Aggregation-Based Bacterial Separation with Gram-Positive Selectivity by Using a Benzoxaborole-Modified Dendrimer
Abstract
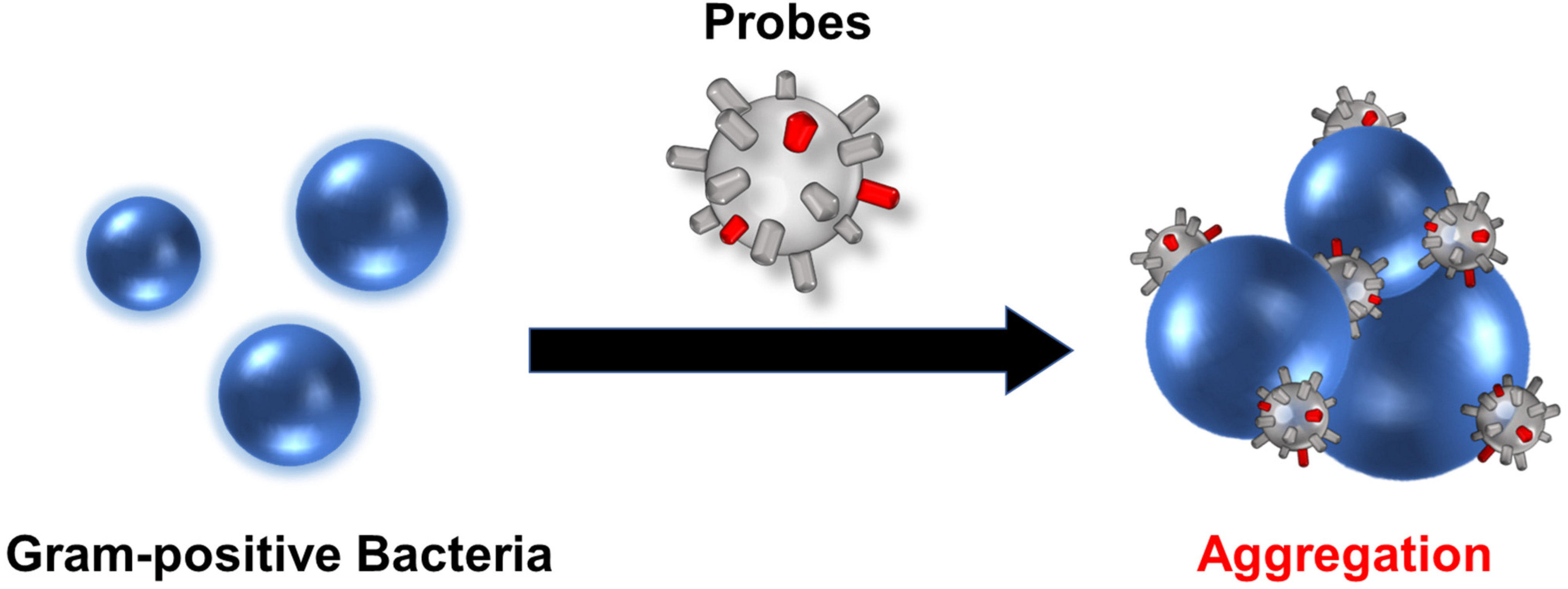
1. Introduction
2. Results and Discussion
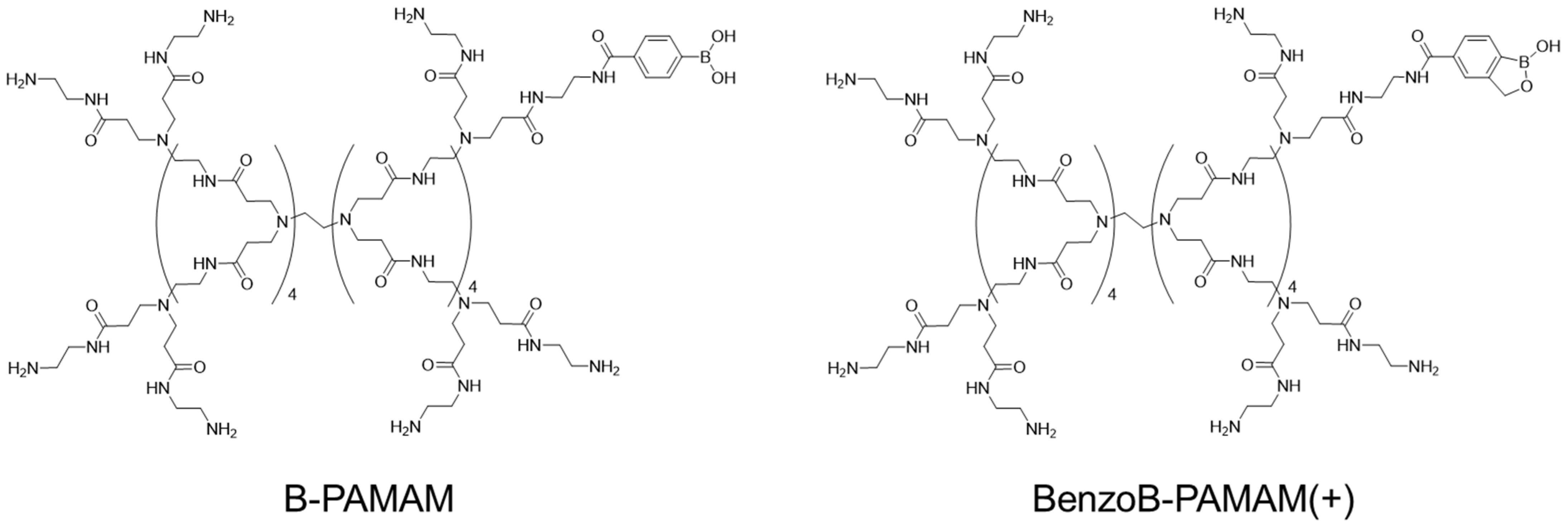
2.1. Characteristics of the Boronic Acid-Based BenzoB-PAMAMs Nanoprobes
