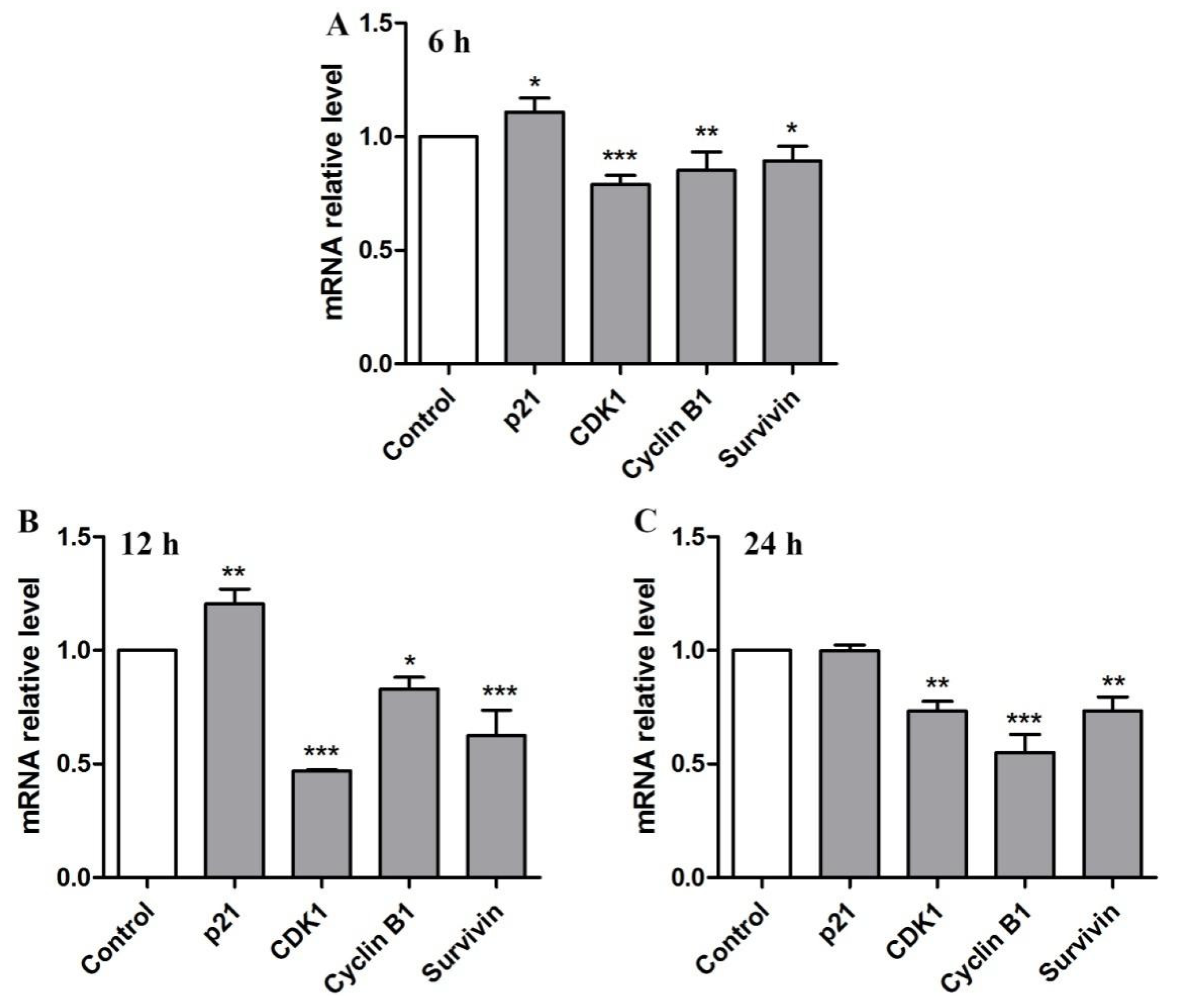
2.5. Quantifying Analysis of RNA-Seq Data
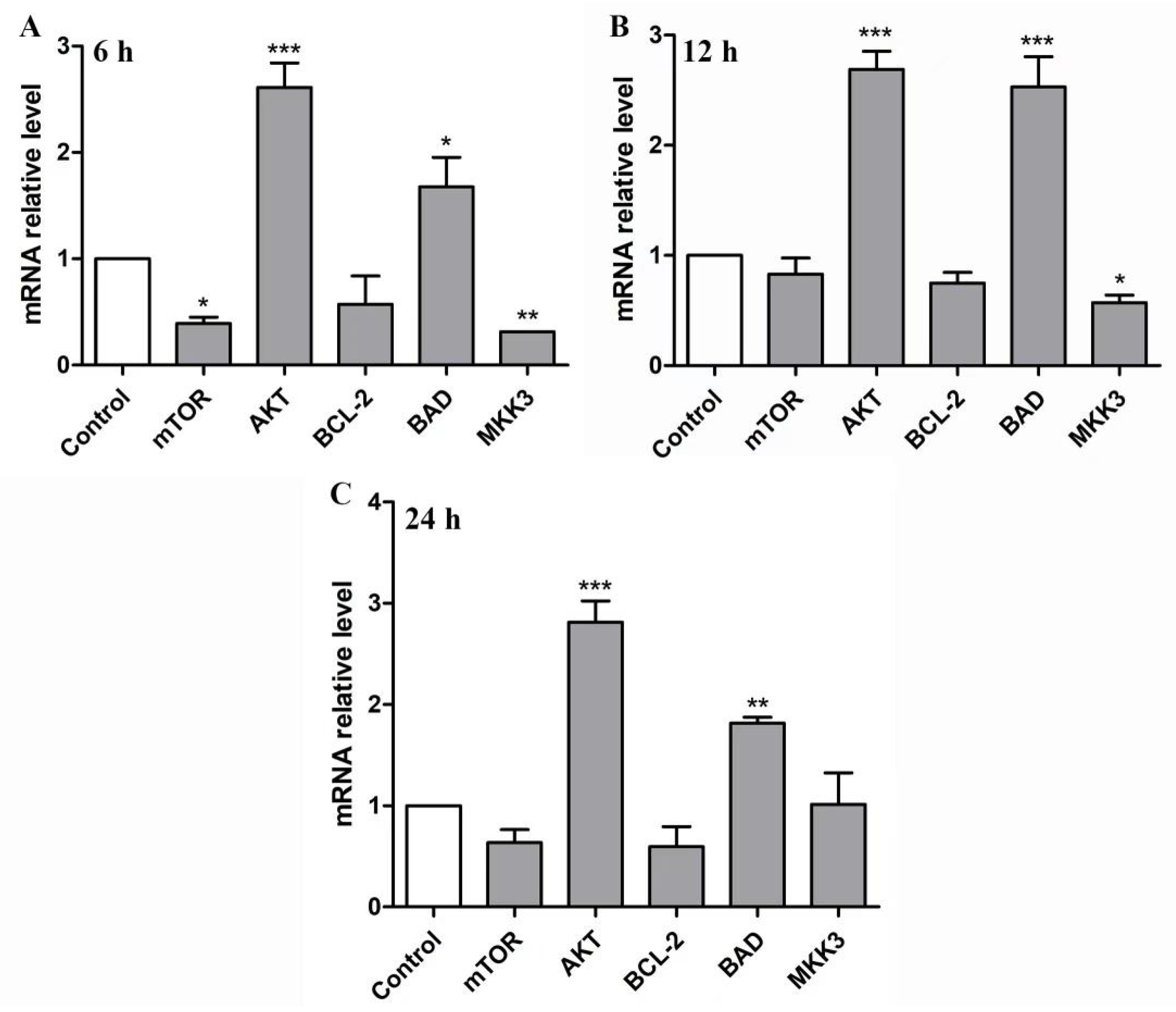
2.6. BBWPW Regulates the PI3K/Akt and MAPK Pathway
3. Materials and Methods
3.1. Experimental Materials and Chemical Reagent
3.2. Cells
3.3. Extraction and Purification of Polysaccharides
3.4. Determination Molecular Weight and Chemical Composition of Polysaccharide
3.5. Monosaccharide Compositions
3.6. Scanning Electron Microscopy and Infrared Spectroscopy Analysis
3.7. Nuclear Magnetic Resonance Detection
3.8. Determination of Cytotoxicity and Anti-Cancer Activity
3.9. Cell Cycle and Cell Apoptotic Detection
3.10. RNA Library Preparation, Sequencing and Data Processing
3.11. Target Prediction Based on the Transcriptomic Data
3.12. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
3.13. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Livshits, Z.; Rao, R.; Smith, S. An approach to chemotherapy-associated toxicity. Emerg. Med. Clin. N. Am. 2014, 32, 167–203. [Google Scholar]
- Atkins, S.; He, F. Chemotherapy and Beyond: Infections in the Era of Old and New Treatments for Hematologic Malignancies. Infect. Dis. Clin. N. Am. 2019, 33, 289–309. [Google Scholar]
- Dasari, S.; Tchounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Magge, R.; DeAngelis, L. The double-edged sword: Neurotoxicity of chemotherapy. Blood Rev. 2015, 29, 93–100. [Google Scholar] [CrossRef]
- Deng, L.; Qi, M.; Li, N.; Lei, Y.; Zhang, D.; Chen, J. Natural products and their derivatives: Promising modulators of tumor immunotherapy. J. Leukoc. Biol. 2020, 108, 493–508. [Google Scholar]
- Lee, M.; Chen, C.; Hwang, B.; Hsu, L. Analysis of saponins from black bean by electrospray ionization and fast atom bombardment tandem mass spectrometry. J. Mass Spectrom. 1999, 34, 804–812. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.; Han, S.; Nam, J.; Na, H.; Ha, T. Black soybean anthocyanins inhibit adipocyte differentiation in 3T3-L1 cells. Nutr. Res. 2012, 32, 770–777. [Google Scholar] [CrossRef]
