In Vivo Incorporation of Photoproteins into GroEL Chaperonin Retaining Major Structural and Functional Properties
Abstract
1. Introduction
2. Results
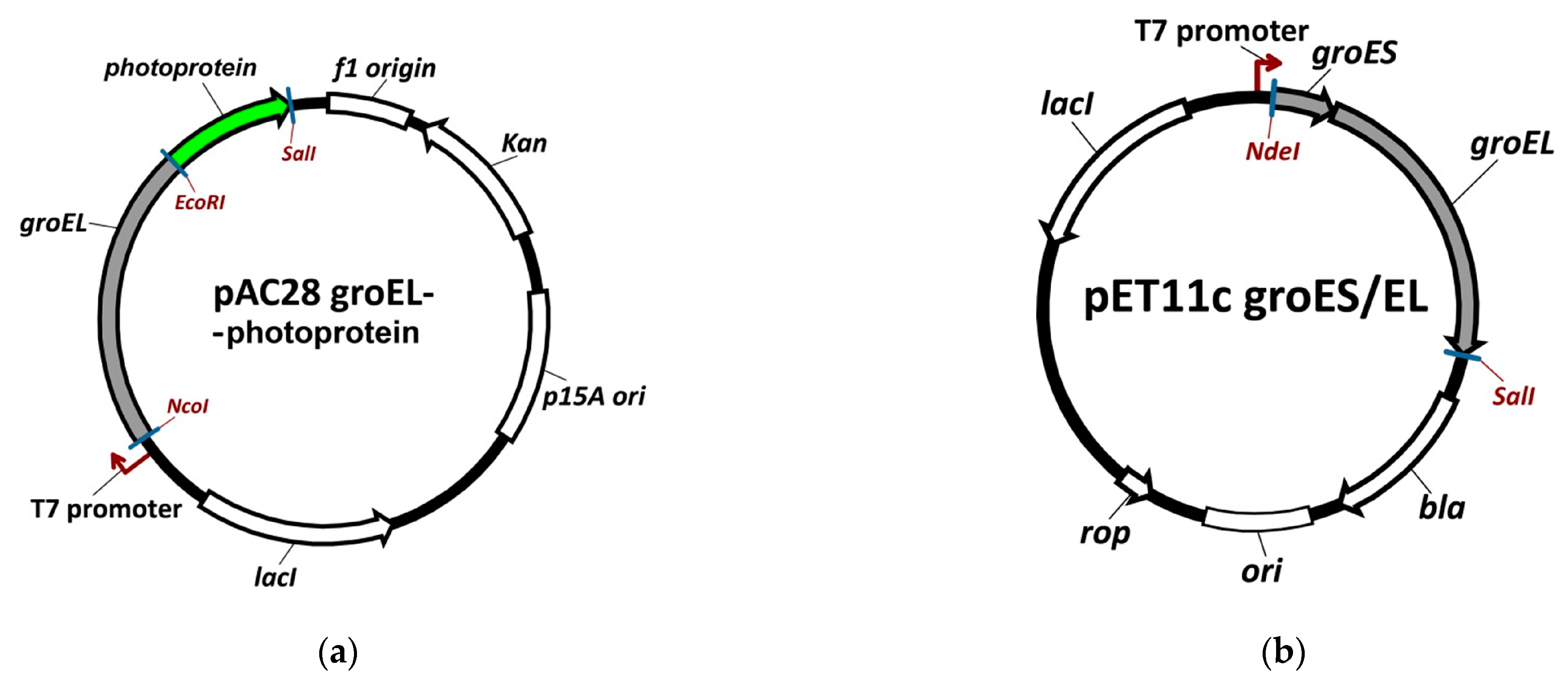
2.1. The Analysis of Plasmid Expression Products
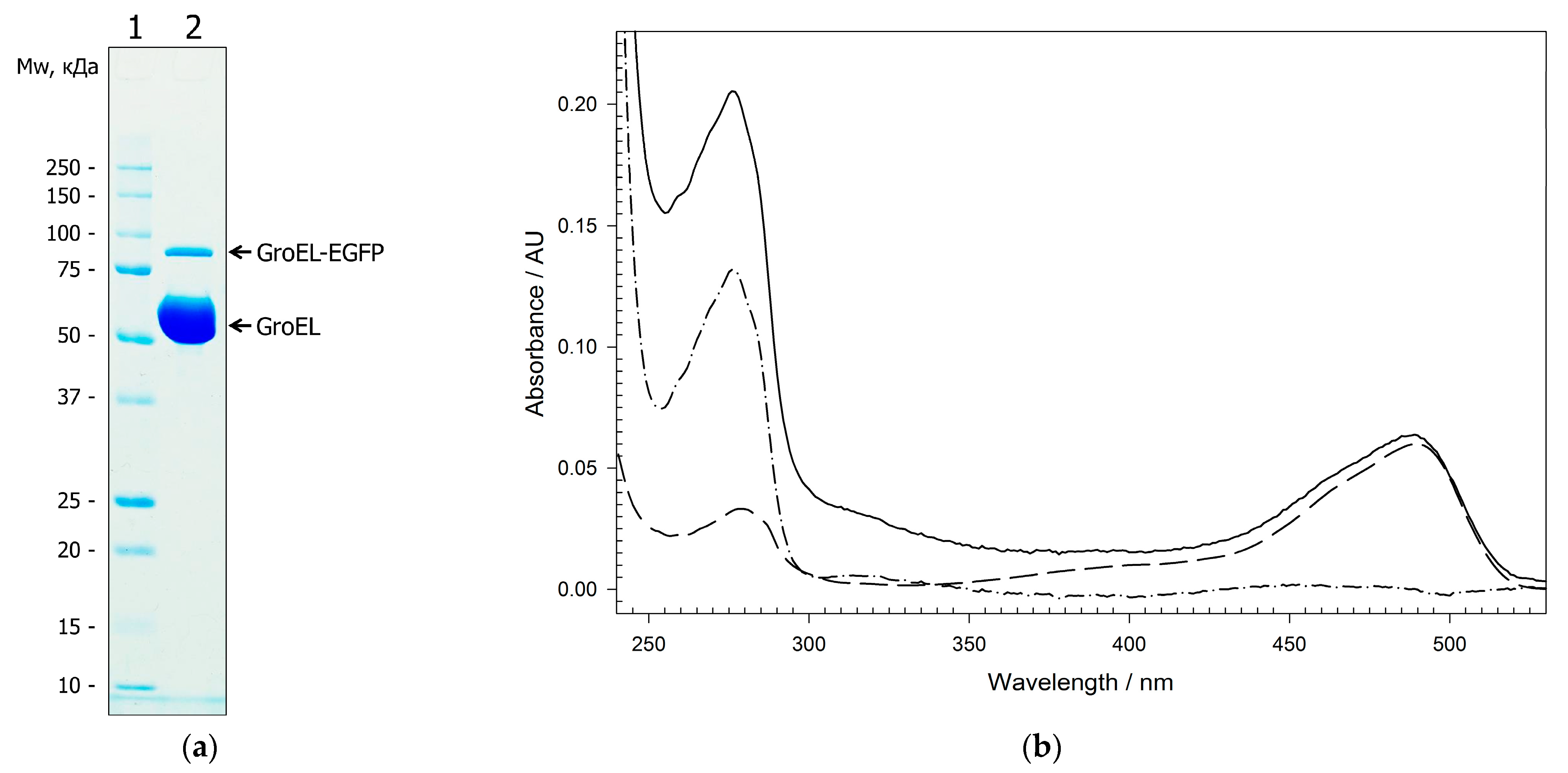
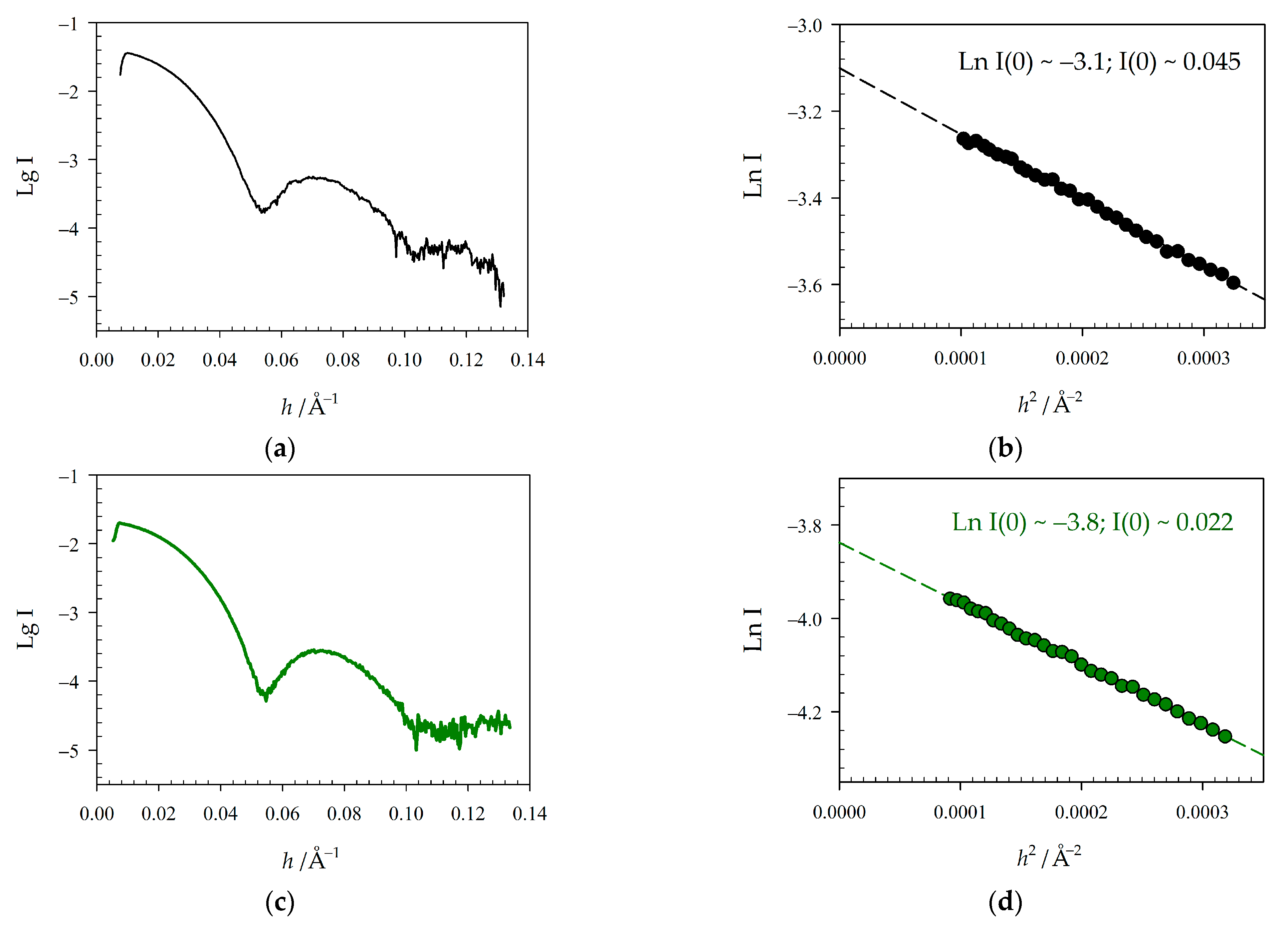
2.2. Physicochemical Properties of GroEL14 with Incorporated EGFP
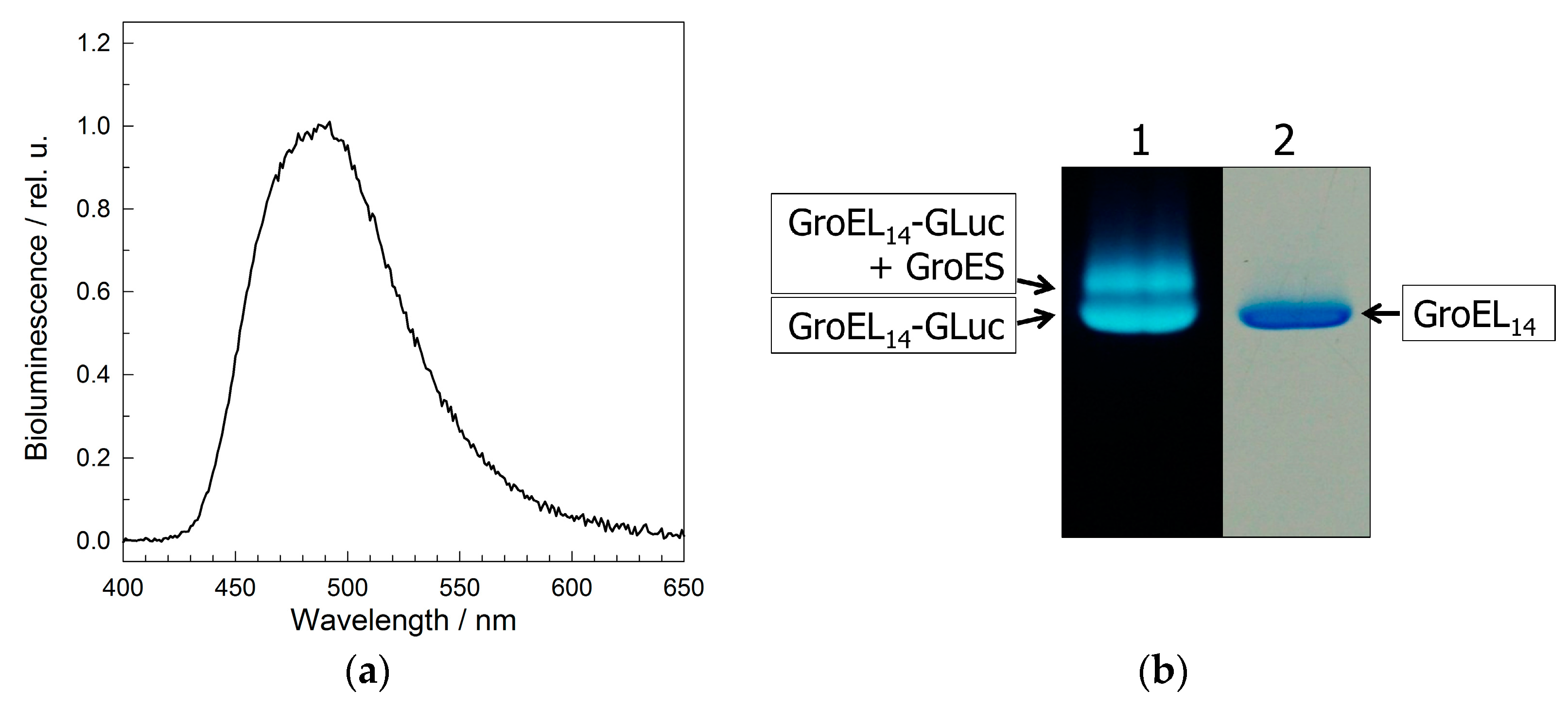
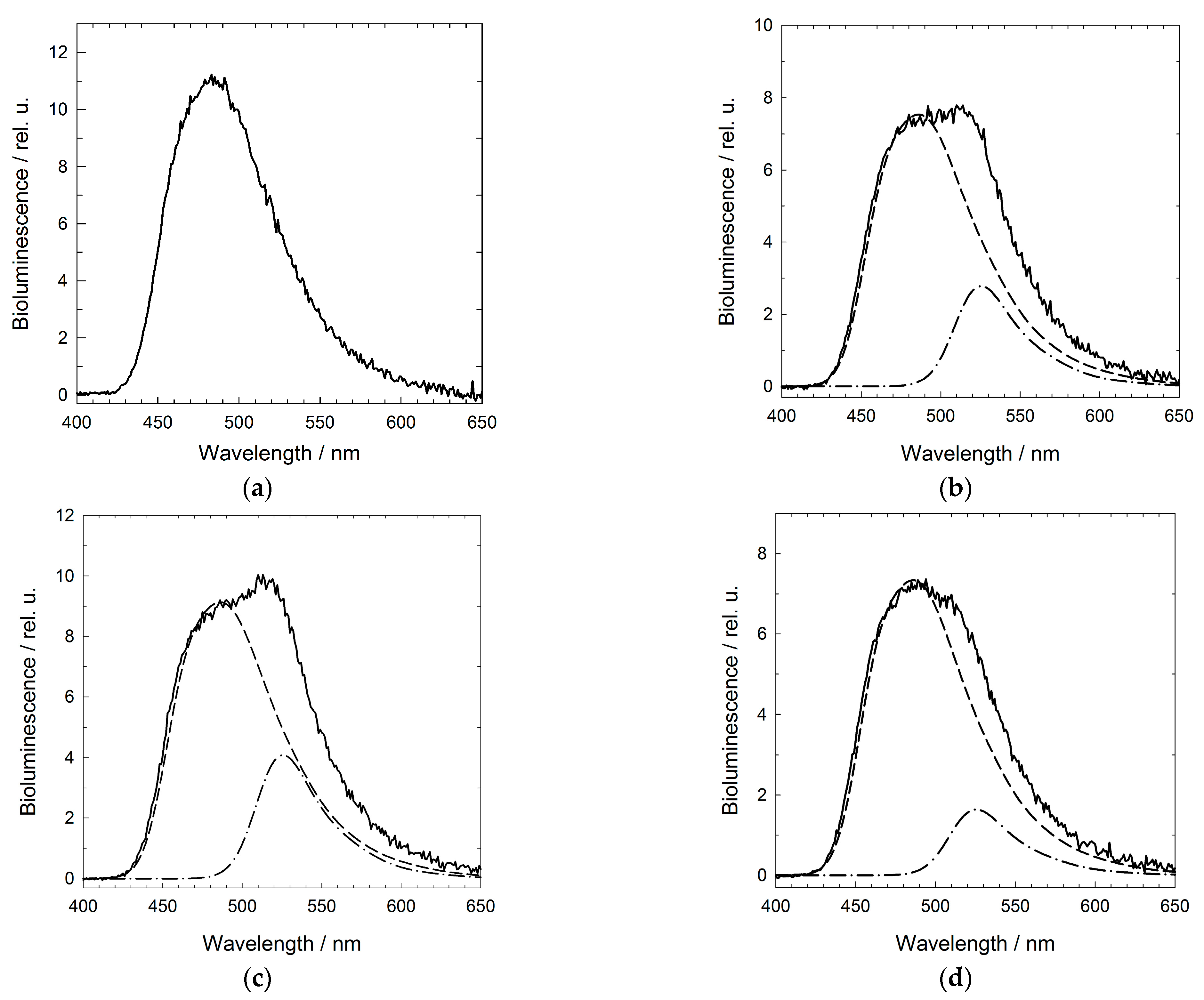
2.3. Physicochemical Properties of the GroEL14 Particle with Incorporated GLuc
3. Discussion
4. Materials and Methods
4.1. Chemicals and Related Enzymes
4.2. Bacterial Strains, Culture Conditions, and Enzymes
4.3. Genetic Manipulations
4.4. Protein Purification and Labeling
4.5. Registration of Fluorescence and Bioluminescence in the Cells, Cell Extracts, Electrophoretic Gels, and Pure Protein Solutions
4.6. SAXS Measurements
4.7. Transmission Electron Microscopy and Images Processing
4.8. Fluorescence Microscopy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef]
- Yoshida, T.; Kawaguchi, R.; Taguchi, H.; Yoshida, M.; Yasunaga, T.; Wakabayashi, T.; Yohda, M.; Maruyama, T. Archaeal group II chaperonin mediates protein folding in the cis-cavity without a detachable GroES-like co-chaperonin. J. Mol. Biol. 2002, 315, 73–85. [Google Scholar] [CrossRef]
