Chemical and Biological Review of Endophytic Fungi Associated with Morus sp. (Moraceae) and In Silico Study of Their Antidiabetic Potential
Abstract
:1. Introduction

2. Literature Review
2.1. Endophytic Fungi Associated with Morus Species
2.2. Chemistry of Endophytic Fungal Metabolites Associated with Morus Species
2.3. Biological Activities of Endophytic Fungi Associated with Morus Species
2.3.1. Reported Biological Studies on Endophytic Fungal Extracts
2.3.2. Reported Biological Studies on Isolated Endophytic Fungal Metabolites
3. Results and Discussion
3.1. Pharmacokinetic Profiling
3.2. Molecular Modelling
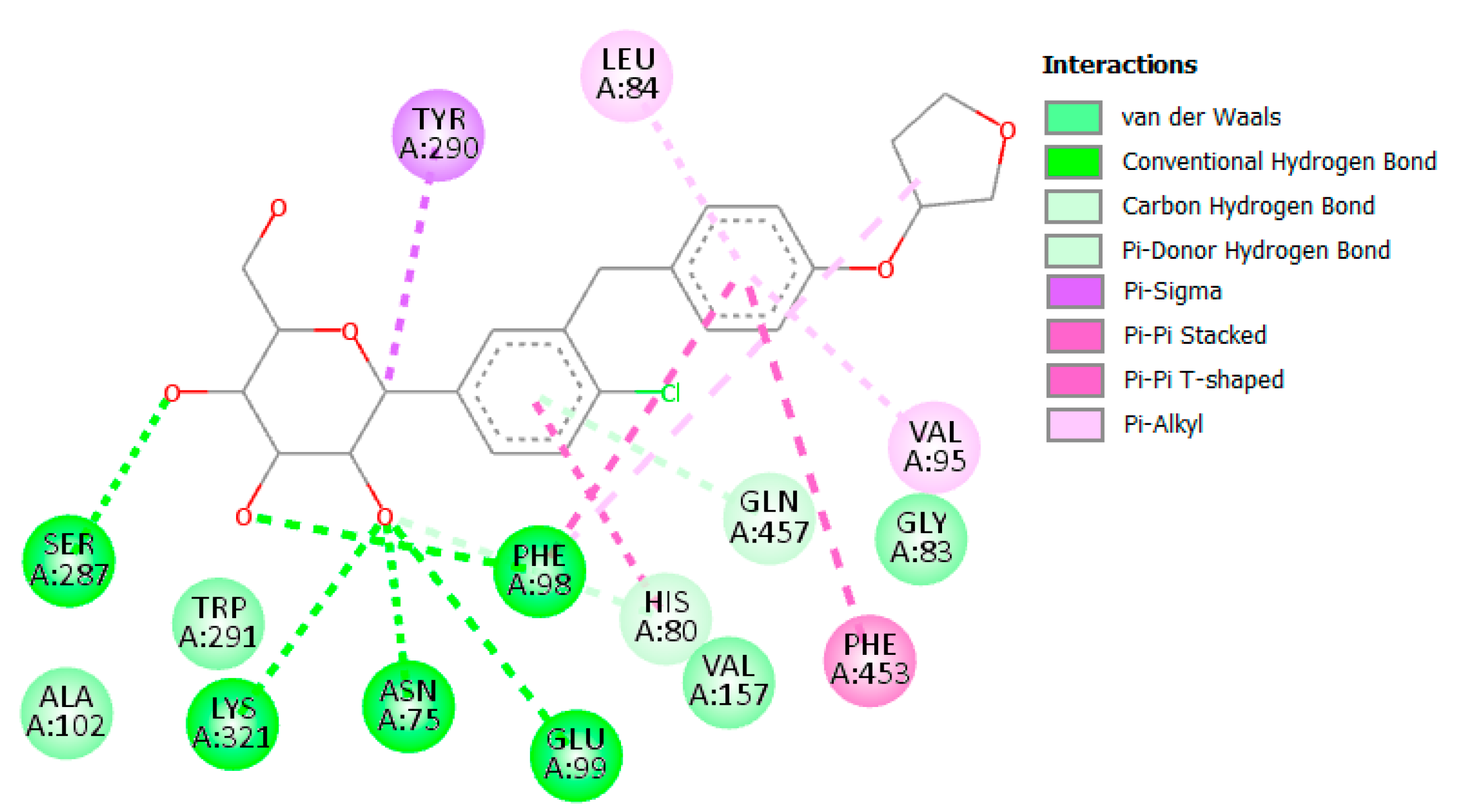
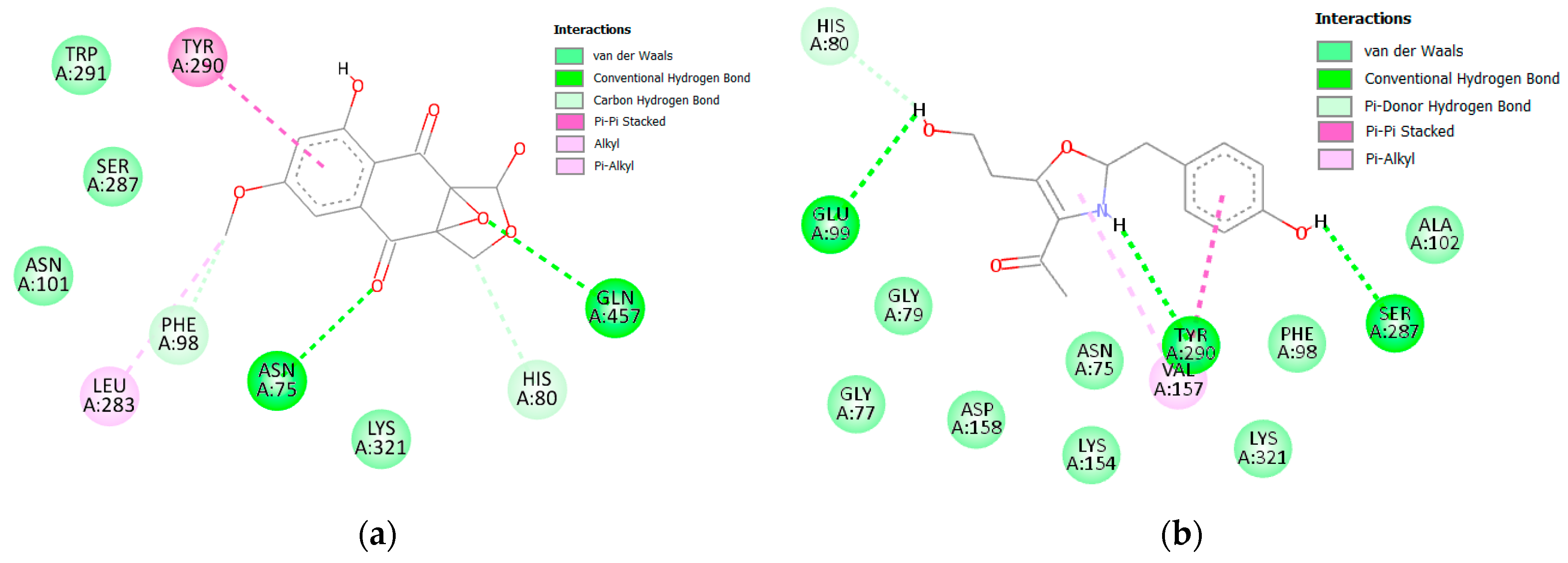
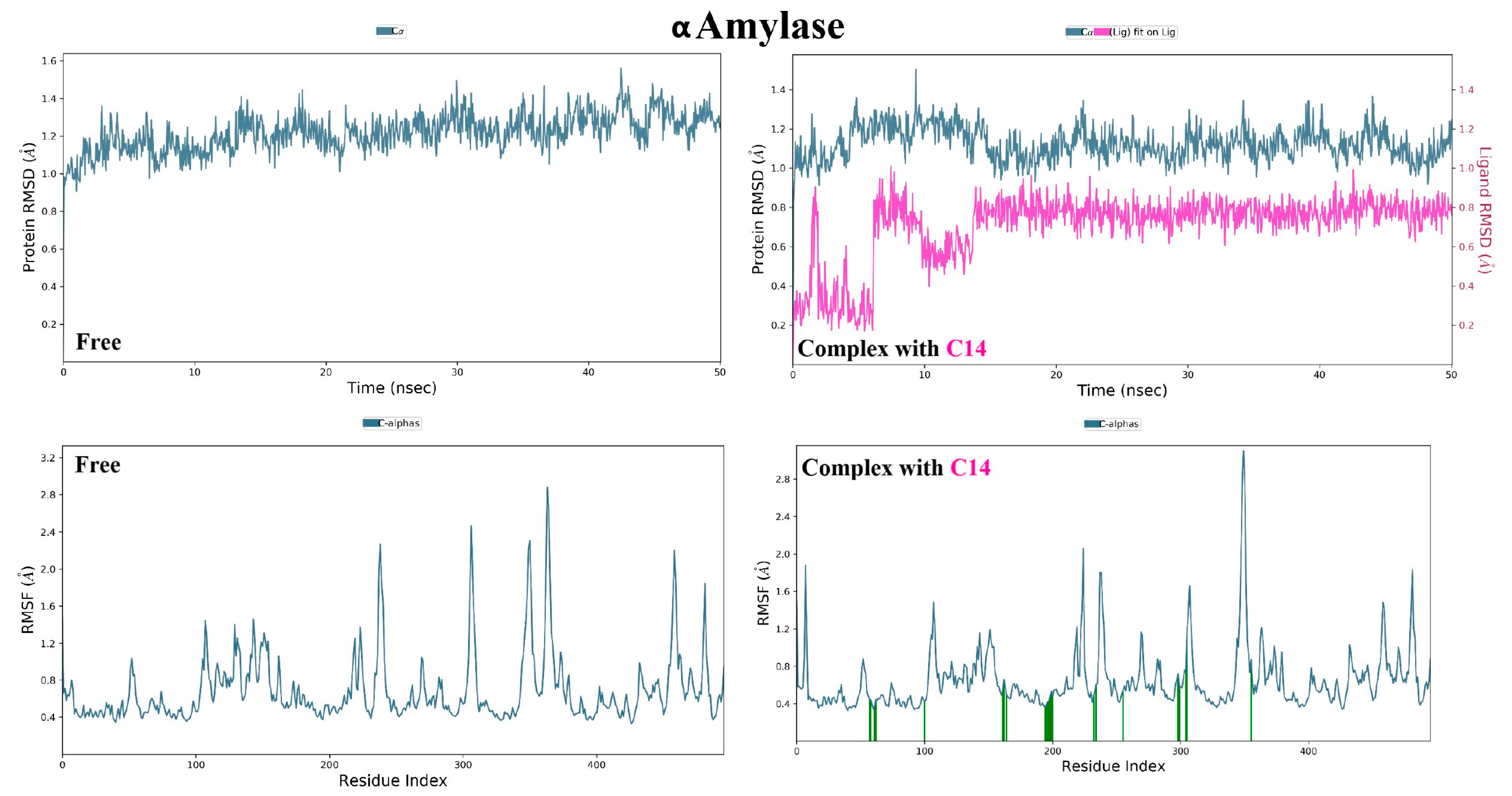
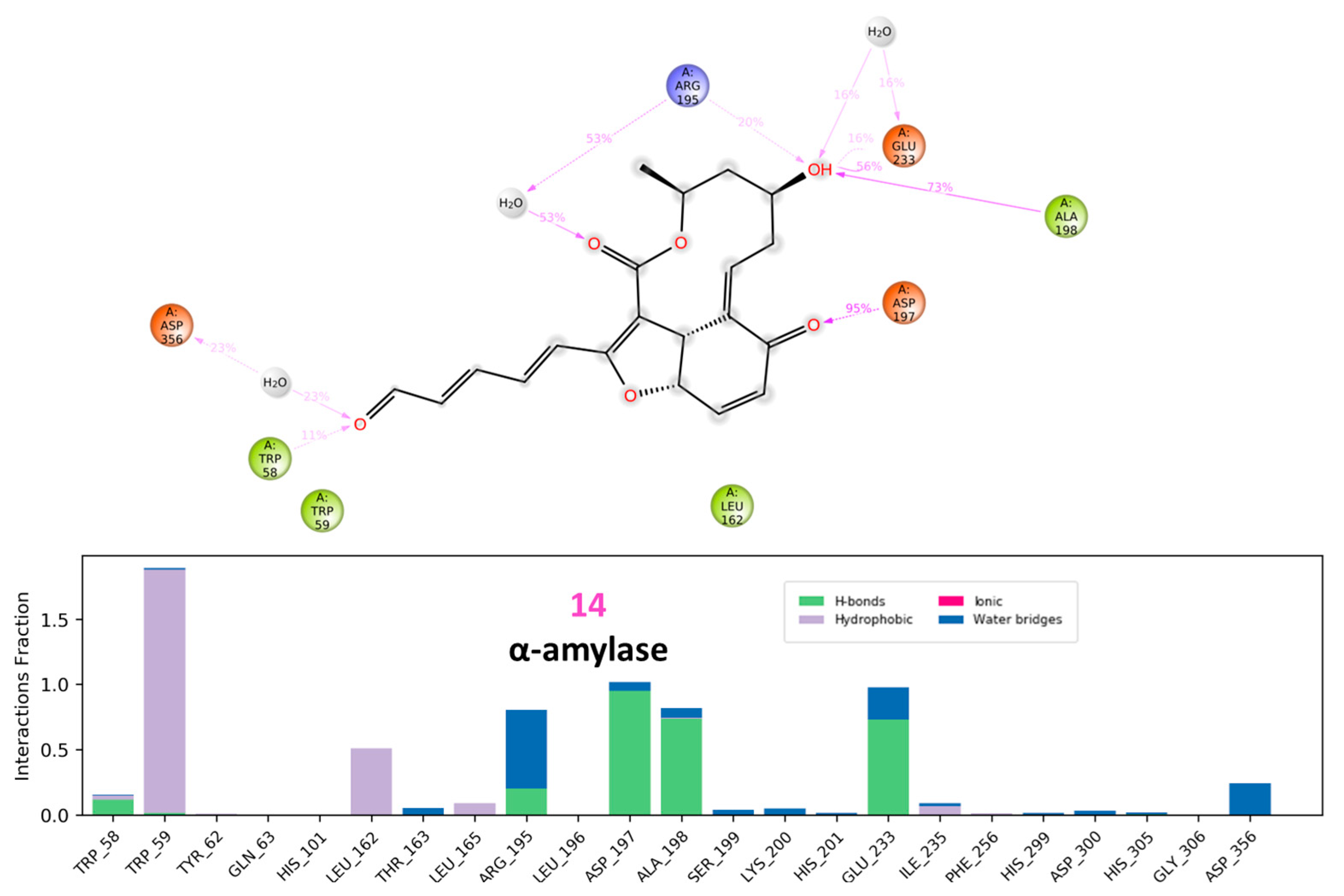
3.2.1. α Amylase Interaction
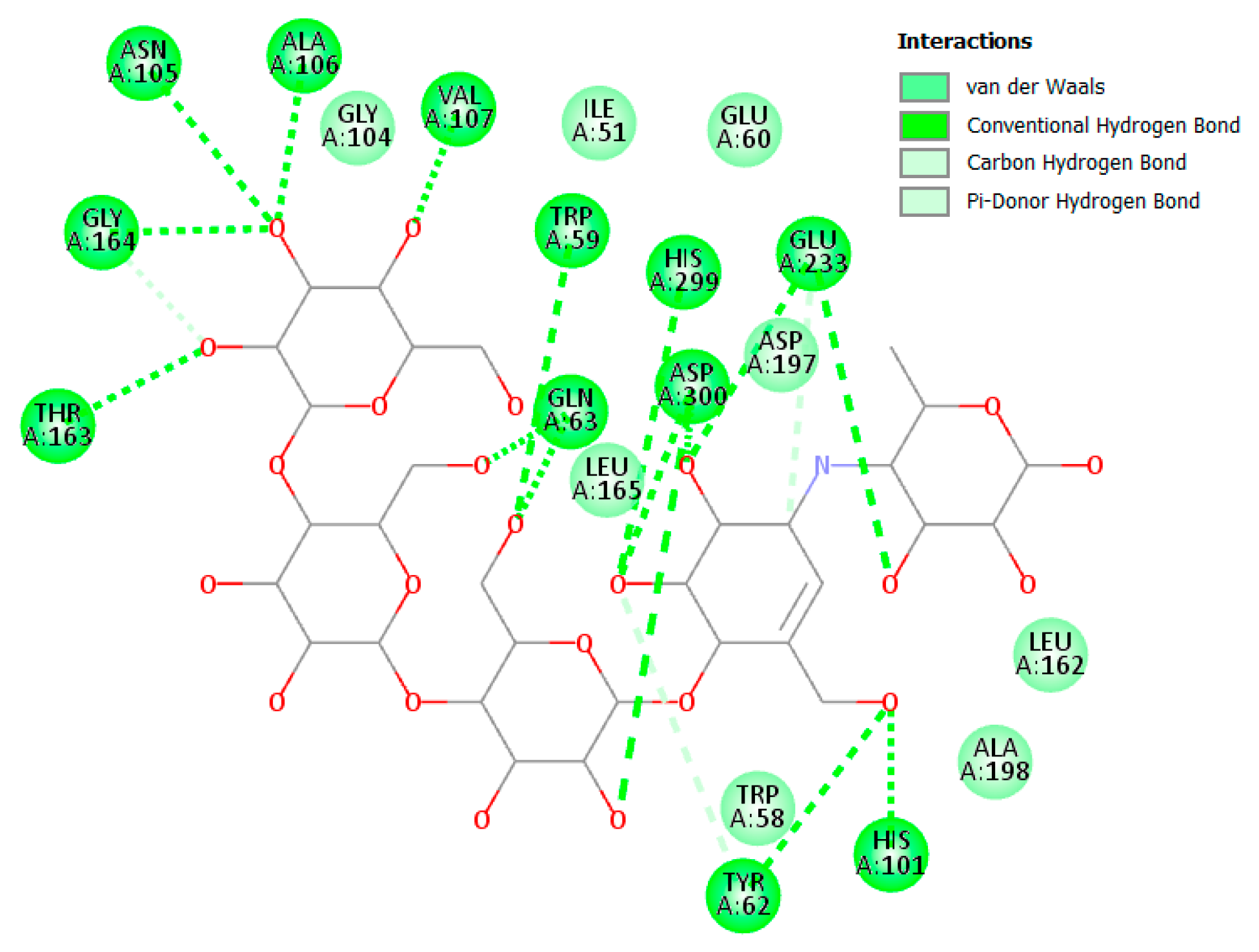
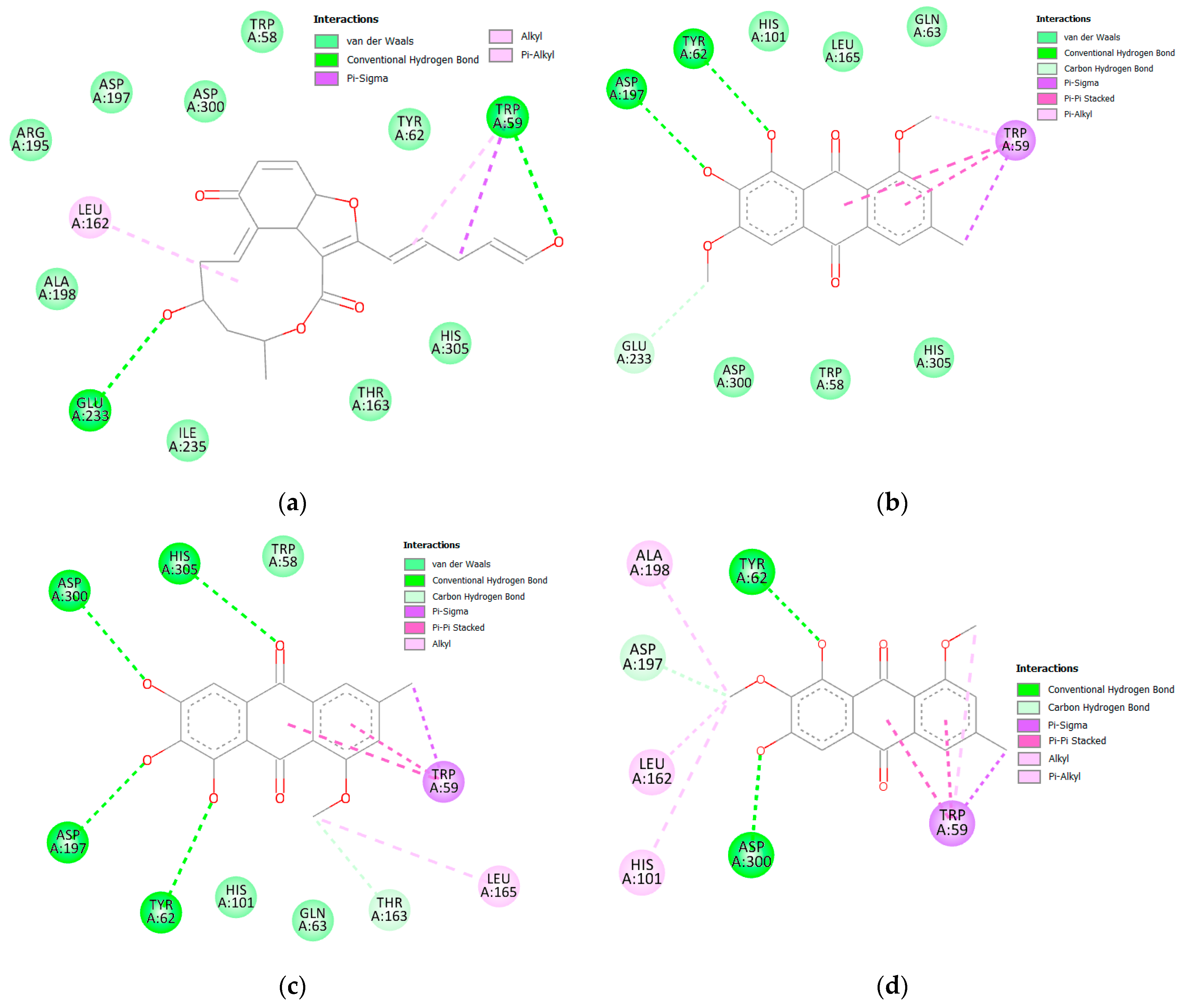
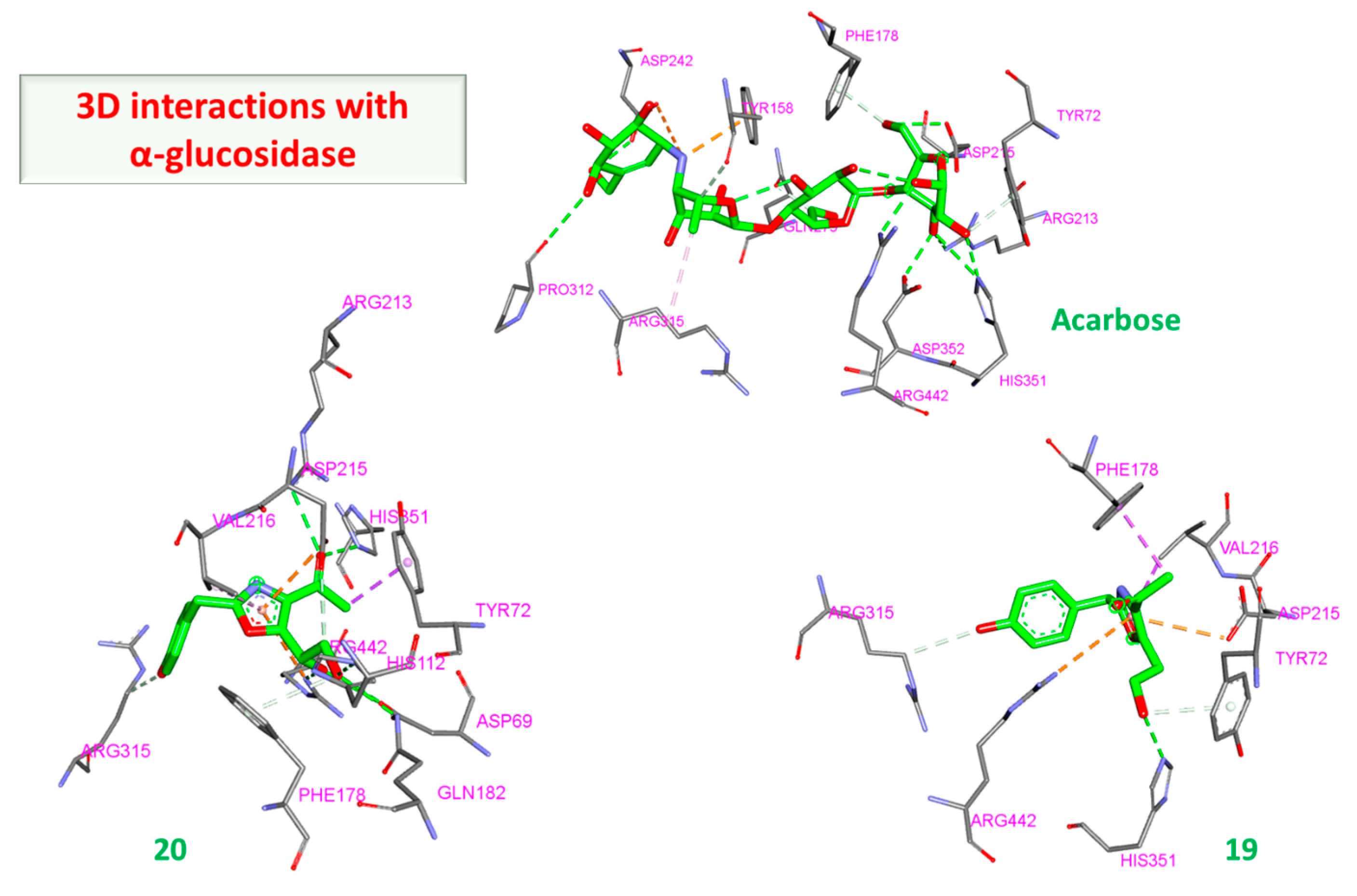
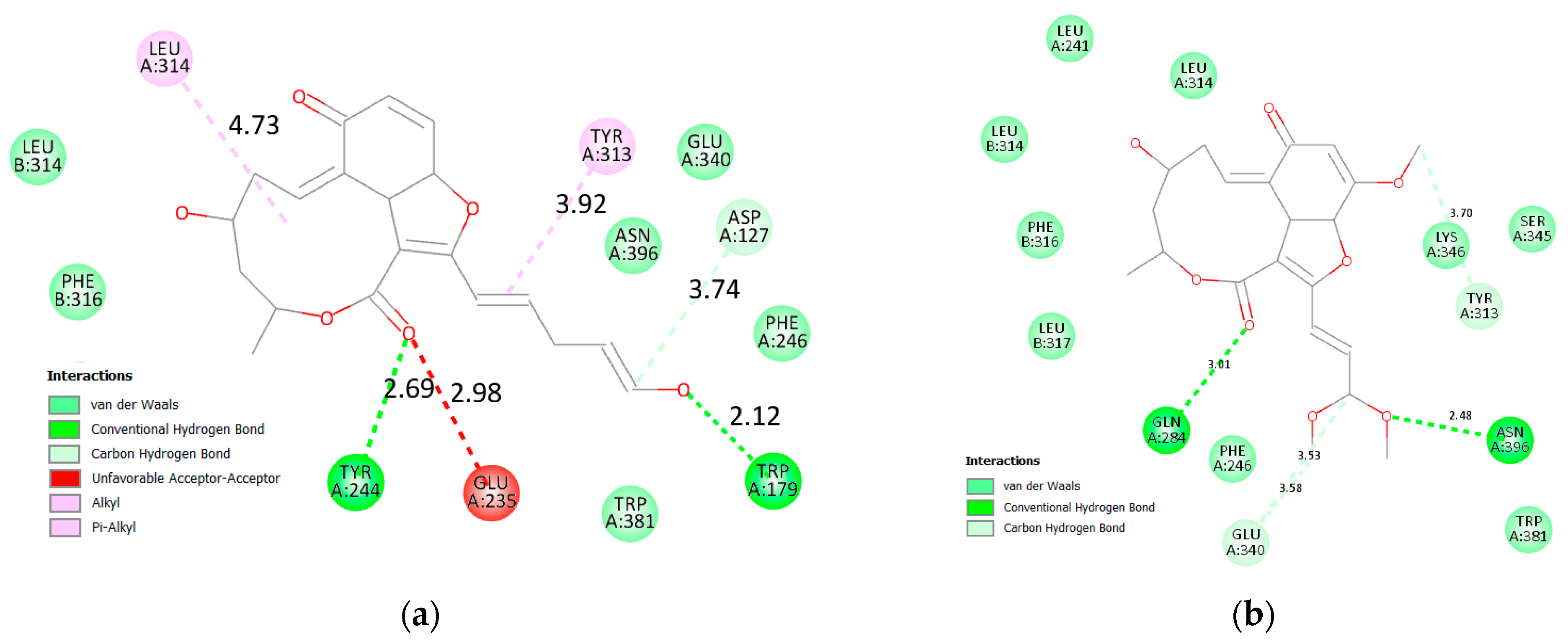
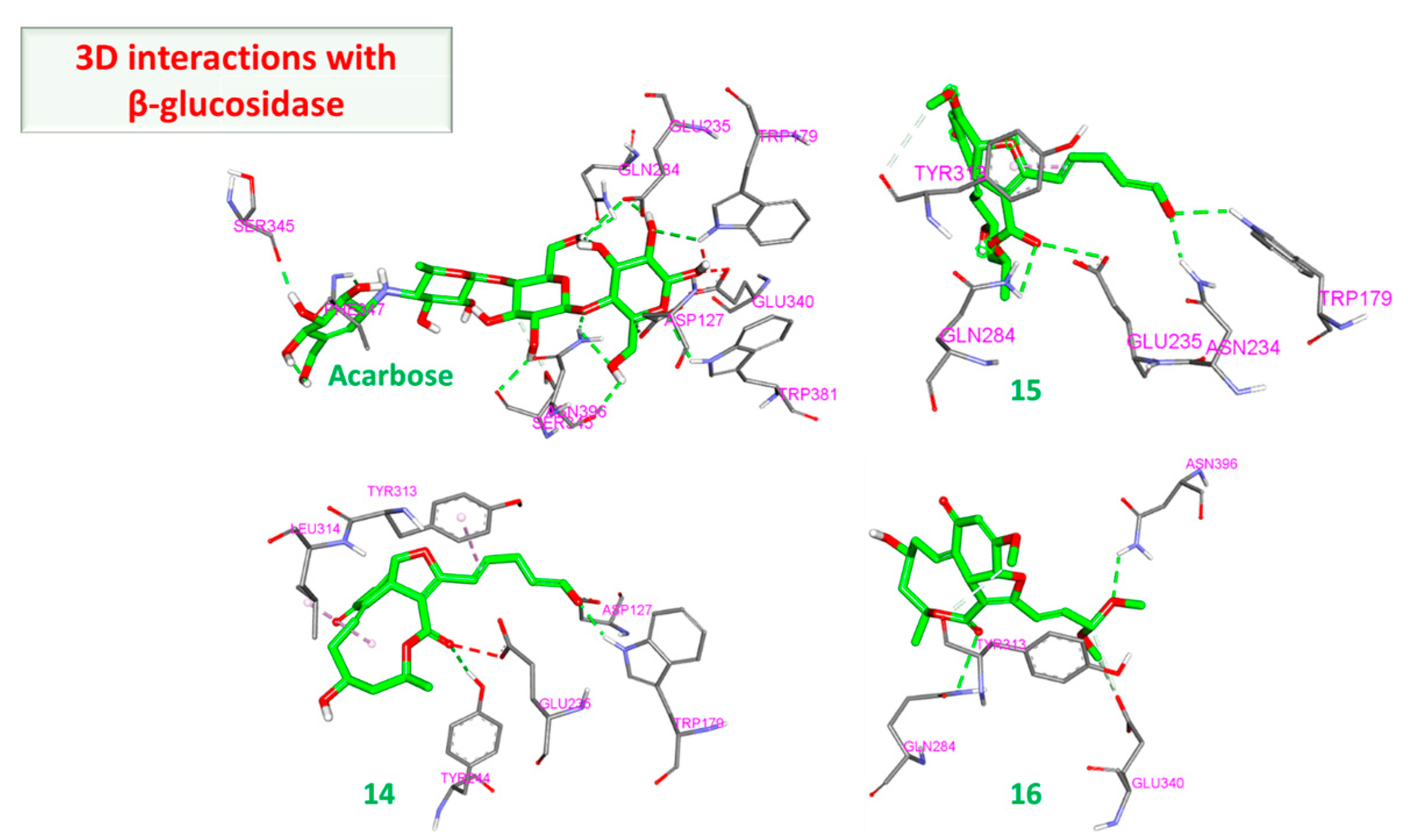
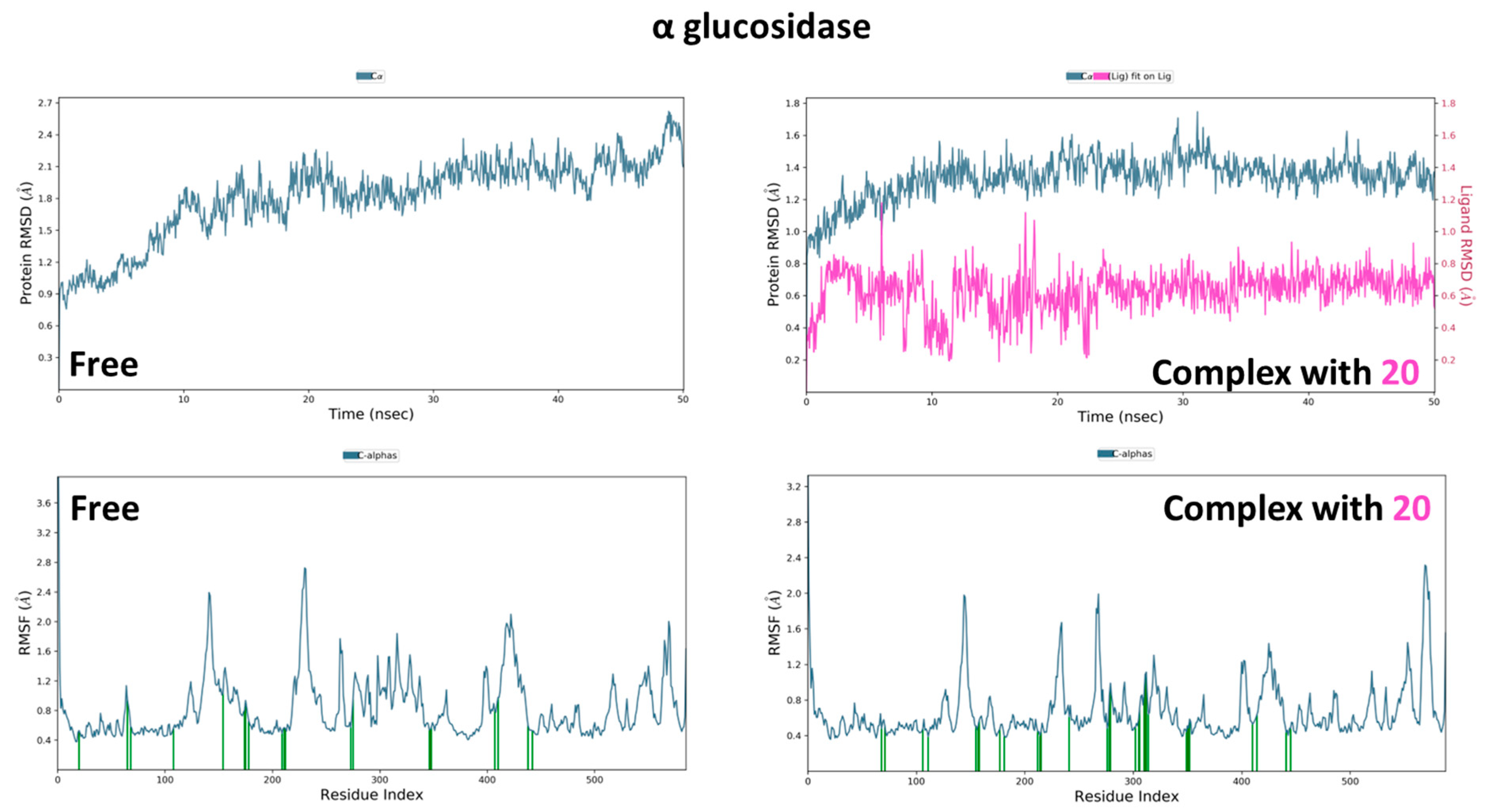
3.2.2. α and β Glucosidase Infarction
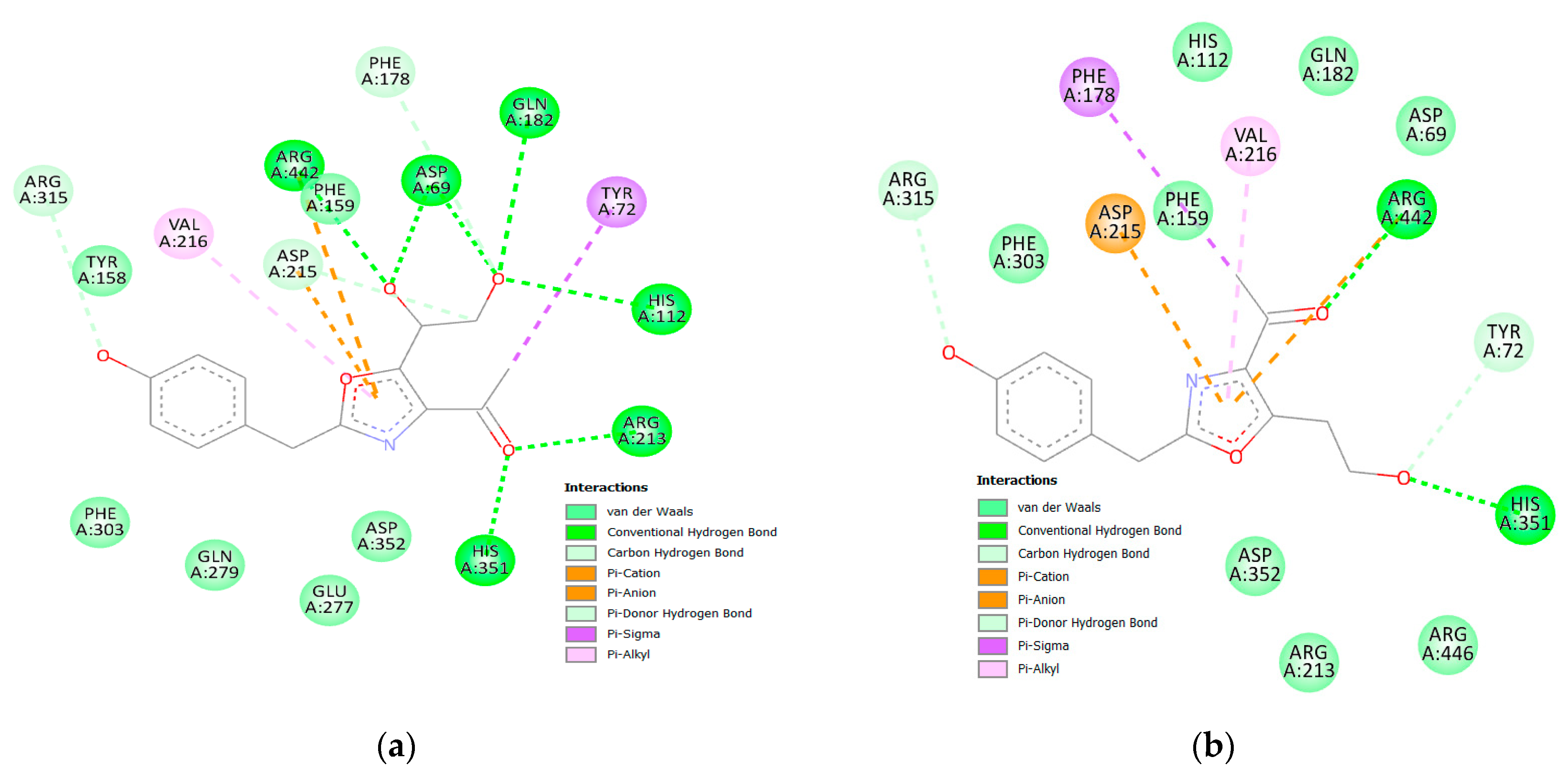
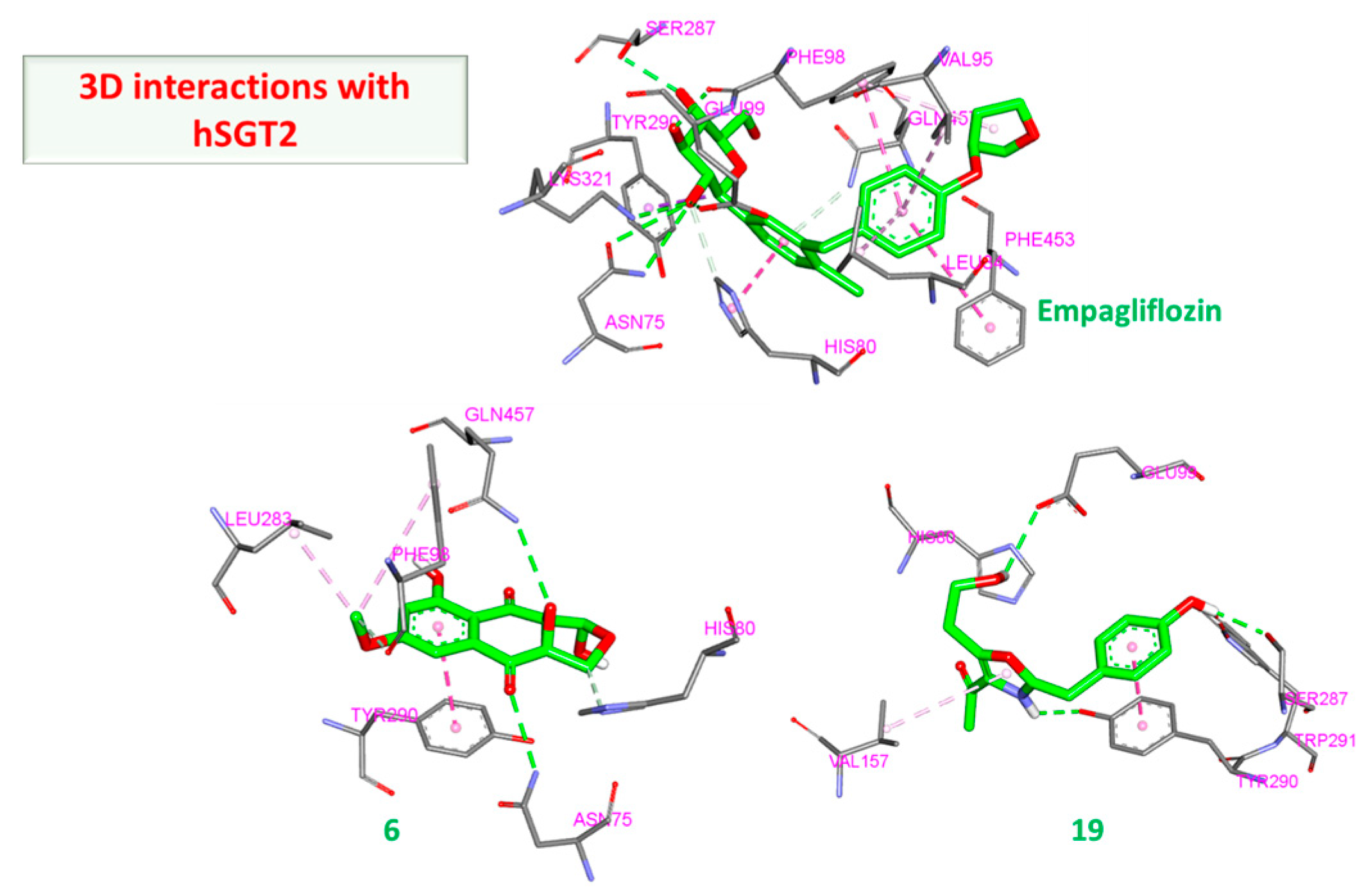
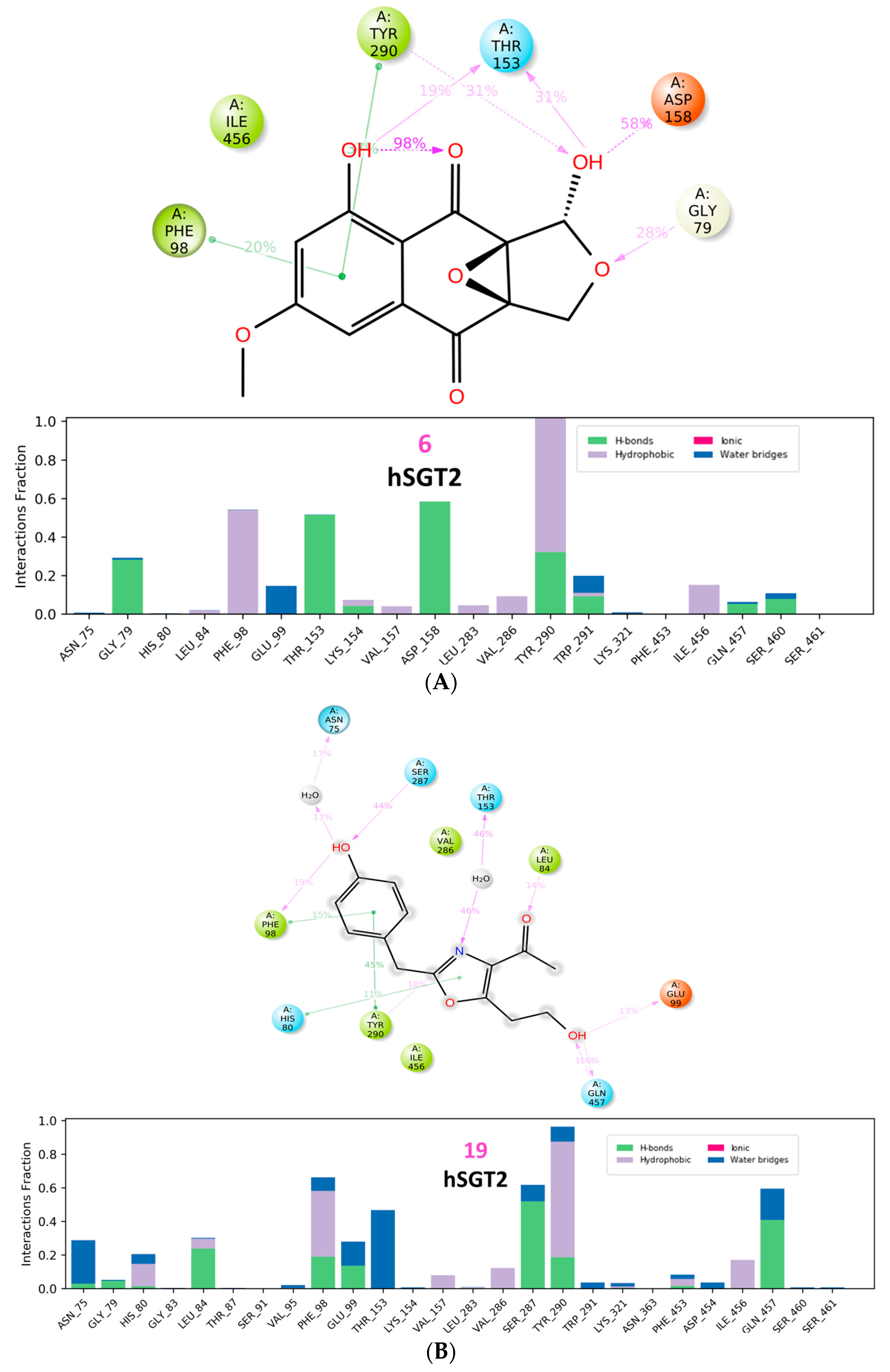
3.2.3. hSGT2 Interaction
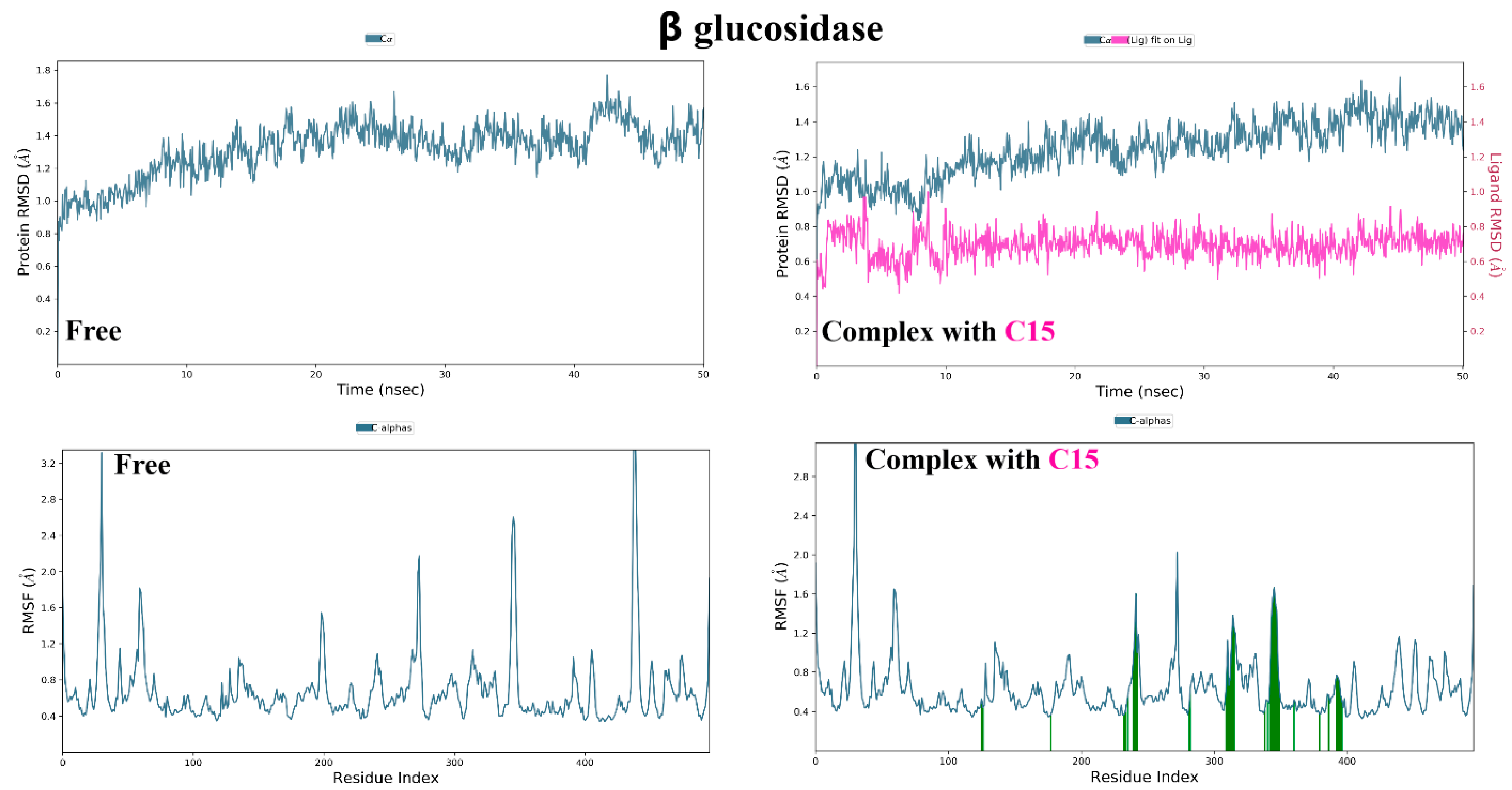
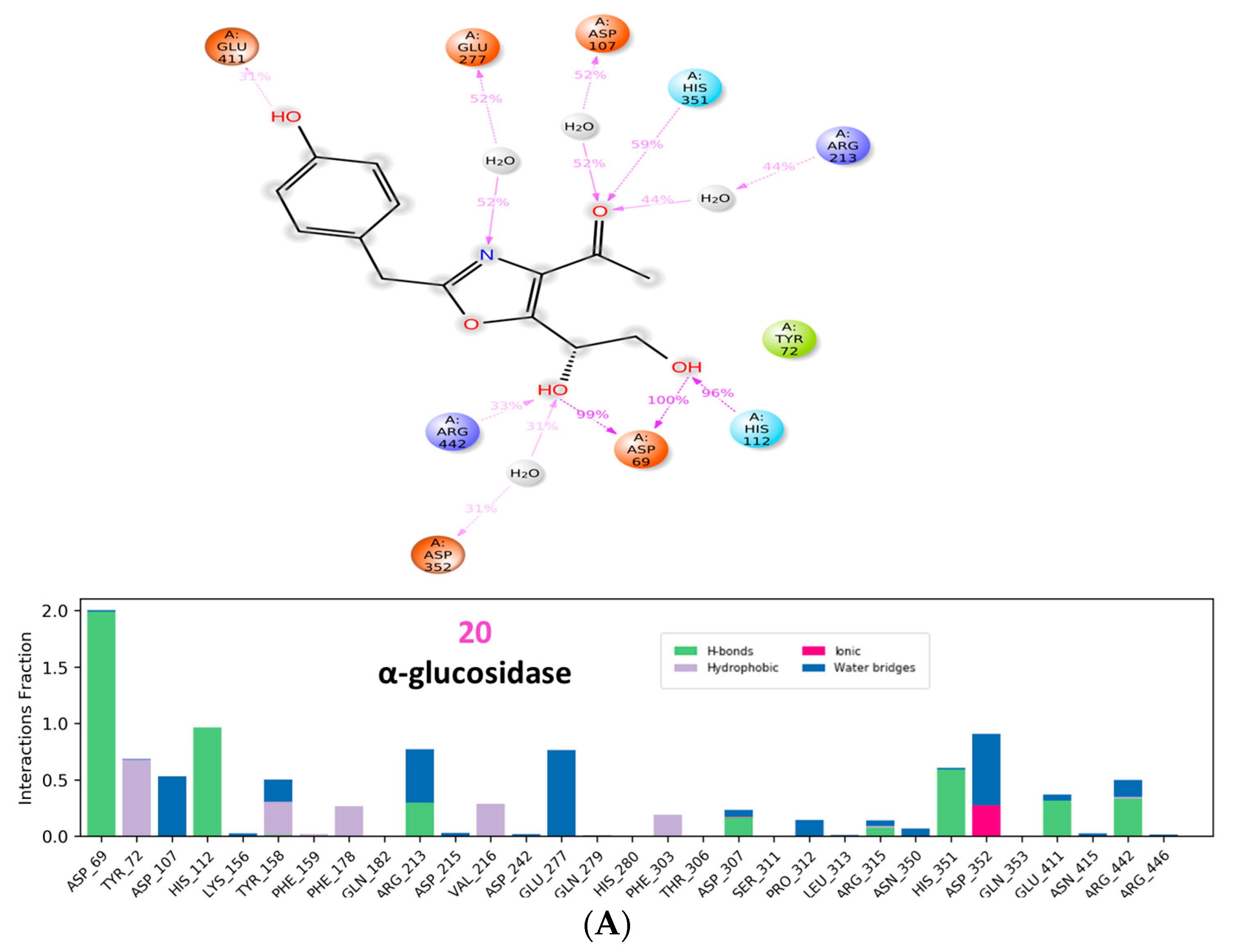
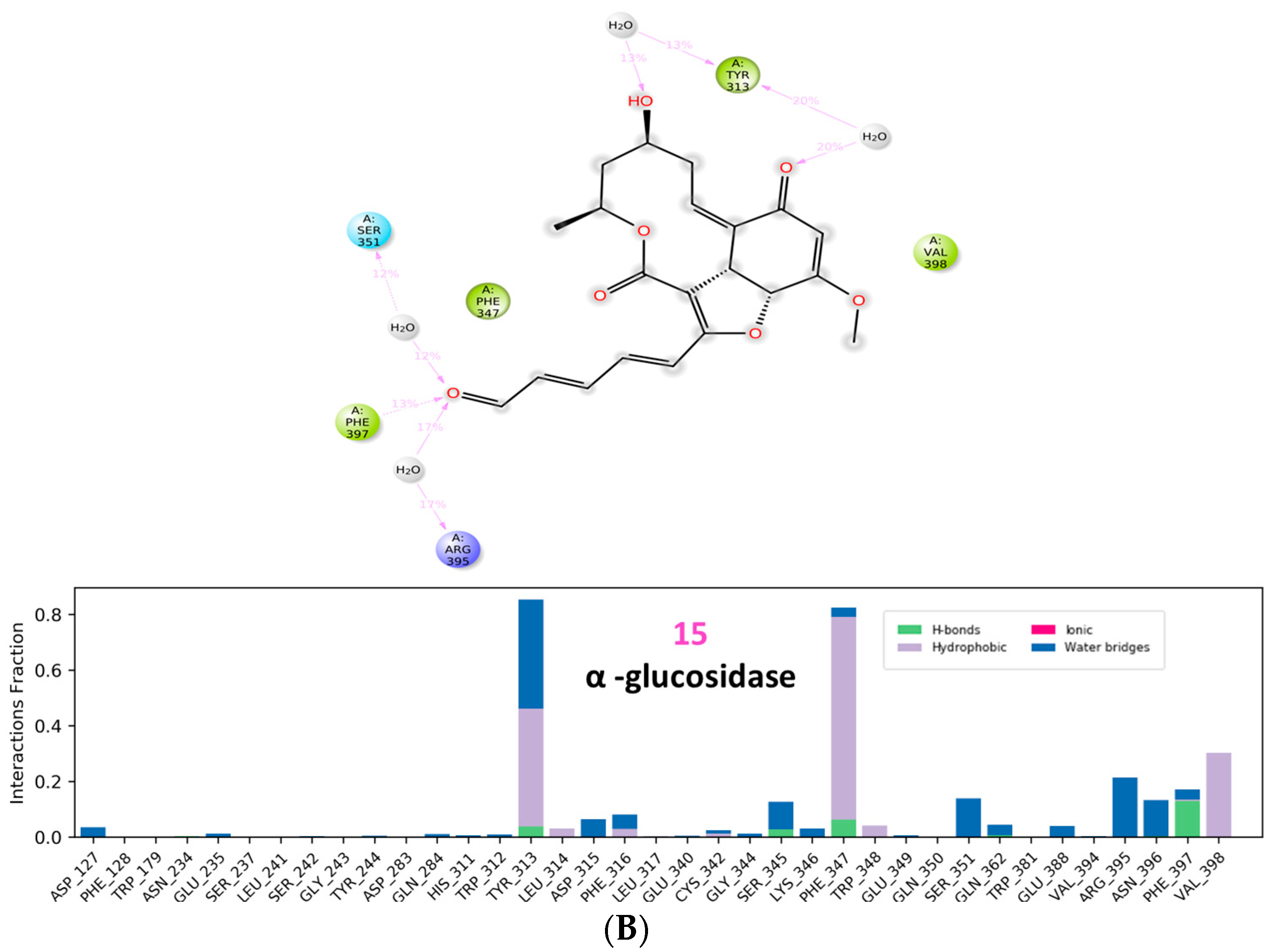
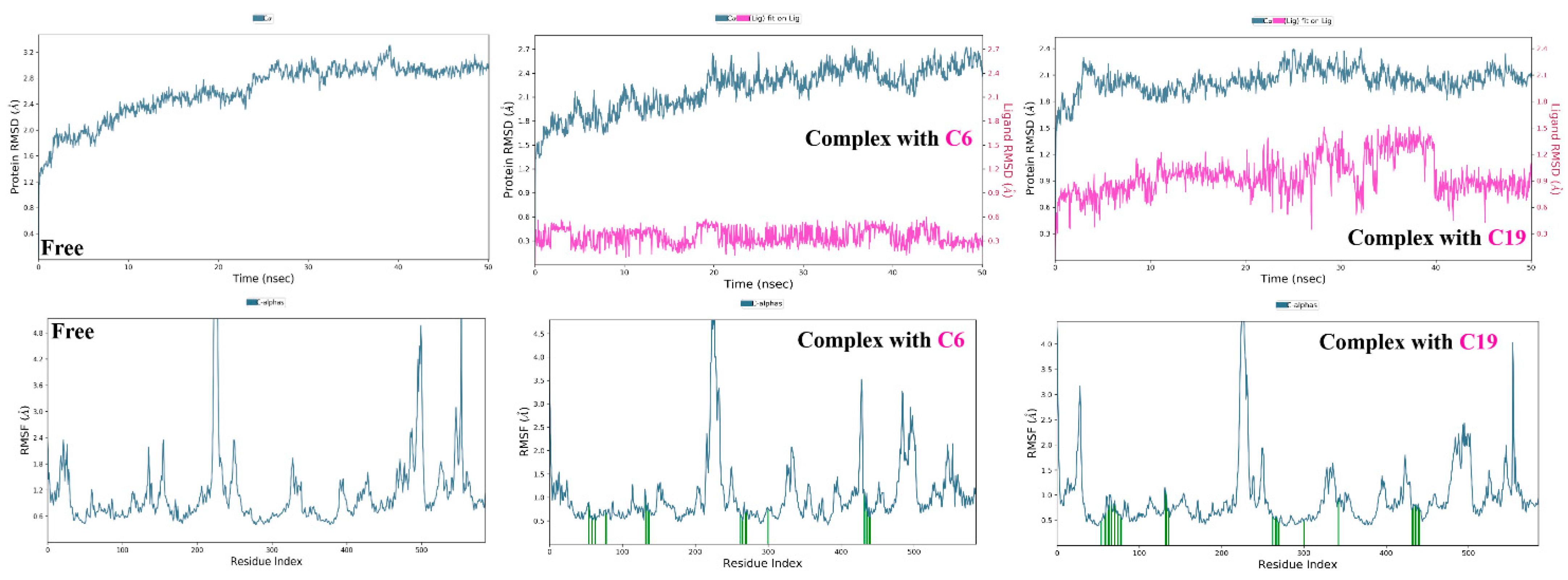
3.3. Molecular Dynamic Simulations and Generalized MMGBSA Calculations
4. Material and Methods
4.1. Eligibility Criteria for the Review
4.2. Pharmacokinetic Profiling
4.3. Molecular Docking
4.4. Molecular Dynamic Simulations and Generalized MMGBSA Calculations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vivó-Barrachina, L.; Rojas-Chacón, M.J.; Navarro-Salazar, R.; Belda-Sanchis, V.; Pérez-Murillo, J.; Peiró-Puig, A.; Herran-González, M.; Pérez-Bermejo, M. The Role of Natural Products on Diabetes Mellitus Treatment: A Systematic Review of Randomized Controlled Trials. Pharmaceutics 2022, 14, 101. [Google Scholar] [CrossRef]
- Hegazi, R.; El-gamal, M.; Abdel-hady, N. Epidemiology of and Risk Factors for Type 2 Diabetes in Egypt. Ann. Glob. Health 2015, 81, 814–820. [Google Scholar] [CrossRef]
- El-Nashar, H.A.S.; Mostafa, N.M.; El-Shazly, M.; Eldahshan, O.A. The Role of Plant-Derived Compounds in Managing Diabetes Mellitus: A Review of Literature from 2014 To 2019. Curr. Med. Chem. 2021, 28, 4694–4730. [Google Scholar] [CrossRef]
- El-Nashar, H.A.S.; Mostafa, N.M.; Eldahshan, O.A.; Singab, A.N.B. A New Antidiabetic and Anti-Inflammatory Biflavonoid from Schinus polygama (Cav.) Cabrera Leaves. Nat. Prod. Res. 2022, 36, 1182–1190. [Google Scholar] [CrossRef]
- Razek, M.M.M.A.; Moussa, A.Y.; El-Shanawany, M.A.; Singab, A.B. Comparative Chemical and Biological Study of Roots and Aerial Parts of Halocnemum Strobilaceum Growing Wildly in Egypt. J. Pharm. Sci. Res. 2019, 11, 3289–3296. [Google Scholar]
- Mostafa, N.M.; Edmond, M.P.; El-Shazly, M.; Fahmy, H.A.; Sherif, N.H.; Singab, A.N.B. Phytoconstituents and Renoprotective Effect of Polyalthia Longifolia Leaves Extract on Radiation-Induced Nephritis in Rats via TGF-β/Smad Pathway. Nat. Prod. Res. 2022, 36, 4187–4192. [Google Scholar] [CrossRef] [PubMed]