- Kwon, S.; Ahn, I.; Kim, S.; Kong, C.; Chung, H.; Do, M.; Park, K. Anti-obesity and hypolipidemic effects of black soybean anthocyanins. J. Med. Food 2007, 10, 552–556. [Google Scholar] [CrossRef]
- Kennedy, A. The evidence for soybean products as cancer preventive agents. J. Nutr. 1995, 125 (Suppl. 3), 733S–743S. [Google Scholar]
- Xu, X.; Chang, Y. Gastric Protective Activities of Sea Cucumber Fucoidans with Different Molecular Weight and Chain Conformations: A Structure-Activity Relationship Investigation. J. Agric. Food Chem. 2018, 66, 8615–8622. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, X. Polysaccharide extracted from Huperzia serrata using response surface methodology and its biological activity. Int. J. Biol. Macromol. 2020, 157, 267–275. [Google Scholar] [CrossRef]
- Jing, Y.; Zhu, J.; Liu, T.; Bi, S.; Hu, X.; Chen, Z.; Song, L.; Lv, W.; Yu, R. Structural characterization and biological activities of a novel polysaccharide from cultured Cordyceps militaris and its sulfated derivative. J. Agric. Food Chem. 2015, 63, 3464–3471. [Google Scholar] [CrossRef]
- Surin, S.; You, S.; Seesuriyachan, P.; Muangrat, R.; Wangtueai, S.; Jambrak, A.; Phongthai, S.; Jantanasakulwong, K.; Chaiyaso, T.; Phimolsiripol, Y. Optimization of ultrasonic-assisted extraction of polysaccharides from purple glutinous rice bran (Oryza sativa L.) and their antioxidant activities. Sci. Rep. 2020, 10, 10410. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.; Chang, L.; Lin, Y.; Lu, H.; Yang, J.; Lee, J.; Chung, J. Morin inhibits the growth of human leukemia HL-60 cells via cell cycle arrest and induction of apoptosis through mitochondria dependent pathway. Anticancer Res. 2007, 27, 395–405. [Google Scholar]
- Liao, Y.; Wang, J.; Jaehnig, E.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Li, P.; Zhang, H.; Zhang, W.; Zhang, Y.; Zhan, L.; Wang, N.; Chen, C.; Fu, B.; Zhao, J.; Zhou, X.; et al. TMNP: A transcriptome-based multi-scale network pharmacology platform for herbal medicine. Brief. Bioinform. 2022, 23, bbab542. [Google Scholar] [CrossRef] [PubMed]