- Vilasi, S.; Bulone, D.; Caruso Bavisotto, C.; Campanella, C.; Marino Gammazza, A.; San Biagio, P.L.; Cappello, F.; de Macario, E.; Macario, A.J.L. Chaperonin of Group I: Oligomeric Spectrum and Biochemical and Biological Implications. Front. Mol. Biosci. 2018, 4, 99. [Google Scholar] [CrossRef]
- Rebeaud, M.E.; Mallik, S.; Goloubinoff, P.; Tawfik, D.S. On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc. Natl. Acad. Sci. USA 2021, 118, e2020885118. [Google Scholar] [CrossRef]
- Horwich, A.L.; Fenton, W.A.; Chapman, E.; Farr, G.W. Two families of chaperonin: Physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007, 23, 115–145. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Braig, K.; Otwinowski, Z.; Hegde, R.; Boisvert, D.C.; Joachimiak, A.; Horwich, A.L.; Sigler, P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 1994, 371, 578–586. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Moiseenko, A.V.; Sokolova, O.S.; Muronetz, V.I.; Kurochkina, L.P. Structural and functional diversity of novel and known bacteriophage-encoded chaperonins. Int. J. Biol. Macromol. 2020, 157, 544–552. [Google Scholar] [CrossRef]
- Qamra, R.; Mande, S.C. Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J. Bacteriol. 2004, 186, 8105–8113. [Google Scholar] [CrossRef]
- Gruber, R.; Horovitz, A. Allosteric Mechanisms in Chaperonin Machines. Chem. Rev. 2016, 116, 6588–6606. [Google Scholar] [CrossRef]
- Lorimer, G.H.; Fei, X.; Ye, X. The GroEL chaperonin: A protein machine with pistons driven by ATP binding and hydrolysis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2018, 373, 20170179. [Google Scholar] [CrossRef]