- Edmond, M.P.; Mostafa, N.M.; El-Shazly, M.; Singab, A.N.B. Two Clerodane Diterpenes Isolated from Polyalthia Longifolia Leaves: Comparative Structural Features, Anti-Histaminic and Anti-Helicobacter Pylori Activities. Nat. Prod. Res. 2021, 35, 5282–5286. [Google Scholar] [CrossRef]
- El-Nashar, H.A.S.; Mostafa, N.M.; El-Badry, M.A.; Eldahshan, O.A.; Singab, A.N.B. Chemical Composition, Antimicrobial and Cytotoxic Activities of Essential Oils from Schinus polygamus (Cav.) Cabrera Leaf and Bark Grown in Egypt. Nat. Prod. Res. 2021, 35, 5369–5372. [Google Scholar] [CrossRef]
- Ghosh, P.; Chatterjee, S.; Das, P.; Banerjee, A.; Karmakar, S.; Mahapatra, S. Natural Habitat, Phytochemistry and Pharmacological Properties of a Medicinal Weed-Cleome Rutidosperma Dc. (Cleomaceae): A Comprehensive Review. Int. J. Pharm. Sci. Res. 2019, 10, 1605. [Google Scholar] [CrossRef]
- Elkhouly, H.I.; Hamed, A.A.; El Hosainy, A.M.; Ghareeb, M.A.; Sidkey, N.M. Bioactive Secondary Metabolite from Endophytic Aspergillus Tubenginses Ash4 Isolated from Hyoscyamus Muticus: Antimicrobial, Antibiofilm, Antioxidant and Anticancer Activity. Pharmacogn. J. 2021, 13, 434–442. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. The Plant Endosphere-Hidden Treasures: A Review of Fungal Endophytes. Biotechnol. Genet. Eng. Rev. 2021, 37, 154–177. [Google Scholar] [CrossRef]
- AbdelRazek, M.M.M.; Moussa, A.Y.; El-Shanawany, M.A.; Singab, A.N.B. New Phenolic Alkaloid from Halocnemum Strobilaceum Endophytes: Antimicrobial, Antioxidant and Biofilm Inhibitory Activities. Chem. Biodivers. 2020, 17, e2000496. [Google Scholar] [CrossRef]
- AbdelRazek, M.M.M.; Moussa, A.Y.; El-Shanawany, M.A.; Singab, A.N.B. Effect of Changing Culture Media on Metabolites of Endophytic Fungi from Halocnemum Strobilaceum. Arch. Pharm. Sci. Ain Shams Univ. 2020, 4, 135–144. [Google Scholar] [CrossRef]
- Caruso, G.; Abdelhamid, M.T.; Kalisz, A.; Composition, M. Linking Endophytic Fungi to Medicinal Plants Therapeutic Activity. A Case Study on Asteraceae. Agriculture 2020, 10, 286. [Google Scholar] [CrossRef]
- Khan, R.; Tahira, S.; Naqvi, Q.; Fatima, N. Study of Antidiabetic Activities of Endophytic Fungi Isolated from Plants. Pure Appl. Biol. 2019, 8, 1287–1295. [Google Scholar] [CrossRef]
- Hussain, H.; Nazir, M.; Saleem, M.; Green, E.I.R. Fruitful Decade of Fungal Metabolites as Anti-Diabetic Agents from 2010 to 2019: Emphasis on α-Glucosidase Inhibitors; Springer: Dordrecht, The Netherlands, 2021; Volume 20, ISBN 0123456789. [Google Scholar]
- Agrawal, S.; Samanta, S.; Deshmukh, S.K. The Antidiabetic Potential of Endophytic Fungi: Future Prospects as Therapeutic Agents. Biotechnol. Appl. Biochem. 2022, 69, 1159–1165. [Google Scholar] [CrossRef]
- Govindappa, M.; Thanuja, V.; Tejashree, S.; Soukhya, C.A.; Suresh, B.; Arthikala, M.; Ravishankar Rai, V. In Vitro and In Silico Antioxidant, Anti-Diabetic, Anti-HIV and Anti- Alzheimer Activity of Endophytic Fungi, Cladosporium Uredinicola Phytochemicals. Int. J.Pharmacol. Phytochem. Ethnomed. 2019, 13, 13–34. [Google Scholar] [CrossRef]