- Amirani, E.; Hallajzadeh, J.; Asemi, Z.; Mansournia, M.; Yousefi, B. Effects of chitosan and oligochitosans on the phosphatidylinositol 3-kinase-AKT pathway in cancer therapy. Int. J. Biol. Macromol. 2020, 164, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Raina, R.; Afroze, N.; Sundaram, M.K.; Haque, S.; Bajbouj, K.; Hamad, M.; Hussain, A. Chrysin inhibits propagation of HeLa cells by attenuating cell survival and inducing apoptotic pathways. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 2206–2220. [Google Scholar]
- Zhang, C.; Yun, Y.; Fan, W.; Liang, Y.; Yu, Y.; Tang, W. Rapid analysis of polysaccharides contents in Glycyrrhiza by near infrared spectroscopy and chemometrics. Int. J. Biol. Macromol. 2015, 79, 983–987. [Google Scholar] [CrossRef]
- Zhang, W.; Song, D.; Xu, D.; Wang, T.; Chen, L.; Duan, J. Characterization of polysaccharides with antioxidant and immunological activities from Rhizoma Acori Tatarinowii. Carbohydr. Polym. 2015, 133, 154–162. [Google Scholar] [CrossRef]
- Liu, L.; Wang, L.; Zonderman, J.; Rouse, J.; Kim, H. Automated, High-Throughput Infrared Spectroscopy for Secondary Structure Analysis of Protein Biopharmaceuticals. J. Pharm. Sci. 2020, 109, 3223–3230. [Google Scholar] [CrossRef]
- Farooqui, A.; Khan, F.; Khan, I.; Ansari, I. Glycyrrhizin induces reactive oxygen species-dependent apoptosis and cell cycle arrest at G0/G1 in HPV18(+) human cervical cancer HeLa cell line. Biomed. Pharmacother. 2018, 97, 752–764. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.; Irizarry, R.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.; Robinson, M. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.; Mukherjee, S.; Ebert, B.; Gillette, M.; Paulovich, A.; Pomeroy, S.; Golub, T.; Lander, E.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, C. Hydralazine inhibits human cervical cancer cell growth in vitro in association with APC demethylation and re-expression. Cancer Chemother. Pharmacol. 2009, 63, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, R.; Feng, Q.; Wang, Y.; Zhang, Y.; Wu, W.; Ge, P.; Qi, J. Safflower polysaccharide induces cervical cancer cell apoptosis via inhibition of the PI3K/Akt pathway. S. Afr. J. Bot. 2018, 118, 209–215. [Google Scholar] [CrossRef]