- Saibil, H.R.; Fenton, W.A.; Clare, D.K.; Horwich, A.L. Structure and Allostery of the Chaperonin GroEL. J. Mol. Biol. 2013, 425, 1476–1487. [Google Scholar] [CrossRef]
- Fei, X.; Yang, D.; LaRonde-LeBlanc, N.; Lorimer, G.H. Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 Å resolution. Proc. Natl. Acad. Sci. USA 2013, 110, E2958–E2966. [Google Scholar] [CrossRef]
- Bracher, A.; Starling-Windhof, A.; Hartl, F.U.; Hayer-Hartl, M. Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco. Nat. Struct. Mol. Biol. 2011, 18, 875–880. [Google Scholar] [CrossRef]
- Chen, L.; Sigler, P.B. The crystal structure of a GroEL/peptide complex: Plasticity as a basis for substrate diversity. Cell 1999, 99, 757–768. [Google Scholar] [CrossRef]
- Chatellier, J.; Hill, F.; Fersht, A.R. From minichaperone to GroEL 2: Importance of avidity of the multisite ring structure. J. Mol. Biol. 2000, 304, 883–896. [Google Scholar] [CrossRef]
- Yuan, Y.; Du, C.; Sun, C.; Zhu, J.; Wu, S.; Zhang, Y.; Ji, T.; Lei, J.; Yang, Y.; Gao, N.; et al. Chaperonin-GroEL as a Smart Hydrophobic Drug Delivery and Tumor Targeting Molecular Machine for Tumor Therapy. Nano Lett. 2018, 18, 921–928. [Google Scholar] [CrossRef]
- Horwich, A.L.; Farr, G.W.; Fenton, W.A. GroEL−GroES-Mediated Protein Folding. Chem. Rev. 2006, 106, 1917–1930. [Google Scholar] [CrossRef]