- Lukša, J.; Servienė, E. White Mulberry (Morus alba L.) Fruit-Associated Bacterial and Fungal Microbiota. J. Environ. Eng. Landsc. Manag. 2022, 28, 183–191. [Google Scholar] [CrossRef]
- Yan, J.; Ruan, J.; Huang, P.; Sun, F.; Zheng, D.; Zhang, Y.; Wang, T. The Structure—Activity Relationship Review of the Main Bioactive Constituents of Morus Genus Plants. J. Nat. Med. 2020, 74, 331–340. [Google Scholar] [CrossRef]
- Singab, A.N.B.; Ayoub, N.A.; Ali, E.N.; Mostafa, N.M. Antioxidant and Hepatoprotective Activities of Egyptian Moraceous Plants against Carbon Tetrachloride-Induced Oxidative Stress and Liver Damage in Rats. Pharm. Biol. 2010, 48, 1255–1264. [Google Scholar] [CrossRef]
- Memon, A.A.; Memon, N.; Luthria, D.L.; Bhanger, M.I.; Pitafi, A.A. Phenolic Acids Profiling and Antioxidant Potential of Mulberry (Morus laevigata W., Morus nigra L., Morus alba L.) Leaves and Fruits Grown in Pakistan. Pol. J. Food Nutr. Sci. 2010, 60, 25–32. [Google Scholar]
- Jan, B.; Zahiruddin, S.; Basist, P.; Irfan, M.; Abass, S.; Ahmad, S. Metabolomic Profiling and Identification of Antioxidant and Antidiabetic Compounds from Leaves of Different Varieties of Morus alba Linn Grown in Kashmir. ACS Omega 2022, 7, 24317–24328. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Kwon, Y.; Jang, H. Mulberry Leaf Extract Reduces Postprandial Hyperglycemia with Few Side Effects by Inhibiting a -Glucosidase in Normal Rats. J. Med. Food 2011, 14, 712–717. [Google Scholar] [CrossRef]
- Santini, A.; Tenore, G.C.; Novellino, E. Nutraceuticals: A Paradigm of proactive Medicine Antonello. Eur. J. Pharm. Sci. 2017, 96, 53–61. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Durazzo, A.; Lucarini, M.; Souto, E.B.; Santini, A.; Imran, M.; Moussa, A.Y.; Mostafa, N.M.; El-Shazly, M. Resveratrol’ Biotechnological Applications: Enlightening Its Antimicrobial and Antioxidant Properties. J. Herb. Med. 2022, 32, 100550. [Google Scholar] [CrossRef]
- Abdallah, S.H.; Mostafa, N.M.; Mohamed, M.A.E.H.; Nada, A.S.; Singab, A.N.B. UPLC-ESI-MS/MS Profiling and Hepatoprotective Activities of Stevia Leaves Extract, Butanol Fraction and Stevioside against Radiation-Induced Toxicity in Rats. Nat Prod Res 2022, 36, 5619–5625. [Google Scholar] [CrossRef]
- Lim, S.H.; Yu, J.S.; Lee, H.S.; Choi, C.; Kim, K.H. Antidiabetic Flavonoids from Fruits of Morus alba Promoting Insulin-Stimulated Glucose Uptake via Akt and AMP-Activated Protein Kinase Activation in 3T3-L1 Adipocytes. Pharmaceutics 2021, 13, 526. [Google Scholar] [CrossRef]
- Su, C.; Tao, X.; Yin, Z.; Zhang, X.; Tian, J.; Chen, R.; Liu, J.; Li, L.; Ye, F.; Zhang, P.; et al. Morusalones A−D, Diels−Alder Adducts with 6/7/6/6/6/6 Hexacyclic Ring Systems as Potential PTP1B Inhibitors from Cell Cultures of Morus alba. Org. Lett. 2019, 21, 9463–9467. [Google Scholar] [CrossRef]
- Xu, L.; Yu, M.; Niu, L.; Huang, C.; Wang, Y. Phenolic Compounds Isolated from Morus nigra and Their α -Glucosidase Inhibitory Activities. Nat. Prod. Res. 2018, 34, 605–612. [Google Scholar] [CrossRef]
- Khedr, S.A. Anti-Diabetic Effect of Black Mulberry Leaves (Morus nigra L.) in Streptozotocin-Induced Diabetic Rats. J. Home Econ. 2016, 26, 159–181. [Google Scholar]