- Peng, R.; Cheng, X.; Zhang, Y.; Lu, X.; Hu, Z. miR-214 down-regulates MKK3 and suppresses malignant phenotypes of cervical cancer cells. Gene 2020, 724, 144146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, F.; Thakur, K.; Wang, J.; Wang, H.; Hu, F.; Zhang, J.; Wei, Z. Molecular mechanism of anti-cancerous potential of Morin extracted from mulberry in Hela cells. Food Chem. Toxicol. 2018, 112, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Date, A.; Chawda, H.; Patel, K. Polysaccharides as potential anticancer agents—A review of their progress. Carbohydr. Polym. 2019, 210, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Wang, Y.; Na, K.; Wang, Y.; Wang, L.; Li, Z.; Guo, C.; Guo, D.; Wang, X. Autophagic flux disruption contributes to Ganoderma lucidum polysaccharide-induced apoptosis in human colorectal cancer cells via MAPK/ERK activation. Cell Death Dis. 2019, 10, 456. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Y.; Li, Z.; Liu, W.; Zhang, H.; Ohizumi, Y.; Nakajima, A.; Xu, J.; Guo, Y. Structure, anti-tumor activity, and potential anti-tumor mechanism of a fungus polysaccharide from Fomes officinalis. Carbohydr. Polym. 2022, 295, 119794. [Google Scholar] [CrossRef] [PubMed]
- Mora-Escobedo, R.; Mdel, C.R.-R.; Ramón-Gallegos, E.; Reza-Alemán, R. Effect of protein hydrolysates from germinated soybean on cancerous cells of the human cervix: An in vitro study. Plant Foods Hum. Nutr. 2009, 64, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Zhang, W.; Cheng, C.; Mo, F.; Chen, L.; Peng, G.; Cai, X.; Wang, J.; Yang, S.; Liu, X. Dioscin inhibits the growth of human osteosarcoma by inducing G2/M-phase arrest, apoptosis, and GSDME-dependent cell death in vitro and in vivo. J. Cell. Physiol. 2020, 235, 2911–2924. [Google Scholar] [CrossRef]