- O’Neil, P.T.; Machen, A.J.; Deatherage, B.C.; Trecazzi, C.; Tischer, A.; Machha, V.R.; Auton, M.T.; Baldwin, M.R.; White, T.A.; Fisher, M.T. The Chaperonin GroEL: A Versatile Tool for Applied Biotechnology Platforms. Front. Mol. Biosci. 2018, 5, 46. [Google Scholar] [CrossRef]
- Furutani, M.; Hata, J.-I.; Shomura, Y.; Itami, K.; Yoshida, T.; Izumoto, Y.; Togi, A.; Ideno, A.; Yasunaga, T.; Miki, K.; et al. An engineered chaperonin caging a guest protein: Structural insights and potential as a protein expression tool. Protein Sci. 2005, 14, 341–350. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Sadykhov, E.G.; Fedorov, A.N. Production of a toxic polypeptide as a fusion inside GroEL cavity. Sci. Rep. 2020, 10, 21024. [Google Scholar] [CrossRef]
- Yurkova, M.S.; Fedorov, A.N. GroEL-A Versatile Chaperone for Engineering and a Plethora of Applications. Biomolecules 2022, 12, 607. [Google Scholar] [CrossRef]
- Kyratsous, C.A.; Silverstein, S.J.; DeLong, C.R.; Panagiotidis, C.A. Chaperone-fusion expression plasmid vectors for improved solubility of recombinant proteins in Escherichia coli. Gene 2009, 440, 9–15. [Google Scholar] [CrossRef]
- Chitradevi, S.T.S.; Kaur, G.; Sivaramakrishna, U.; Singh, D.; Bansal, A. Development of recombinant vaccine candidate molecule against Shigella infection. Vaccine 2016, 34, 5376–5383. [Google Scholar] [CrossRef]
- Kyratsous, C.A.; Panagiotidis, C.A. Heat-Shock Protein Fusion Vectors for Improved Expression of Soluble Recombinant Proteins in Escherichia coli BT—Recombinant Gene Expression. In Recombinant Gene Expression; Lorence, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 109–129. ISBN 978-1-61779-433-9. [Google Scholar]