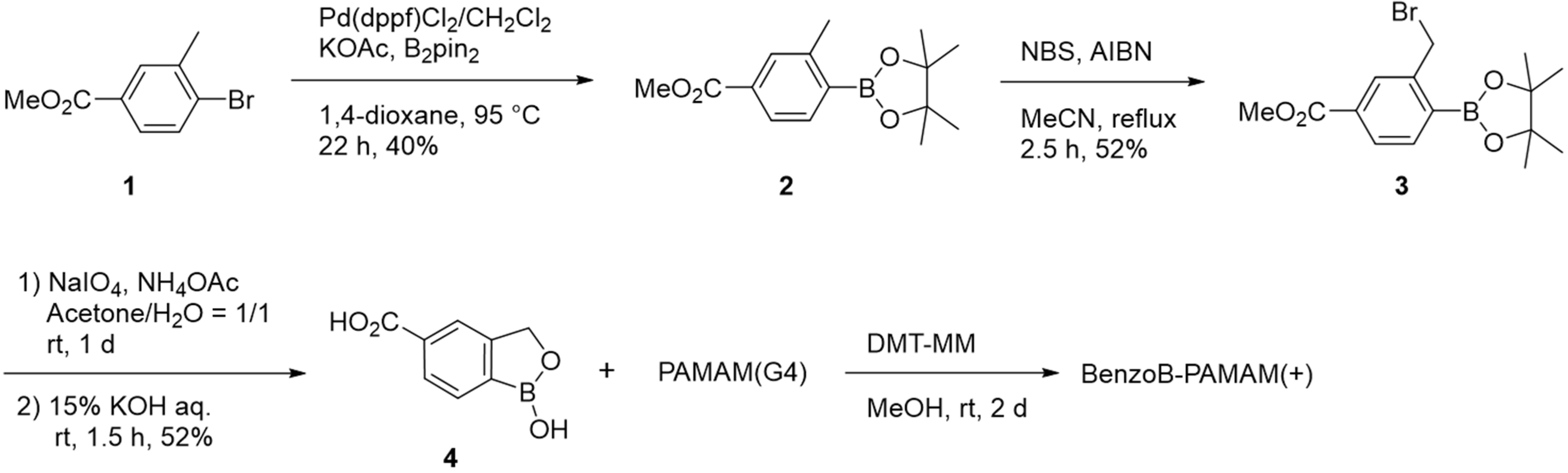
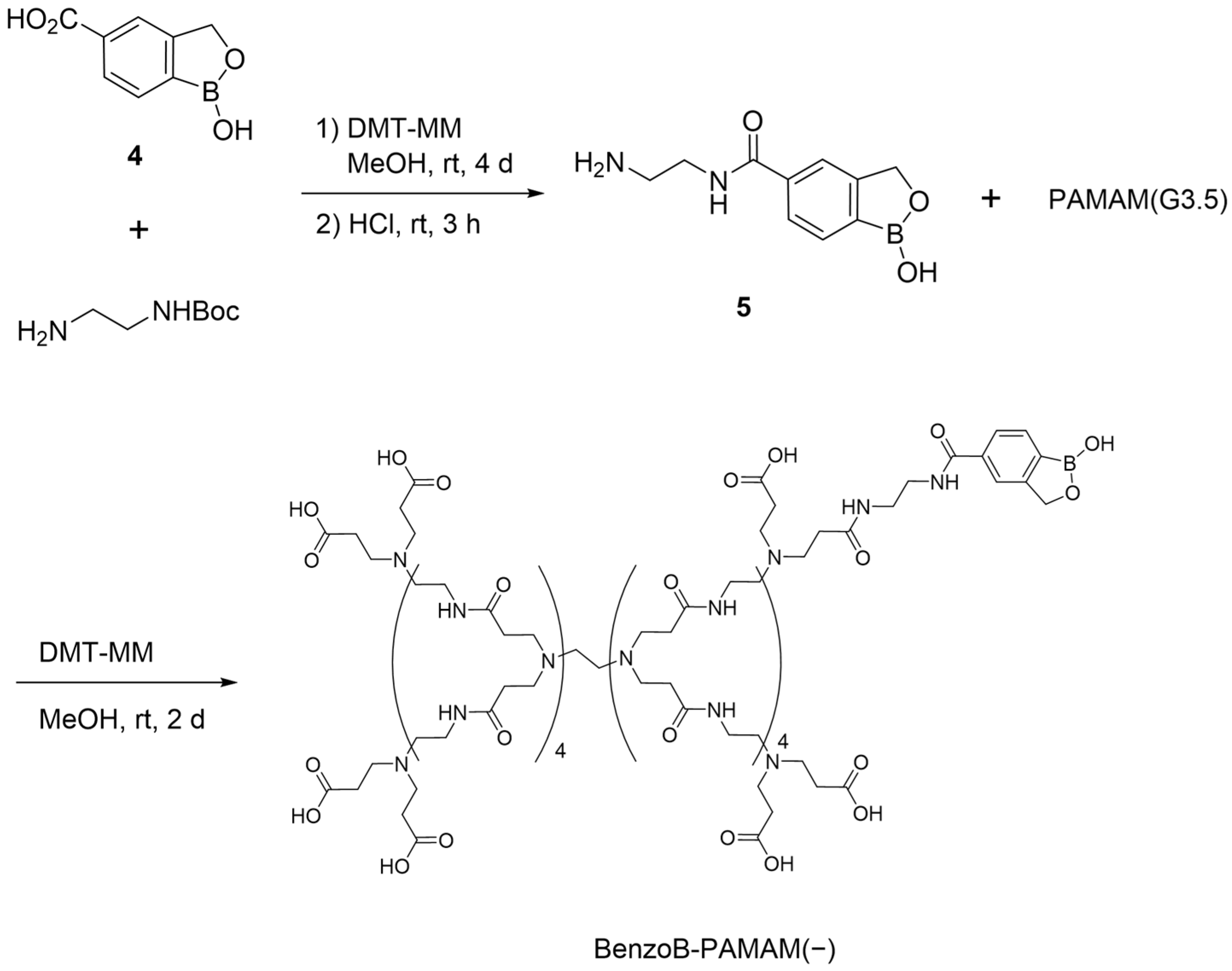
2.1.1. Structure and Synthesis of BenzoB-PAMAMs
2.1.2. Surface Properties of BenzoB-PAMAMs
2.2. Bacterial Recognition by BenzoB-PAMAMs
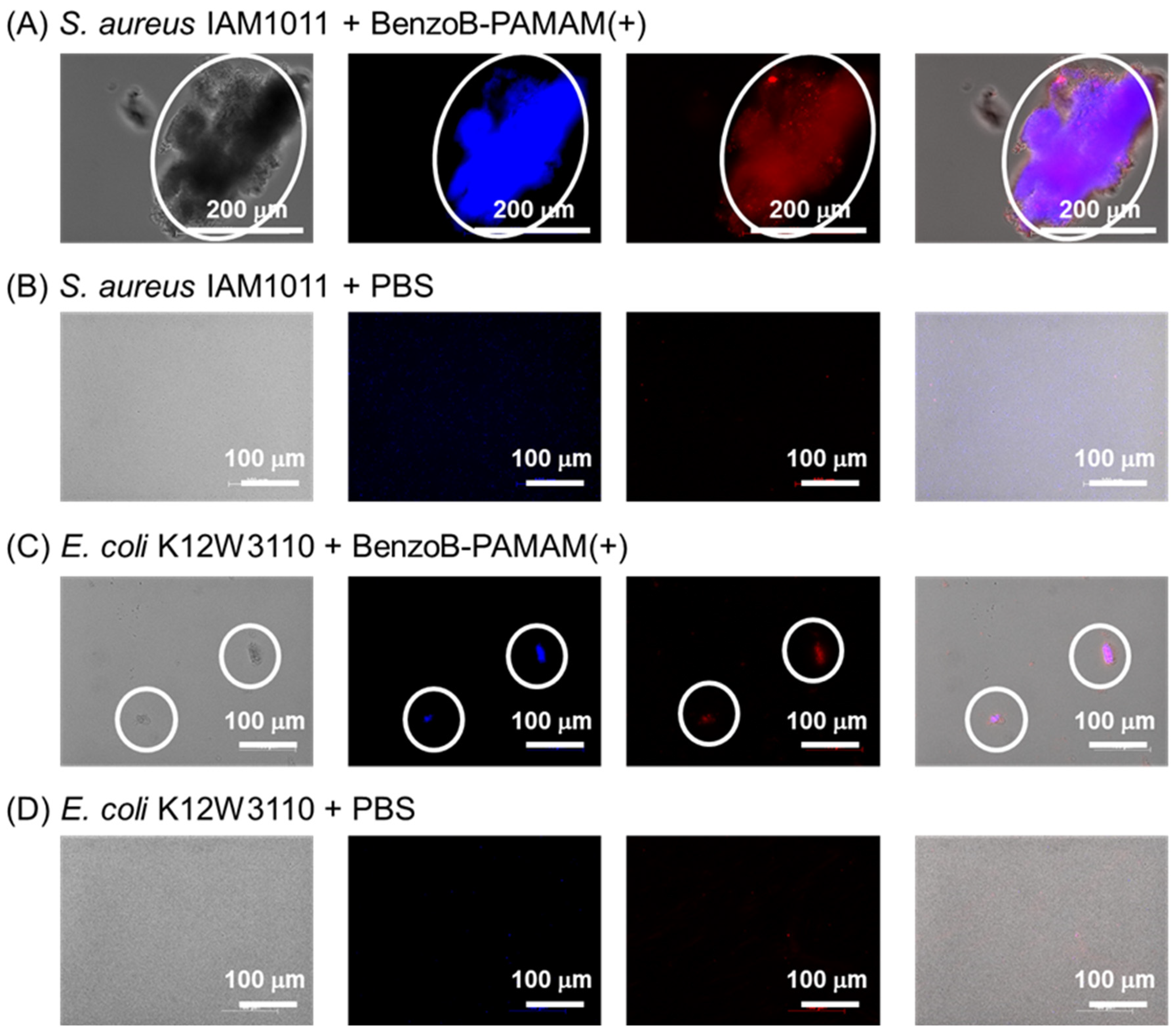
2.2.1. Recognition Confirmed by a Turbidity Measurement and Direct Observation