- Zong, A.; Cao, H.; Wang, F. Anticancer polysaccharides from natural resources: A review of recent research. Carbohydr. Polym. 2012, 90, 1395–1410. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zheng, X.; Ye, J.; Wu, T.; Wang, J.; Chen, W. Potential anticancer activity of myricetin in human T24 bladder cancer cells both in vitro and in vivo. Nutr. Cancer 2012, 64, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Wang, C.; Li, L.; Liang, H.; Dai, J.; Ling, X.; Tang, H. Luteoloside Inhibits Proliferation and Promotes Intrinsic and Extrinsic Pathway-Mediated Apoptosis Involving MAPK and mTOR Signaling Pathways in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1664. [Google Scholar] [CrossRef]
- Martinou, J.; Youle, R. Mitochondria in apoptosis: BCL-2 family members and mitochondrial dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, J.; Feng, Y.; Weng, L.; Zhao, G.; Xiang, J.; Zhang, M.; Xing, D. Targeting of growth factors in the treatment of hepatocellular carcinoma: The potentials of polysaccharides. Oncol. Lett. 2017, 13, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kim, W.J.; Jiang, J.; Lee, S.H.; Youn, H.S.; Moon, E.Y.; Kim, T.J.; Ye, S.K.; Ryu, J.H.; Kang, T.B.; et al. MyD88-dependent Toll-like receptor signaling is required for murine macrophages response to IS2. Int. Immunopharmacol. 2011, 11, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wu, X.; Giovanni, V.; Meng, X. Effects of soybean oligosaccharides on intestinal microbial communities and immune modulation in mice. Saudi J. Biol. Sci. 2017, 24, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Hwang, J.; Zhang, W.; Go, S.; Kim, J. Polysaccharide from Codium fragile Induces Anti-Cancer Immunity by Activating Natural Killer Cells. Mar. Drugs 2020, 18, 626. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hu, X.; Wang, S.; Jiao, Z.; Sun, T.; Liu, T.; Song, K. Characterization and anti-tumor bioactivity of astragalus polysacharides by immunomodulation. Int. J. Biol. Macromol. 2020, 145, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Parkin, J.; Cohen, B. An overview of the immune system. Lancet 2001, 357, 1777–1789. [Google Scholar] [CrossRef]
- Bruni, D.; Angell, H. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef]