- Yurkova, M.S.; Savvin, O.I.; Zenin, V.A.; Fedorov, A.N. Design and Characterization of a Methionineless Variant of Thermostable Chaperon GroEL from Thermus thermophilus. Appl. Biochem. Microbiol. 2019, 55, 112–116. [Google Scholar] [CrossRef]
- Zenin, V.; Yurkova, M.; Tsedilin, A.; Fedorov, A. Enfuvirtide biosynthesis in thermostable chaperone-based fusion. Biotechnol. Rep. 2022, 35, e00734. [Google Scholar] [CrossRef]
- Charbon, G.; Wang, J.; Brustad, E.; Schultz, P.G.; Horwich, A.L.; Jacobs-Wagner, C.; Chapman, E. Localization of GroEL determined by in vivo incorporation of a fluorescent amino acid. Bioorg. Med. Chem. Lett. 2011, 21, 6067–6070. [Google Scholar] [CrossRef]
- Glatter, O.; Kratky, O. Small Angle X-ray Scattering; Kratky, O., Glatter, O., Eds.; Academic Press: London, UK, 1982; ISBN 0122862805. [Google Scholar]
- Piiadov, V.; Ares de Araújo, E.; Oliveira Neto, M.; Craievich, A.F.; Polikarpov, I. SAXSMoW 2.0: Online calculator of the molecular weight of proteins in dilute solution from experimental SAXS data measured on a relative scale. Protein Sci. 2019, 28, 454–463. [Google Scholar] [CrossRef]
- de Oliveira Neto, M.; de Freitas Fernandes, A.; Piiadov, V.; Craievich, A.F.; de Araújo, E.A.; Polikarpov, I. SAXSMoW 3.0: New advances in the determination of the molecular weight of proteins in dilute solutions from SAXS intensity data on a relative scale. Protein Sci. 2022, 31, 251–258. [Google Scholar] [CrossRef]