- Kwon, R.; Thaku, N.; Timalsina, B.; Park, S.; Choi, J. Inhibition Mechanism of Components Isolated from Morus alba Branches on Diabetes and Diabetic Complications via Experimental and Molecular Docking Analyses. Antioxidants 2022, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, N.; Singab, A.N.; Mostafa, N.; Schultze, W. Volatile Constituents of Leaves of Ficus Carica Linn. Grown in Egypt. J. Essent. Oil-Bear. Plants 2010, 13, 316–321. [Google Scholar] [CrossRef]
- Jayant, K.K.; Vijayakumar, B.S. In-Vitro Anti-Oxidant and Anti-Diabetic Potential of Endophytic Fungi Associated with Ficus Religiosa. Ital. J. Mycol. 2021, 50, 10–20. [Google Scholar] [CrossRef]
- Brumshtein, B.; Aguilar-Moncayo, M.; Benito, J.M.; García Fernandez, J.M.; Silman, I.; Shaaltiel, Y.; Aviezer, D.; Sussman, J.L.; Futerman, A.H.; Ortiz Mellet, C. Cyclodextrin-Mediated Crystallization of Acid β-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues. Org. Biomol. Chem. 2011, 9, 4160. [Google Scholar] [CrossRef] [PubMed]
- Bindu, J.; Narendhirakannan, R.T. Role of Medicinal Plants in the Management of Diabetes Mellitus: A Review. 3 Biotech 2019, 9, 4. [Google Scholar] [CrossRef]
- Swilam, N.; Nawwar, M.A.M.; Radwan, R.A.; Mostafa, E.S. Antidiabetic Activity and In Silico Molecular Docking of Polyphenols from Ammannia baccifera L. Subsp. Aegyptiaca (Willd.) Koehne Waste: Structure Elucidation of Undescribed Acylated Flavonol Diglucoside. Plants 2022, 11, 452. [Google Scholar] [CrossRef]
- Melo, E.; De, B.; Carvalho, I. Alpha- and Beta-Glucosidase Inhibitors: Chemical Structure and Biological Activity. Tetrahedron 2006, 62, 10277–10302. [Google Scholar] [CrossRef]
- Sakulkeo, O.; Wattanapiromsakul, C.; Pitakbut, T.; Dej-adisai, S. Alpha-Glucosidase Inhibition and Molecular Docking of Isolated Compounds from Traditional Thai Medicinal Plant, Neuropeltis Racemosa Wall. Molecules 2022, 27, 639. [Google Scholar] [CrossRef]
- Feng, R.; Dong, L.; Wang, L.; Xu, Y.; Lu, H.; Zhang, J. Development of Sodium Glucose Co-Transporter 2 (SGLT2) Inhibitors with Novel Structure by Molecular Docking and Dynamics Simulation. J. Mol. Model. 2019, 25, 175. [Google Scholar] [CrossRef]
- Norton, L.; Shannon, C.E.; Fourcaudot, M.; Hu, C.; Wang, N.; Ren, W.; Song, J.; Abdul-Ghani, M.; Defronzo, R.A.; Ren, J.; et al. Sodium-Glucose Co-Transporter (SGLT) and Glucose Transporter ( GLUT ) Expression in the Kidney of Type 2 Diabetic Subjects. Diabetes Obes. Metab. 2017, 19, 1322–1326. [Google Scholar] [CrossRef]
- Sato, S.; Takeo, J.; Aoyama, C.; Kawahara, H. Na+-Glucose Cotransporter (SGLT) Inhibitory Flavonoids from the Roots of Sophora flavescens. Bioorg. Med. Chem. 2007, 15, 3445–3449. [Google Scholar] [CrossRef]
- Choi, C.-I. Sodium-Glucose Cotransporter 2 (SGLT2) Inhibitors from Natural Products: Discovery of Next-Generation Antihyperglycemic Agents. Molecules 2016, 21, 1136. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Lee, C.; Lee, D.; Kim, S.; Bang, S.; Shin, M.-S.; Lee, J.; Kang, K.S.; Shim, S.H. Neuroprotective Compound from an Endophytic Fungus, Colletotrichum Sp JS-0367. J. Nat. Prod. 2018, 81, 1411–1416. [Google Scholar] [CrossRef]