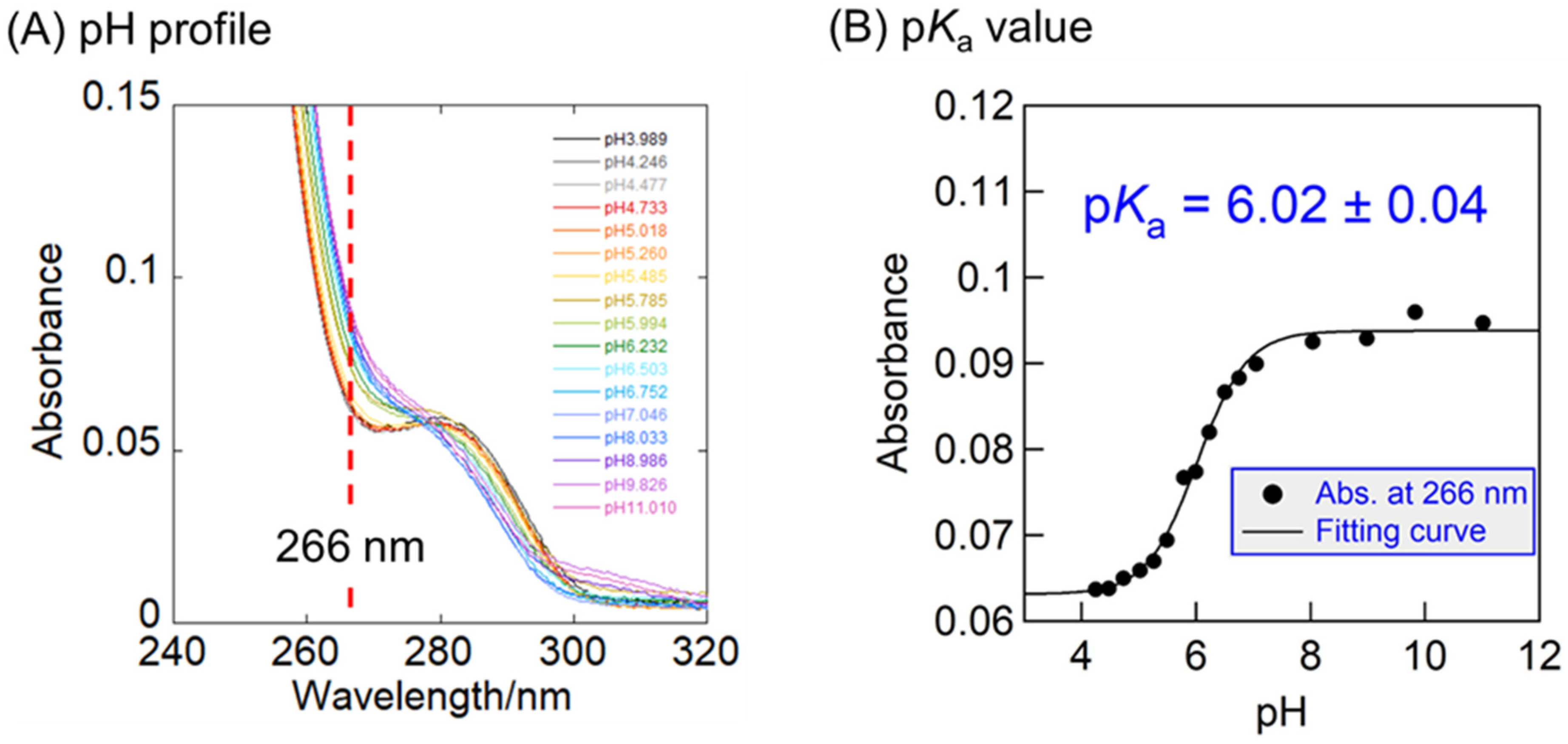
2.2.2. Improved Recognition in Association with the Desirable pKa Value of Benzoxaborole
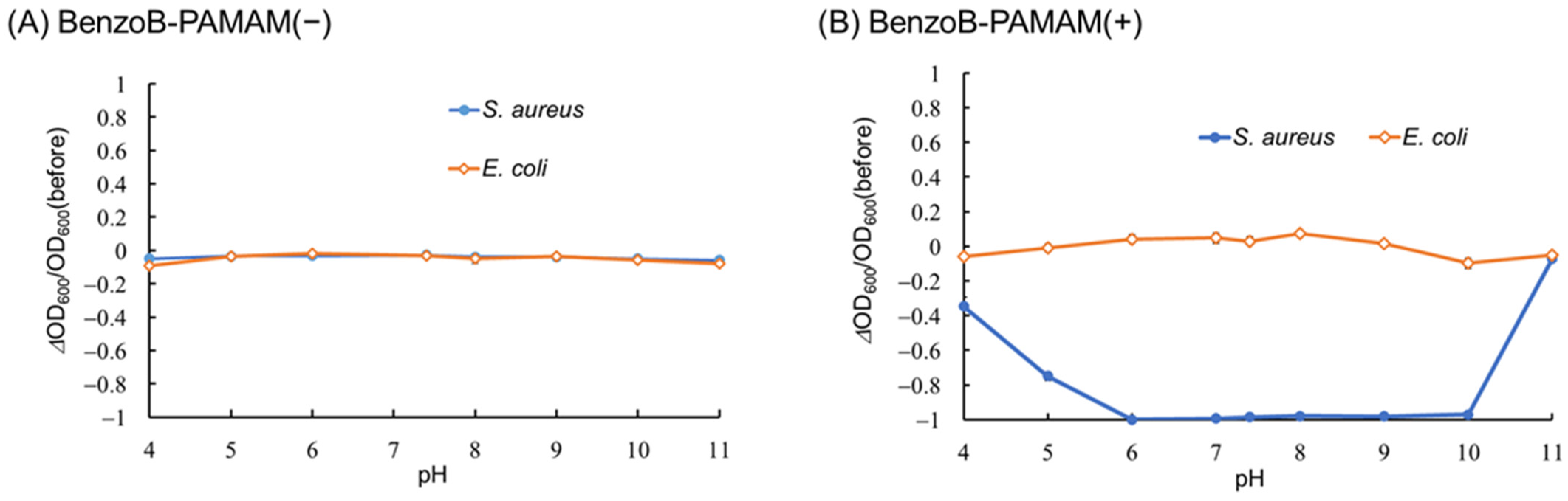
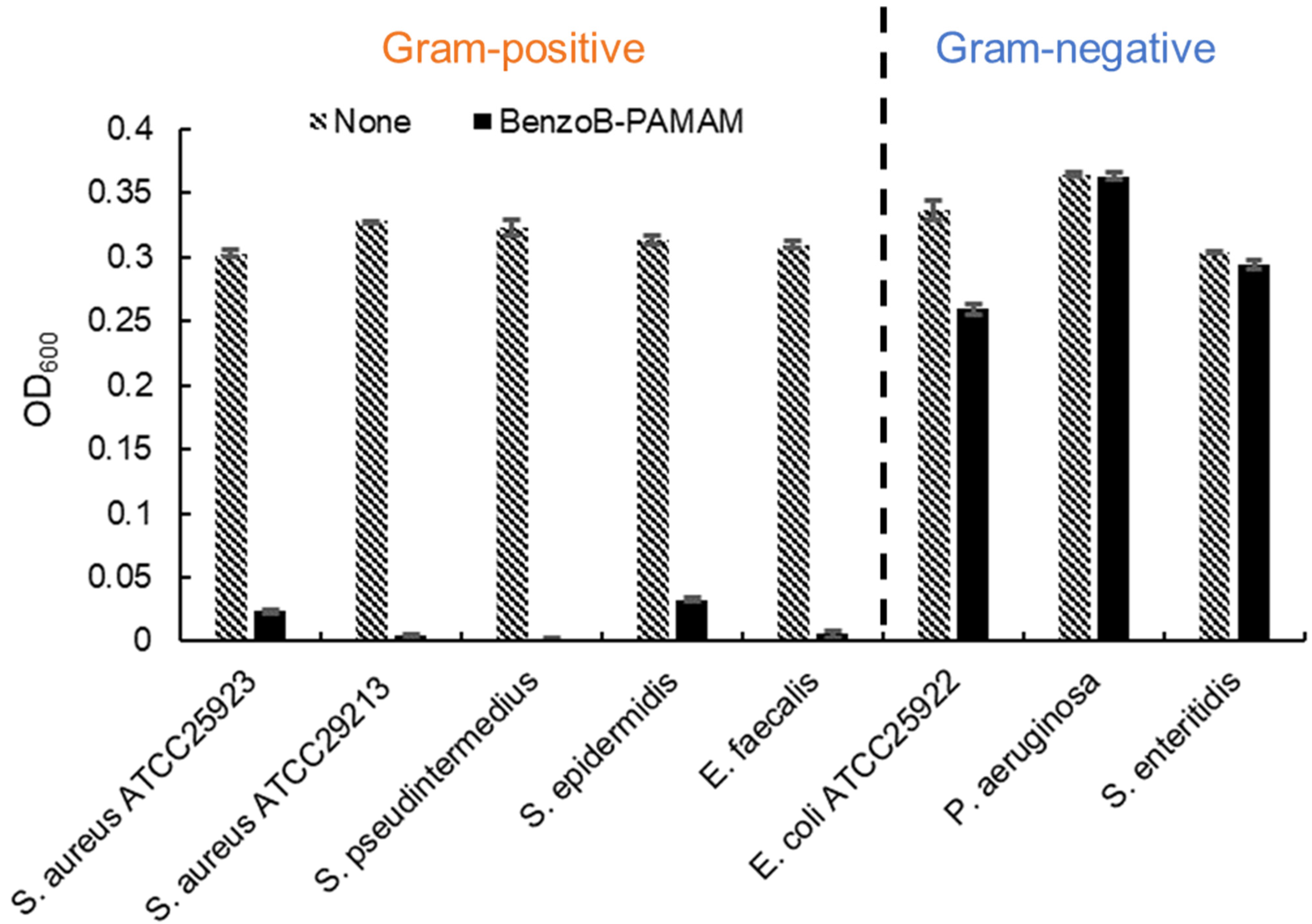
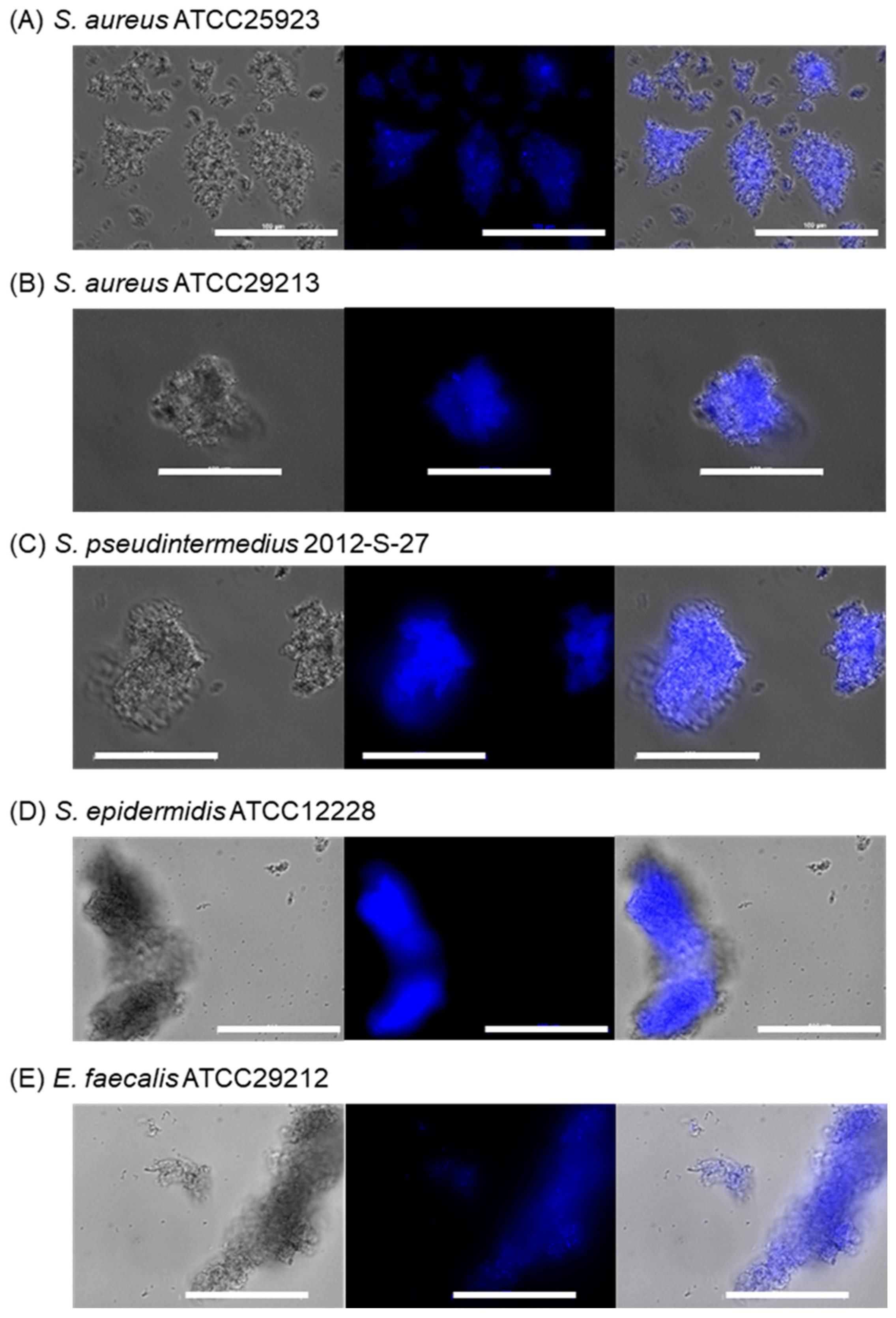
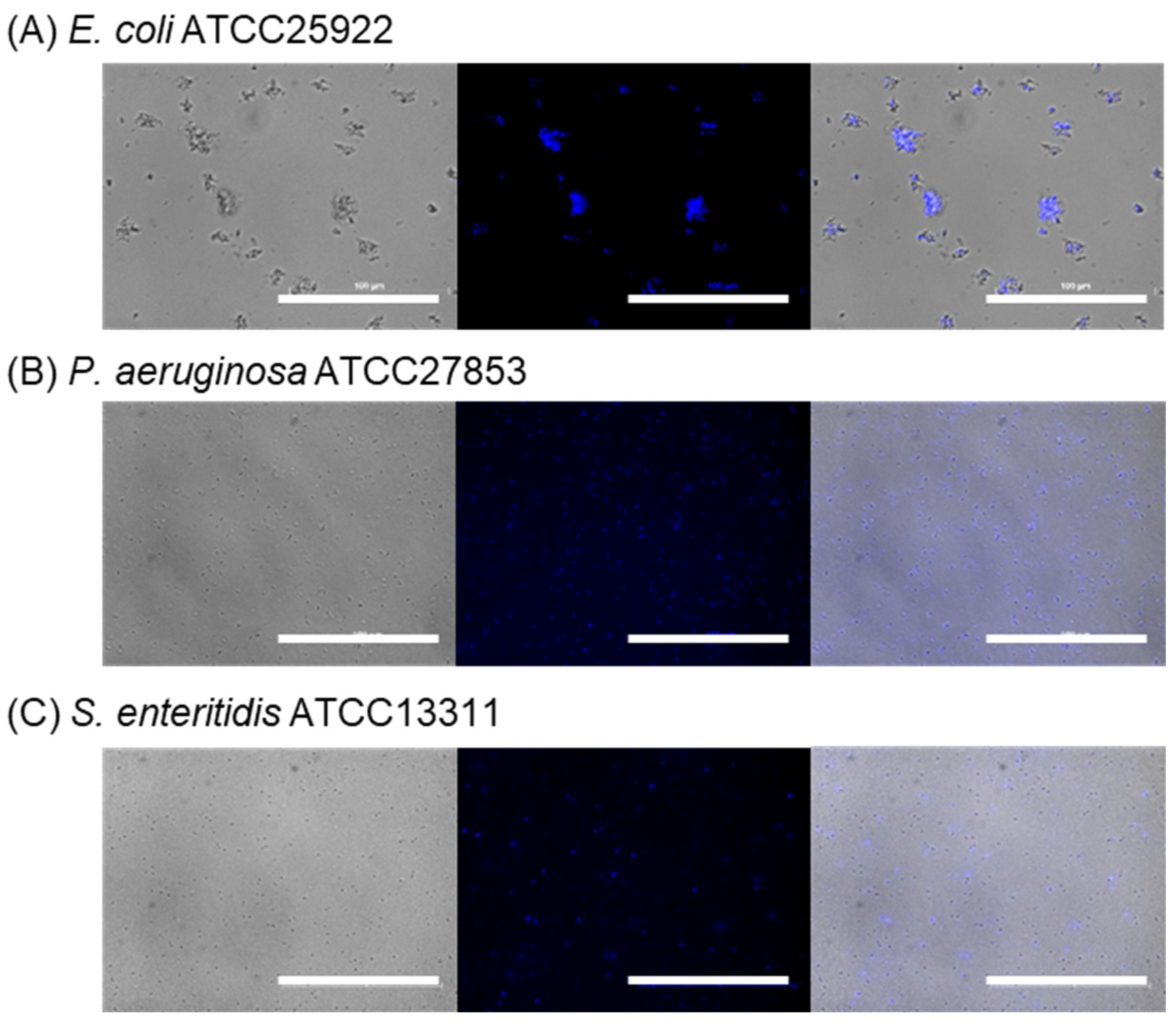