- Lissin, N.M.; Venyaminov, S.Y.; Girshovich, A.S. (Mg-ATP)-dependent self-assembly of molecular chaperone GroEL. Nature 1990, 348, 339–342. [Google Scholar] [CrossRef]
- Bell, J.M.; Chen, M.; Baldwin, P.R.; Ludtke, S.J. High resolution single particle refinement in EMAN2.1. Methods 2016, 100, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Marchenko, N.Y.; Sikorskaya, E.V.; Marchenkov, V.V.; Kashparov, I.A.; Semisotnov, G.V. Affinity chromatography of chaperones based on denatured proteins: Analysis of cell lysates of different origin. Protein Expr. Purif. 2016, 119, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Marchenkov, V.; Gorokhovatsky, A.; Marchenko, N.; Ivashina, T.; Semisotnov, G. Back to GroEL-assisted protein folding: GroES binding-induced displacement of denatured proteins from GroEL to bulk solution. Biomolecules 2020, 10, 162. [Google Scholar] [CrossRef]
- Marchenkov, V.; Ivashina, T.; Marchenko, N.; Ksenzenko, V.; Semisotnov, G. Bioluminescence Resonance Energy Transfer (BRET) Allows Monitoring the Barnase-Barstar Complex In Vivo. Biophysica 2022, 2, 7. [Google Scholar] [CrossRef]
- Braig, K.; Adams, P.D.; Brünger, A.T. Conformational variability in the refined structure of the chaperonin groel at 2.8 Â resolution. Nat. Struct. Biol. 1995, 2, 1095–1101. [Google Scholar] [CrossRef]