- Zheng, L.P.; Zhang, Z.; Xie, L.Q.; Yuan, H.Y.; Zhang, Y.Q. Antifungal Activity of Endophyte Cultures of Morus alba L. against Phytopathogenic Fungi. Adv. Mat. Res. 2013, 642, 615–618. [Google Scholar] [CrossRef]
- Bang, S.; Eun, H.; Yun, J.; Sik, D.; Kim, S.; Nam, S.; Lee, D.; Sung, K.; Hee, S. Colletotrichalactones A-Ca, Unusual 5/6/10-Fused Tricyclic Polyketides Produced by an Endophytic Fungus, Colletotrichum sp. JS-0361. Bioorg. Chem. 2020, 105, 104449. [Google Scholar] [CrossRef]
- Choi, H.G.; Song, J.H.; Park, M.; Kim, S.; Kim, C.; Kang, K.S.; Shim, S.H. Neuroprotective γ-Pyrones from Fusarium Solani JS-0169: Cell-Based Identification of Active Compounds and an Informatics Approach to Predict the Mechanism of Action. Biomolecules 2020, 10, 91. [Google Scholar] [CrossRef]
- Ku, H.; Baek, J.; Kang, K.S.; Shim, S.H. A New Anti-Proliferative Compound from an Endophytic Fungus, Phoma sp. Nat. Prod. Res. 2021, 36, 5584–5590. [Google Scholar] [CrossRef]
- Vora, J.; Velhal, S.; Sinha, S.; Patel, V.; Shrivastava, N. Bioactive Phytocompound Mulberroside C and Endophytes of Morus alba as Potential Inhibitors of HIV-1 Replication: A Mechanistic Evaluation. HIV Med. 2021, 22, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Ayesha, R.; Iftikhar, T. New Fungal Records on Morus alba from Faisalabad Pakistan I. Pak. J. Bot. 2010, 42, 583–592. [Google Scholar]
- Aparecida, A.; Polonio, J.C.; Bulla, A.M.; Polli, D.; Castro, J.C.; Soares, L.C.; De, V.A.; Elisa, V.; Vicentini, P.; José, A.; et al. Antimicrobial and Antioxidant Activities of Secondary Metabolites from Endophytic Fungus Botryosphaeria Fabicerciana (MGN23-3) Associated to Morus nigra L. Antimicrobial and Antioxidant Activities of Secondary Fabicerciana (MGN23-3) Associated to Morus. Nat. Prod. Res. 2021, 36, 3158–3162. [Google Scholar] [CrossRef]
- Hermawati, E.; Juliawaty, L.D.; Hakim, E.H. A Quinone Derivative from an Endophytic Fungus Phomopsis sp. From Morus cathayana. Rec. Nat. Prod. 2017, 11, 315–317. [Google Scholar]
- Hermawati, E.; Ellita, S.D.; Juliawaty, L.D.; Hakim, E.H.; Syah, Y.M.; Ishikawa, H. Epoxyquinophomopsins A and B from Endophytic Fungus Phomopsis sp. and Their Activity against Tyrosine Kinase. J. Nat. Med. 2021, 75, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, J.; Motohashi, K.; Sakamoto, K.; Hashimoto, S.; Yamanouchi, M.; Tanaka, H.; Takahashi, T.; Takagi, M.; Shin-Ya, K. Screening and Evaluation of New Inhibitors of Hepatic Glucose Production. J. Antibiot. 2009, 62, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Nguyen, Q.N.; Phung, H.M.; Shim, S.H.; Kim, D.; Hwang, G.S.; Kang, K.S. Preventive Effects of Anthraquinones Isolated from an Endophytic Fungus, Colletotrichum sp. JS-0367 in Tumor Necrosis Factor-α-Stimulated Damage of Human Dermal Fibroblasts. Antioxidants 2021, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Pranay Kumar, K.; Javvaji, K.; Poornachandra, Y.; Allanki, A.D.; Misra, S. Antimicrobial, Anti-Plasmodial and Cytotoxicity Properties of Bioactive Compounds from Fusarium sp. USNPF102. J. Microbiol. Res. 2017, 2017, 23–30. [Google Scholar] [CrossRef]