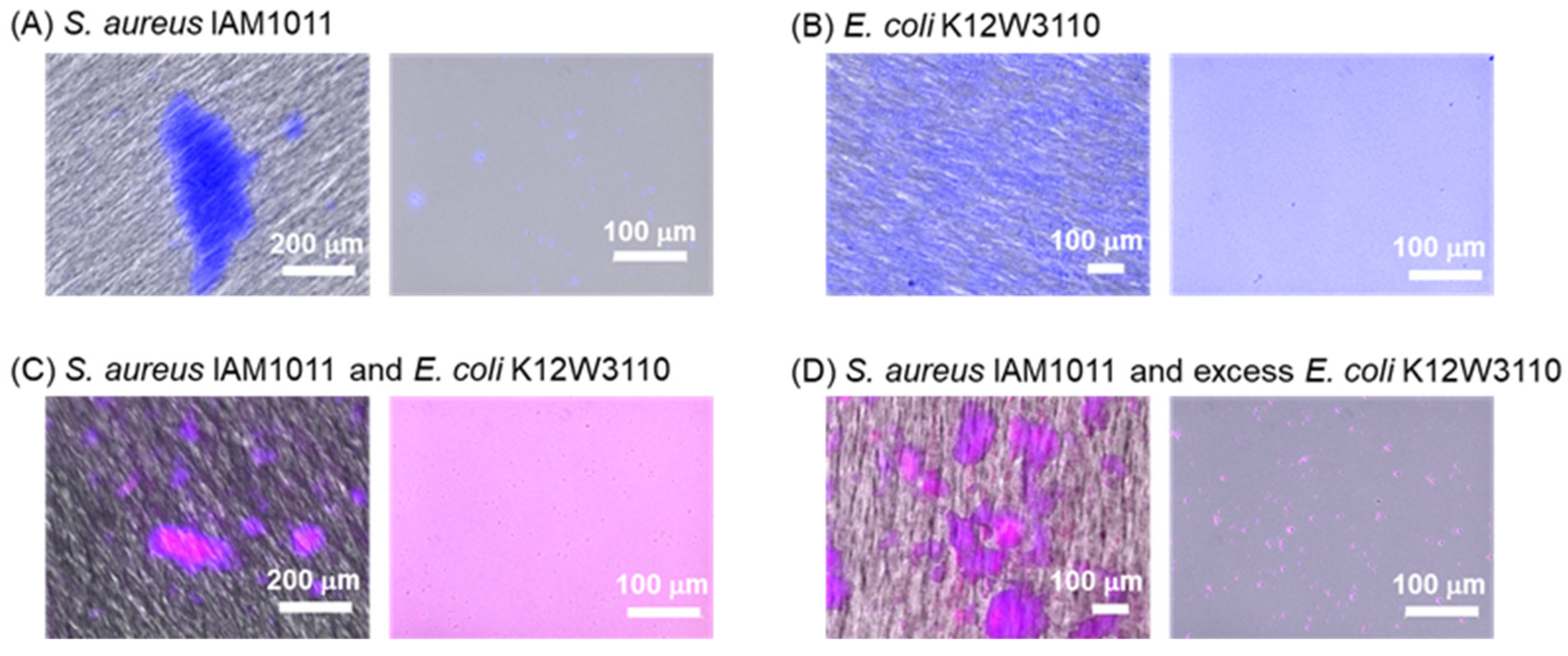
2.2.3. Bacterial Selectivity Using BenzoB-PAMAM(+)
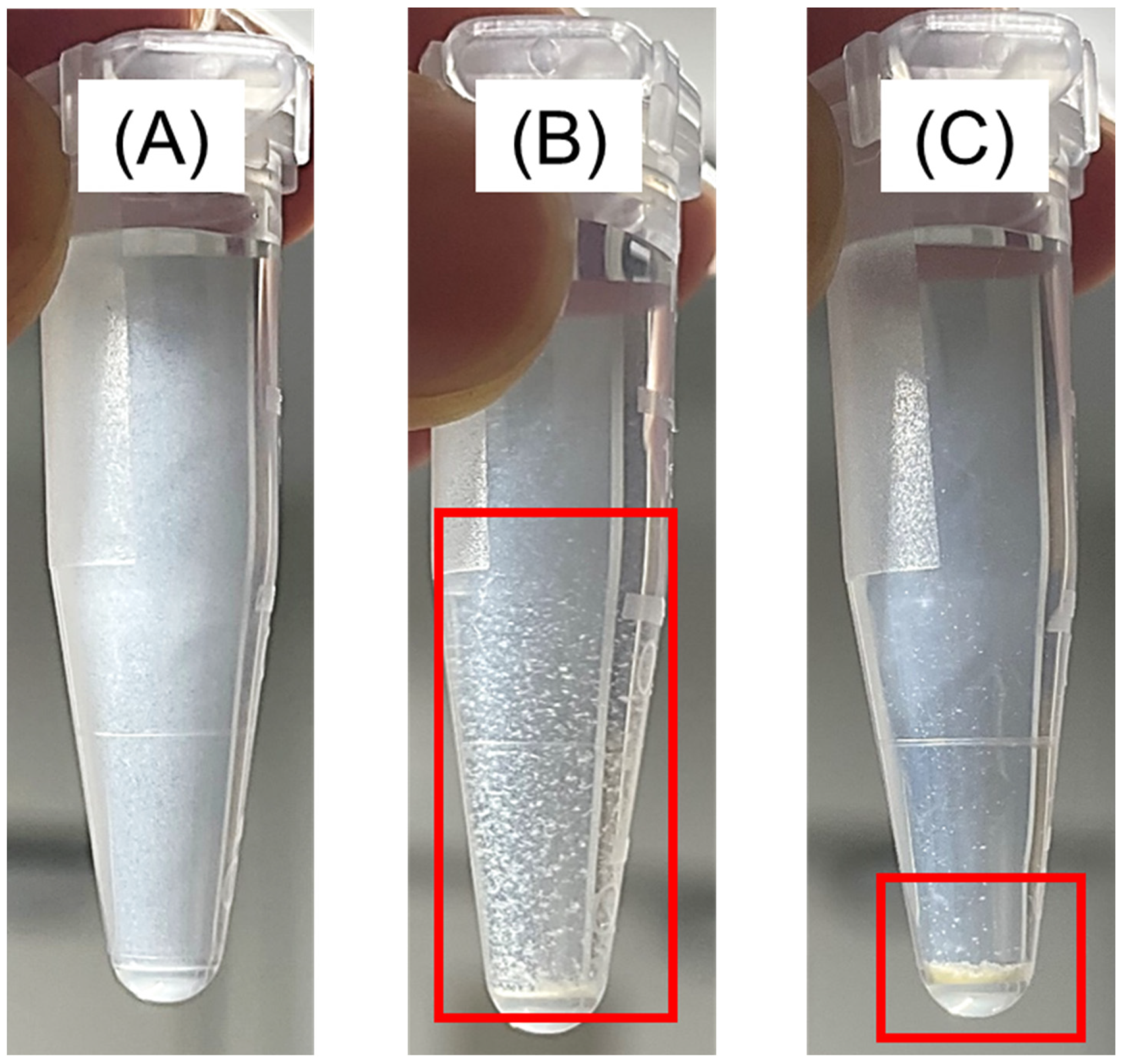
2.2.4. Filtration for Separating Aggregations
3. Materials and Methods
3.1. Reagents and Apparatus
3.1.1. Chemical Reagents
3.1.2. Apparatus