- Royant, A.; Noirclerc-Savoye, M. Stabilizing role of glutamic acid 222 in the structure of Enhanced Green Fluorescent Protein. J. Struct. Biol. 2011, 174, 385–390. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Ogino, H.; Wachi, M.; Ishii, A.; Iwai, N.; Nishida, T.; Yamada, S.; Nagai, K.; Sugai, M. FtsZ-dependent localization of GroEL protein at possible division sites. Genes Cells 2004, 9, 765–771. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 0-87969-577-3. [Google Scholar]
- Kholod, N.; Mustelin, T. Novel vectors for co-expression of two proteins in E. coli. Biotechniques 2001, 31, 322–328. [Google Scholar] [CrossRef]
- Gorokhovatsky, A.Y.; Rudenko, N.V.; Marchenkov, V.V.; Skosyrev, V.S.; Arzhanov, M.A.; Burkhardt, N.; Zakharov, M.V.; Semisotnov, G.V.; Vinokurov, L.M.; Alakhov, Y.B. Homogeneous assay for biotin based on Aequorea victoria bioluminescence resonance energy transfer system. Anal. Biochem. 2003, 313, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.; Klunker, D.; Ang, D.; Naylor, D.J.; Kerner, M.J.; Georgopoulos, C.; Hartl, F.U.; Hayer-Hartl, M. Functional characterization of an archaeal GroEL/GroES chaperonin system: Significance of substrate encapsulation. J. Biol. Chem. 2004, 279, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Ward, W.W. Biochemical and physical properties of green fluorescent protein. Methods Biochem. Anal. 2006, 47, 39–65. [Google Scholar] [PubMed]