- Hridoy, M.; Gorapi, M.Z.H.; Noor, S.; Chowdhury, N.S.; Rahman, M.M.; Muscari, I.; Masia, F.; Adorisio, S.; Delfino, D.V.; Mazid, M.A. Putative Anticancer Compounds from Plant-Derived Endophytic Fungi: A Review. Molecules 2022, 27, 296. [Google Scholar] [CrossRef]
- Kemkuignou, B.M.; Treiber, L.; Zeng, H.; Schrey, H.; Schobert, R.; Stadler, M. Macrooxazoles a–d, New 2,5-Disubstituted Oxazole-4-Carboxylic Acid Derivatives from the Plant Pathogenic Fungus Phoma Macrostoma. Molecules 2020, 25, 5497. [Google Scholar] [CrossRef]
- Parizadeh, H.; Garampalli, R.H. Evaluation of Some Lichen Extracts for β-Glucosidase Inhibitory as a Possible Source of Herbal Anti-Diabetic Drugs. Am. J. Biochem. 2016, 6, 46–50. [Google Scholar] [CrossRef]
- Ferrannini, E. Sodium-Glucose Co-Transporters and Their Inhibition: Clinical Physiology. Cell Metab. 2017, 26, 27–38. [Google Scholar] [CrossRef]
- Frampton, J.E. Empagliflozin: A Review in Type 2 Diabetes. Drugs 2018, 78, 1037–1048. [Google Scholar] [CrossRef]
- Ongaro, A.; Oselladore, E.; Memo, M.; Ribaudo, G.; Gianoncelli, A. Insight into the LFA-1/SARS-CoV-2 Orf7a Complex by Protein–Protein Docking, Molecular Dynamics, and MM-GBSA Calculations. J. Chem. Inf. Model. 2021, 61, 2780–2787. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. ILOGP: A Simple, Robust, and Efficient Description of n -Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Sun, X.; Belal, A.; Elanany, M.A.; Alsantali, R.I.; Alrooqi, M.M.; Mohamed, A.R.; Hasabelnaby, S. Identification of Some Promising Heterocycles Useful in Treatment of Allergic Rhinitis: Virtual Screening, Pharmacophore Mapping, Molecular Docking, and Molecular Dynamics. Russ. J. Bioorg. Chem. 2022, 48, 438–456. [Google Scholar] [CrossRef]
- Maurus, R.; Begum, A.; Williams, L.K.; Fredriksen, J.R.; Zhang, R.; Withers, S.G.; Brayer, G.D. Alternative Catalytic Anions Differentially Modulate Human α-Amylase Activity and Specificity. Biochemistry 2008, 47, 3332–3344. [Google Scholar] [CrossRef]
- Niu, Y.; Liu, R.; Guan, C.; Zhang, Y.; Chen, Z.; Hoerer, S.; Nar, H.; Chen, L. Structural Basis of Inhibition of the Human SGLT2–MAP17 Glucose Transporter. Nature 2022, 601, 280–284. [Google Scholar] [CrossRef]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal Structures of Isomaltase from Saccharomyces Cerevisiae and in Complex with Its Competitive Inhibitor Maltose. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef]
- Mostafa, N.M. Antibacterial Activity of Ginger (Zingiber Officinale) Leaves Essential Oil Nanoemulsion against the Cariogenic Streptococcus Mutans. J. Appl. Pharm. Sci. 2018, 8, 34–41. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Methods in Molecular Biology; Springer: Clifton, NJ, USA, 2015; Volume 1263, pp. 243–250. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Chemical Computing Group Molecular Operating Environment (MOE) 2008.10. Available online: www.chemcomp.com (accessed on 10 September 2022).
- Dassault Systèmes BIOVIA Discovery Studio Visualizer 2021. Available online: www.3ds.com/products-services/biovia/ (accessed on 10 September 2022).