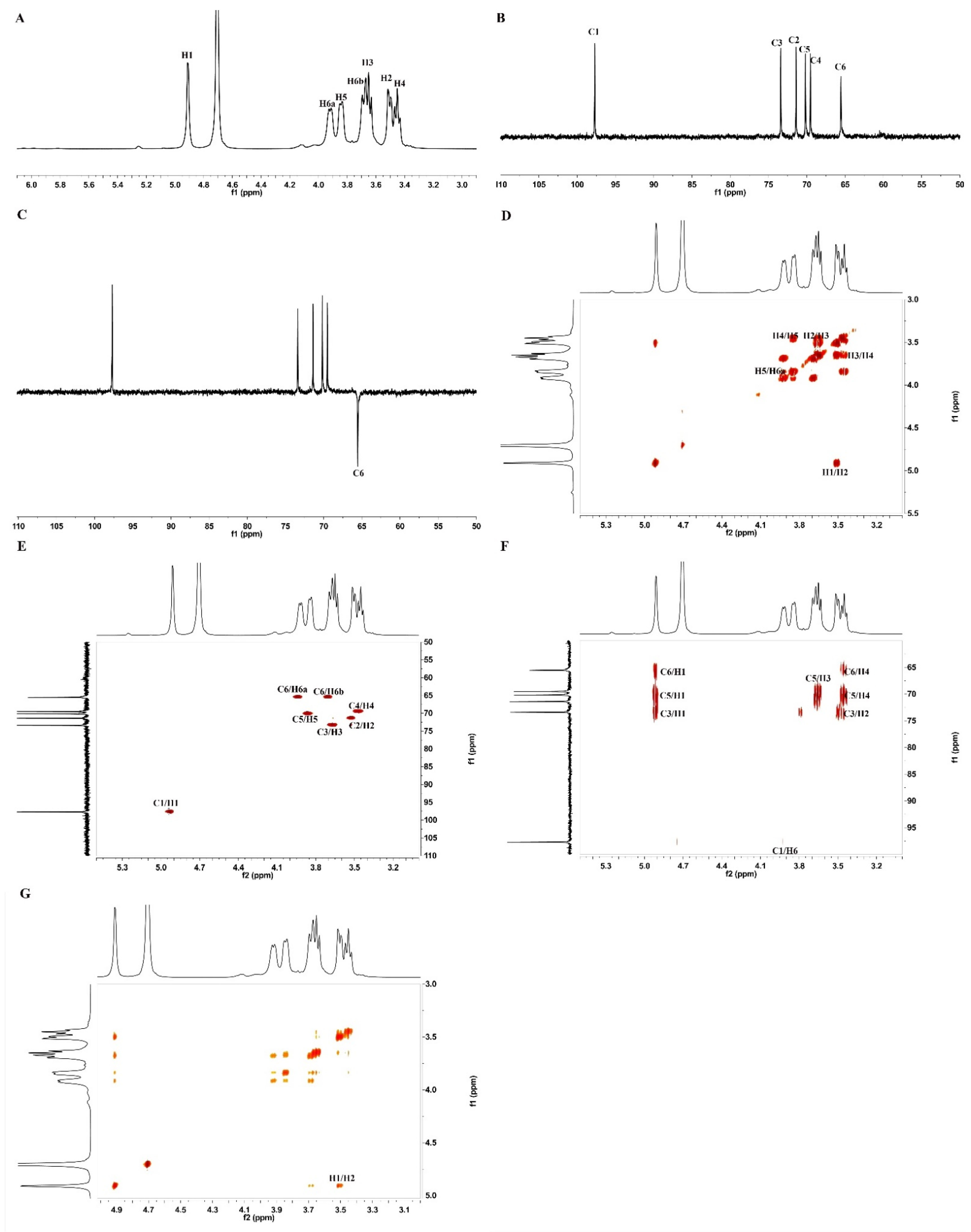

| Sugar Residues | C1 H1 | C2 H2 | C3 H3 | C4 H4 | C5 H5 | C6 H6a | H6b |
|---|---|---|---|---|---|---|---|
| α-1,6-Glc | 97.77 4.91 | 71.41 3.52 | 73.40 3.65 | 69.57 3.45 | 70.22 3.85 | 65.60 3.93 | 3.69 |
| Genes | Primer Sequences (5′−3′) |
|---|---|
| GAPDH | F: CATGAGAAGTATGACAACAGCCT; R: AGTCCTTCCACGATACCAAAGT |
| p21 | F: GCGGAACAAGGAGTCAGACA; R: GAACCAGGACACATGGGGAG |
| CDK1 | F: TTGAAACTGCTCGCACTTGG; R: TCCCGGCTTATTATTCCGCG |
| Cyclin B1 | F: CTGCTGGGTGTAGGTCCTTG; R: TGCCATGTTGATCTTCGCCT |
| Survivin | F: GGCCCAGTGTTTCTTCTGCT; R: ATGAGGGTGGAAAGCAACCC |
| mTOR | F: TCCGAGAGATGAGTCAAGAGGAGTC; R: GCTGGAAACCAATTCAAAAATGTG |
| BAD | F: CGGAGGATGAGTGACGAGTTTGT; R: ATCCCACCAGGACTGGAAGACTC |
| AKT | F: ACCTTCCATGTGGAGACTCCTGAG; R: GTCCATCTCCTCCTCCTCCTGC |
| BCL-2 | F: CATGTGTGTGGAGAGCGTCAAC; R: CTTCAGAGACAGCCAGGAGAAATC |
| MKK3 | F: CGCGGATCCAAAGAAGCAGACC; R: CGGAATTCAAAGCCGGGATAGAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Hu, Y.; Zhan, L.; He, J.; Lu, J.; Gao, C.; Du, W.; Yue, A.; Zhao, J.; Zhang, W. A Natural Glucan from Black Bean Inhibits Cancer Cell Proliferation via PI3K-Akt and MAPK Pathway. Molecules 2023, 28, 1971. https://doi.org/10.3390/molecules28041971
Li P, Hu Y, Zhan L, He J, Lu J, Gao C, Du W, Yue A, Zhao J, Zhang W. A Natural Glucan from Black Bean Inhibits Cancer Cell Proliferation via PI3K-Akt and MAPK Pathway. Molecules. 2023; 28(4):1971. https://doi.org/10.3390/molecules28041971
Chicago/Turabian StyleLi, Peng, Yihua Hu, Lingmin Zhan, Jiaqi He, Jingwu Lu, Chunyan Gao, Weijun Du, Aiqin Yue, Jinzhong Zhao, and Wuxia Zhang. 2023. "A Natural Glucan from Black Bean Inhibits Cancer Cell Proliferation via PI3K-Akt and MAPK Pathway" Molecules 28, no. 4: 1971. https://doi.org/10.3390/molecules28041971
APA StyleLi, P., Hu, Y., Zhan, L., He, J., Lu, J., Gao, C., Du, W., Yue, A., Zhao, J., & Zhang, W. (2023). A Natural Glucan from Black Bean Inhibits Cancer Cell Proliferation via PI3K-Akt and MAPK Pathway. Molecules, 28(4), 1971. https://doi.org/10.3390/molecules28041971