- Amemiya, Y.; Wakabayashi, K.; Hamanaka, T.; Wakabayashi, T.; Matsushita, T.; Hashizume, H. Design of small-angle X-ray diffractometer using synchrotron radiation at the photon factory. Nucl. Instrum. Methods Phys. Res. 1983, 208, 471–477. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchenkov, V.; Ivashina, T.; Marchenko, N.; Ryabova, N.; Selivanova, O.; Timchenko, A.; Kihara, H.; Ksenzenko, V.; Semisotnov, G. In Vivo Incorporation of Photoproteins into GroEL Chaperonin Retaining Major Structural and Functional Properties. Molecules 2023, 28, 1901. https://doi.org/10.3390/molecules28041901
Marchenkov V, Ivashina T, Marchenko N, Ryabova N, Selivanova O, Timchenko A, Kihara H, Ksenzenko V, Semisotnov G. In Vivo Incorporation of Photoproteins into GroEL Chaperonin Retaining Major Structural and Functional Properties. Molecules. 2023; 28(4):1901. https://doi.org/10.3390/molecules28041901
Chicago/Turabian StyleMarchenkov, Victor, Tanya Ivashina, Natalia Marchenko, Natalya Ryabova, Olga Selivanova, Alexander Timchenko, Hiroshi Kihara, Vladimir Ksenzenko, and Gennady Semisotnov. 2023. "In Vivo Incorporation of Photoproteins into GroEL Chaperonin Retaining Major Structural and Functional Properties" Molecules 28, no. 4: 1901. https://doi.org/10.3390/molecules28041901
APA StyleMarchenkov, V., Ivashina, T., Marchenko, N., Ryabova, N., Selivanova, O., Timchenko, A., Kihara, H., Ksenzenko, V., & Semisotnov, G. (2023). In Vivo Incorporation of Photoproteins into GroEL Chaperonin Retaining Major Structural and Functional Properties. Molecules, 28(4), 1901. https://doi.org/10.3390/molecules28041901