- Younis, M.M.; Ayoub, I.M.; Mostafa, N.M.; El Hassab, M.A.; Eldehna, W.M.; Al-Rashood, S.T.; Eldahshan, O.A. GC/MS Profiling, Anti-Collagenase, Anti-Elastase, Anti-Tyrosinase and Anti-Hyaluronidase Activities of a Stenocarpus sinuatus Leaves Extract. Plants 2022, 11, 918. [Google Scholar] [CrossRef]
- Mostafa, N.M.; Mostafa, A.M.; Ashour, M.L.; Elhady, S.S. Neuroprotective Effects of Black Pepper Cold-Pressed Oil on Scopolamine-Induced Oxidative Stress and Memory Impairment in Rats. Antioxidants 2021, 10, 1993. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, N.M. β-Amyrin Rich Bombax Ceiba Leaf Extract with Potential Neuroprotective Activity against Scopolamine-Induced Memory Impairment in Rats. Rec. Nat. Prod. 2018, 12, 480. [Google Scholar] [CrossRef]
- Moussa, A.Y.; Mostafa, N.M.; Singab, A.N.B. Pulchranin A: First Report of Isolation from an Endophytic Fungus and Its Inhibitory Activity on Cyclin Dependent Kinases. Nat. Prod. Res. 2020, 34, 2715–2722. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Vanden Broeck, A.; Lotz, C.; Drillien, R.; Haas, L.; Bedez, C.; Lamour, V. Structural Basis for Allosteric Regulation of Human Topoisomerase IIα. Nat. Commun. 2021, 12, 2962. [Google Scholar] [CrossRef]
- Belal, A.; Elanany, M.A.; Santali, E.Y.; Al-Karmalawy, A.A.; Aboelez, M.O.; Amin, A.H.; Abdellattif, M.H.; Mehany, A.B.M.; Elkady, H. Screening a Panel of Topical Ophthalmic Medications against MMP-2 and MMP-9 to Investigate Their Potential in Keratoconus Management. Molecules 2022, 27, 3584. [Google Scholar] [CrossRef]

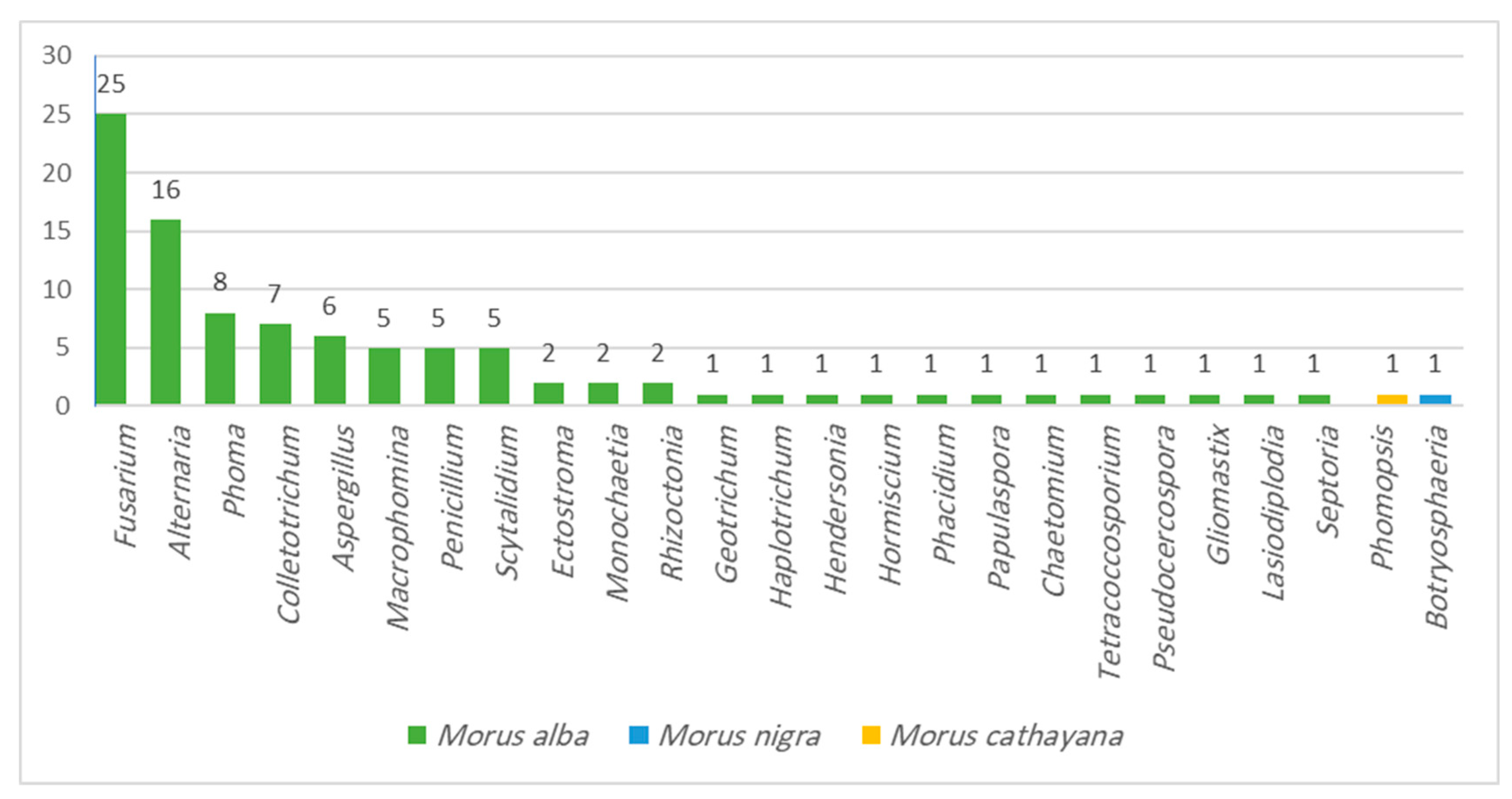
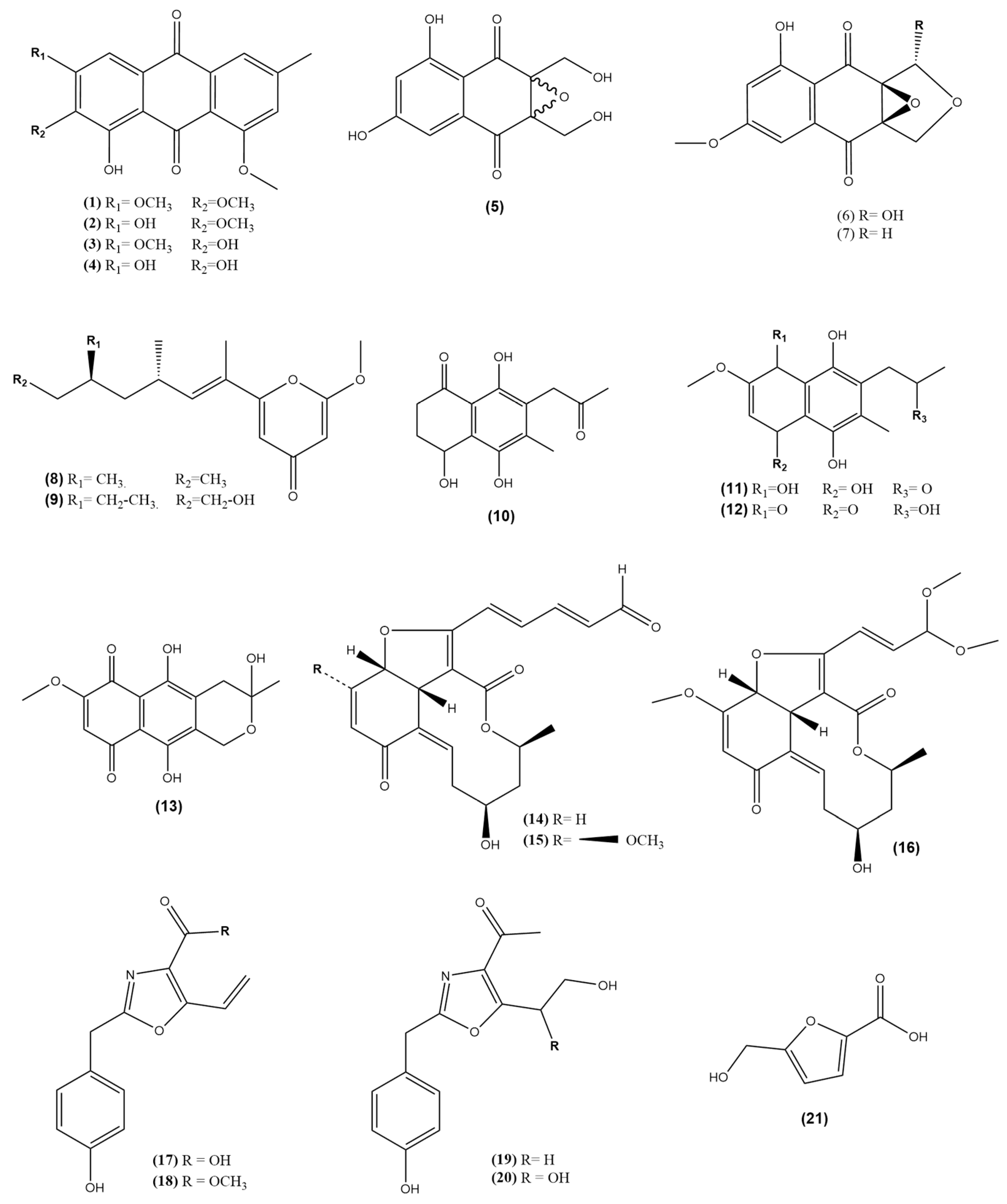
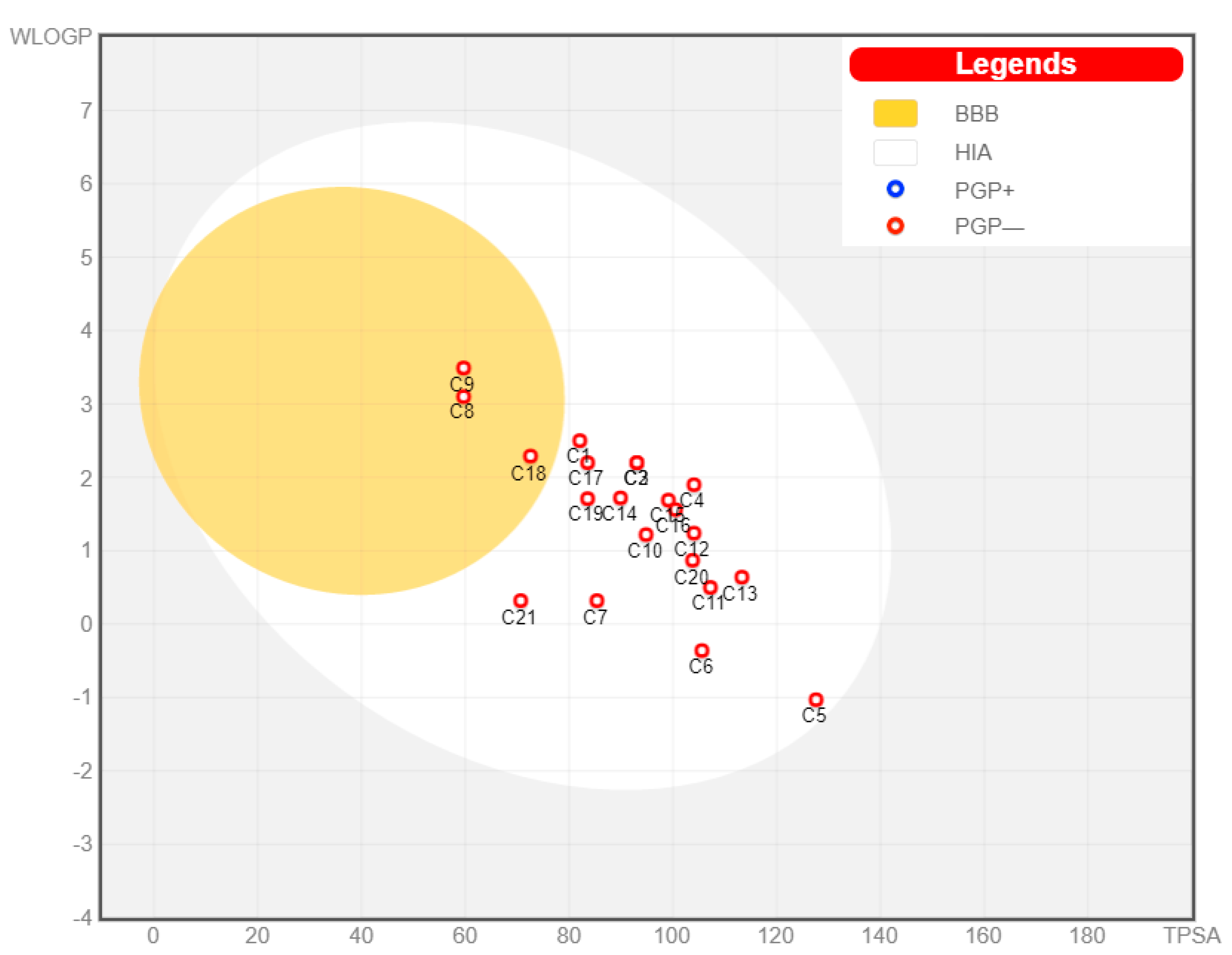
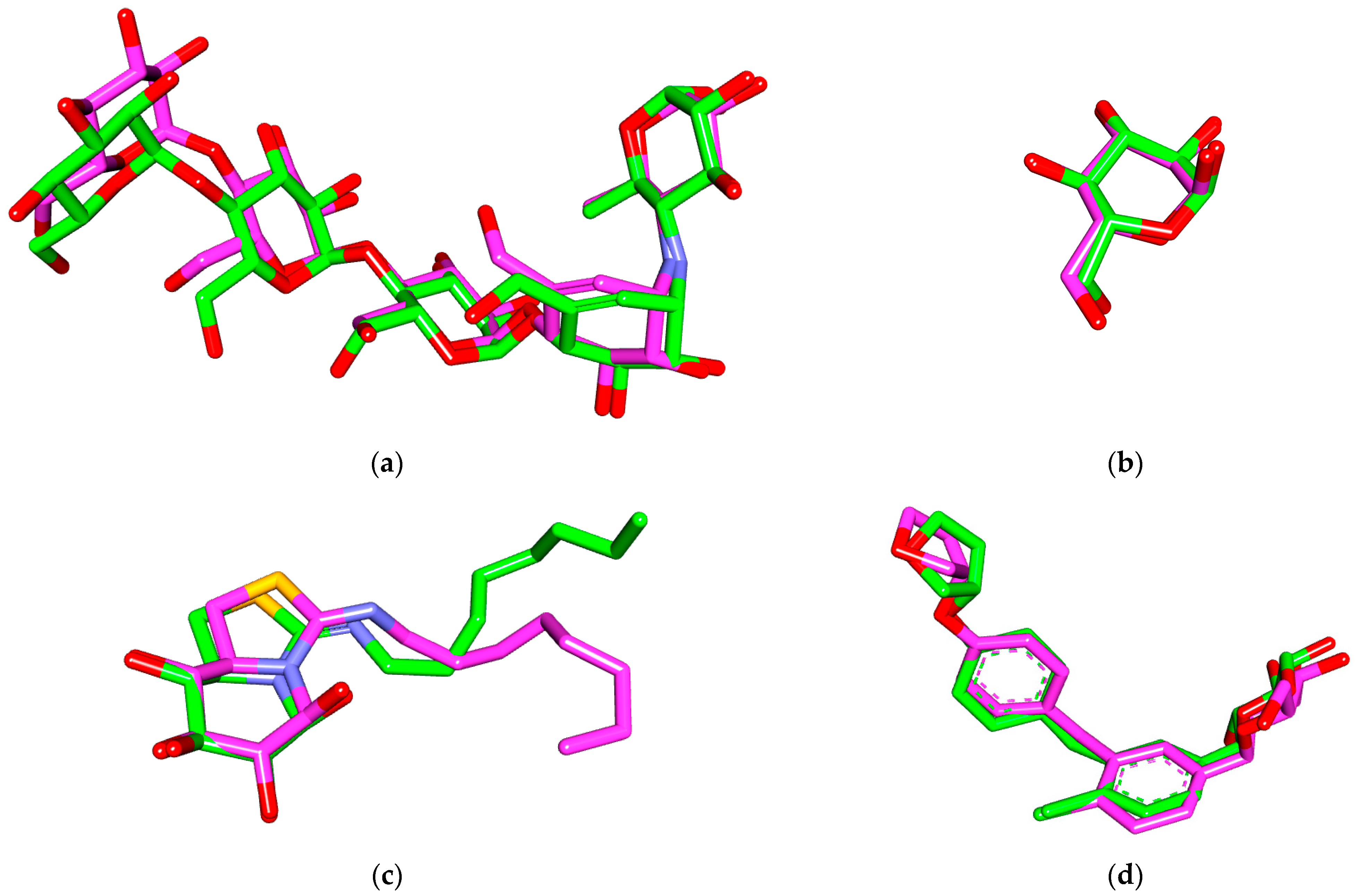



| No | Compound Class/Name | M.W. | Reported Biological Assay | Biological Activity | Source | References |
|---|---|---|---|---|---|---|
| Anthraquinone | ||||||
| 1 | 1-hydroxy-2,3,8- trimethoxy-6-methyl anthraquinone | 328.32 | ROS (EC20) NO (EC50) PGE2 (EC20) Neuroprotective (HT22 Viability%) | 100 µM >100 µM >100 µM 75% at conc. 12 µM | Colletotrichum sp. JS-0367 associated with M. alba Leaves (South Korea) | [44,55] |
| 2 | 1,3-dihydroxy-2,8-dimethoxy-6-methyl anthraquinone | 314.29 | ROS (EC20) NO (EC50) PGE2 (EC20) Neuroprotective (HT22 Viability%) | 51.1 µM 27 µM 49.5 µM 42% at conc. 12 µM | ||
| 3 | 1,2-dihydroxy-3,8- dimethoxy-6-methyl anthraquinone | 314.29 | ROS (EC20) NO (EC50) PGE2 (EC20) Neuroprotective (HT22 Viability%) | >100 µM >100 µM 75.3 µM 30% at conc. 12 µM | ||
| 4 | Evariquinone | 300.27 | ROS (EC20) NO (EC50) PGE2 (EC20) Neuroprotective (HT22 Viability%) Antioxidant DPPH (IC50) | 71.2 µM >100 µM >100 µM 50% at conc. 12 µM 42.2 μM | ||
| Quinone | ||||||
| 5 | Epoxyquinophomopsin | 266.21 | - | - | Phomopsis sp. AZ1a associated with M. cathayana Twigs (Indonesia) | [52,53] |
| 6 | Epoxyquinophomopsin A | 278.22 | TK inhibition (%) | 16–20% | ||
| 7 | Epoxyquinophomopsin B | 262.22 | TK inhibition (%) | 19–20% | ||
| Pyrone | ||||||
| 8 | 6-((9‵R,11‵R, E)-13-hydroxy-9,11-dimethyloct-7-en-7-yl)-2-methoxy-4H-pyran-4-one | 280.36 | Hepatoprotective (HT22 Viability%) | 41% at conc. 12.5 µM | Fusarium Solani JS-0169 associated with M. alba leaves (South Korea) | [47,54,56] |
| 9 | Fusarester D | 294.39 | Neuroprotective (HT22 Viability%) | <1% at conc. 12.5 µM | ||
| Naphthoquinones | ||||||
| 10 | Karuquinone B | 264.28 | Neuroprotective (HT22 Viability%) | <1% at conc. 12.5 µM | ||
| 11 | Javanicin | 294.30 | Neuroprotective (HT22 Viability%) Glucose production inhibition (IC50) Antimicrobial Activity Antimalarial activity Hemolytic Activity Cytotoxicity (IC50) | 50% at conc. 12.5 µM 3.8 µM 25 μg/mL (S. aureus, P. aeroginosa, S. epidermidis, E. coli) 50 μg/mL (K. pneumoniae) 290 µM IC50 = 1389 µM (14% at 200 µM) 37.1 µM (MCF7) >100 µM (DU145) 23.1 µM (HeLa) >100 µM (A549) 39.1 µM (B16F10) 13 µM (MDA-MB321) 3.3 µM (H4IIE-C3) | ||
| 12 | Solaniol | 292.29 | Neuroprotective (HT22 Viability%) Glucose production inhibition (IC50) Cytotoxicity (IC50) | <1% at conc. 12.5 µM 4.4 µM 9.5 µM (H4IIE-C3) | ||
| 13 | Fusarubin | 306.27 | Neuroprotective (HT22 Viability%) Antimicrobial Activity Antimalarial activity Hemolytic Activity DPPH (IC50) Cytotoxicity (IC50) | 90% at conc. 12.5 µM 1.56 μg/mL (S. aureus, E. coli, P. aeruginosa), 3.125 μg/mL (S. epidermidis), 12.5 μg/mL (K. pneumoniae) IC50 = 176 µM IC50 = 1914 µM (11.3% at 200 µM) 60 µM 7.7 µM (MCF7) 4.2 µM (DU145) 15.6 µM (HeLa) 10.3 µM (A549) 1.5 µM (B16F10) 16 µM (MDA-MB321) | ||
| Polyketides | ||||||
| 14 | Colletotrichalactone A | 356.37 | Cytotoxicity (IC50) | 35 µM (MCF7) | Colletotrichum sp. JS-0361 associated with M. alba leaves (South Korea) | [46] |
| 15 | Colletotrichalactone B | 386.40 | Cytotoxicity (IC50) | >100 µM (MCF7) | ||
| 16 | Colletotrichalactone 3A | 406.43 | Cytotoxicity (IC50) | 25 µM (MCF7) | ||
| Oxazole | ||||||
| 17 | Macrooxazole E | 245.23 | Cytotoxicity (IC50) | No activity on (MCF7) and (LNCaP) | Phoma sp. JS0228 associated with M. alba leaves (South Korea) | [48,57,58] |
| 18 | Macrooxazole C | 259.26 | Biofilm inhibitory% Biofilm destructive% Cytotoxicity (IC50) | 59% (125 μg/mL) against S. aureus 48% (125 μg/mL) against S. aureus 29 µM (MCF7), 36 µM (LNCaP) | ||
| 19 | Macrooxazole A | 261.28 | Biofilm inhibitory% Biofilm destructive% | No activity against S. aureus No activity against S. aureus | ||
| 20 | Macrooxazole B | 277.28 | Biofilm inhibitory% Biofilm destructive% | 43% (125 μg/mL) against S. aureus 31% (125 μg/mL) against S. aureus | ||
| Furoic acid derivative | ||||||
| 21 | 5-hydroxymethyl-2-furan carboxylic acid | 142.03 | - | - |
| Compound | TPSA | Log P | Solubility | GI Absorption | BBB Permeability | CYP2D6 Inhibition |
|---|---|---|---|---|---|---|
| 1 | 82.06 | 2.57 | Moderately | High | No | No |
| 2 | 93.06 | 2.22 | Moderately | High | No | No |
| 3 | 93.06 | 2.22 | Moderately | High | No | No |
| 4 | 104.06 | 1.86 | Moderately | High | No | No |
| 5 | 127.59 | −0.3 | Soluble | High | No | No |
| 6 | 105.59 | 0.25 | Soluble | High | No | No |
| 7 | 85.36 | 0.77 | Soluble | High | No | No |
| 8 | 59.67 | 3 | Moderately | High | Yes | No |
| 9 | 59.67 | 3.32 | Moderately | High | Yes | Yes |
| 10 | 94.83 | 1.22 | Soluble | High | No | No |
| 11 | 107.22 | 0.66 | Soluble | High | No | No |
| 12 | 104.06 | 1.53 | Soluble | High | No | No |
| 13 | 113.29 | 0.92 | Soluble | High | No | No |
| 14 | 89.9 | 1.73 | Soluble | High | No | No |
| 15 | 99.13 | 1.59 | Soluble | High | No | No |
| 16 | 100.52 | 1.44 | Soluble | High | No | No |
| 17 | 83.56 | 1.97 | Soluble | High | No | No |
| 18 | 72.56 | 2.36 | Moderately | High | Yes | No |
| 19 | 83.56 | 1.67 | Moderately | High | No | No |
| 20 | 103.79 | 0.91 | Soluble | High | No | No |
| 21 | 70.67 | 0.16 | Soluble | High | No | No |
| Compound | α-Amylase | α Glucosidase | β Glucosidase | hSGT2 |
|---|---|---|---|---|
| 1 | −8.10 | −4.52 | −7.60 | −6.10 |
| 2 | −8.40 | −4.44 | −7.80 | −6.20 |
| 3 | −8.50 | −3.72 | −7.60 | −6.80 |
| 4 | −8.50 | −3.91 | −8.00 | −7.90 |
| 5 | −6.90 | −6.06 | −6.50 | −7.80 |
| 6 | −7.30 | −5.73 | −7.30 | −8.80 |
| 7 | −7.10 | −5.65 | −7.30 | −8.30 |
| 8 | −6.90 | −5.79 | −7.20 | −8.30 |
| 9 | −7.00 | −6.04 | −7.30 | −7.70 |
| 10 | −7.20 | −5.69 | −7.00 | −8.30 |
| 11 | −7.10 | −5.65 | −7.30 | −7.90 |
| 12 | −7.40 | −4.42 | −7.30 | −7.50 |
| 13 | −8.00 | −5.52 | −7.70 | −8.10 |
| 14 | −8.80 | −3.34 | −8.60 | −8.50 |
| 15 | −8.00 | −3.38 | −9.10 | −6.60 |
| 16 | −7.70 | −2.57 | −8.50 | −7.50 |
| 17 | −6.90 | −5.79 | −7.40 | −8.70 |
| 18 | −6.70 | −6.05 | −7.50 | −8.70 |
| 19 | −7.10 | −6.58 | −7.30 | −8.80 |
| 20 | −7.10 | −6.96 | −7.40 | −8.70 |
| 21 | −5.10 | −5.46 | −5.40 | −6.00 |
| Acarbose | −9.70 | −8.97 | −8.70 | ---- |
| Empagliflozin | ---- | ---- | ---- | −11.60 |
| Complex | Compound | dG Binding | dG Binding Coulomb | dG Binding (NS) | dG Binding (NS) Coulomb | |
|---|---|---|---|---|---|---|
| α-amylase | 14 | Start | −63.40 | −36.48 | −64.65 | −36.54 |
| End | −67.56 | −30.09 | −69.59 | −30.46 | ||
| α glucosidase | 20 | Start | −38.54 | −22.09 | −42.82 | −25.58 |
| End | −36.46 | −17.85 | −41.39 | −23.17 | ||
| β glucosidase | 15 | Start | −31.37 | −9.02 | −32.49 | −9.22 |
| End | −38.47 | −9.47 | −39.29 | −9.41 | ||
| hSGT2 | 6 | Start | −61.81 | −22.73 | −64.53 | −24.44 |
| End | −50.556 | −8.38 | −51.09 | −8.81 | ||
| 19 | Start | −51.94 | −22.66 | −53.70 | −21.56 | |
| End | −47.63 | −16.95 | −50.73 | −17.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AbdelRazek, M.M.M.; Elissawy, A.M.; Mostafa, N.M.; Moussa, A.Y.; Elanany, M.A.; Elshanawany, M.A.; Singab, A.N.B. Chemical and Biological Review of Endophytic Fungi Associated with Morus sp. (Moraceae) and In Silico Study of Their Antidiabetic Potential. Molecules 2023, 28, 1718. https://doi.org/10.3390/molecules28041718
AbdelRazek MMM, Elissawy AM, Mostafa NM, Moussa AY, Elanany MA, Elshanawany MA, Singab ANB. Chemical and Biological Review of Endophytic Fungi Associated with Morus sp. (Moraceae) and In Silico Study of Their Antidiabetic Potential. Molecules. 2023; 28(4):1718. https://doi.org/10.3390/molecules28041718
Chicago/Turabian StyleAbdelRazek, Mohamed M. M., Ahmed M. Elissawy, Nada M. Mostafa, Ashaimaa Y. Moussa, Mohamed A. Elanany, Mohamed A. Elshanawany, and Abdel Nasser B. Singab. 2023. "Chemical and Biological Review of Endophytic Fungi Associated with Morus sp. (Moraceae) and In Silico Study of Their Antidiabetic Potential" Molecules 28, no. 4: 1718. https://doi.org/10.3390/molecules28041718
APA StyleAbdelRazek, M. M. M., Elissawy, A. M., Mostafa, N. M., Moussa, A. Y., Elanany, M. A., Elshanawany, M. A., & Singab, A. N. B. (2023). Chemical and Biological Review of Endophytic Fungi Associated with Morus sp. (Moraceae) and In Silico Study of Their Antidiabetic Potential. Molecules, 28(4), 1718. https://doi.org/10.3390/molecules28041718