3.2. Preparation of Dendrimer Probes
3.2.1. Synthesis of Methyl 3-Methyl-4-(4,4,5,5-Tetramethyl-1,3,2-Dioxaborolan-2-Yl)Benzoate (2)
3.2.2. Synthesis of Methyl 3-(Bromomethyl)-4-(4,4,5,5-Tetramethyl-1,3,2-Dioxaborolan-2-Yl)Benzoate (3)
3.2.3. Synthesis of 1-Hydroxy-1,3-Dihydro-2,1-Benzoxaborole-5-Carboxylic Acid (4)
3.2.4. Synthesis of BenzoB-PAMAM(+)
3.2.5. Synthesis of N-(2-Aminoethyl)-1-Hydroxy-1,3-Dihydro-2,1-Benzoxaborole-5-Carboxamide (5)
3.2.6. Synthesis of BenzoB-PAMAM(−)
3.3. Biological Experiments
3.3.1. Bacterial Culture
3.3.2. Bacterial Recognition
- ΔOD600 = OD600 (after) − OD600 (before).
- Turbidity change = ΔOD600/OD600 (before).
3.3.3. Bacterial Separation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis. Pharmacol. Ther. 2015, 40, 277–283. [Google Scholar]
- Reardon, S. Antibiotic resistance sweeping developing world. Nature 2014, 509, 141–142. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A.; Archer, G.L. New mechanisms of bacterial resistance to antimicrobial agents. N. Engl. J. Med. 1991, 324, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 8 January 2023).
- Hamner, S.; Brown, B.L.; Hasan, N.A.; Franklin, M.J.; Doyle, J.; Eggers, M.J.; Colwell, R.R.; Ford, T.E. Metagenomic profiling of microbial pathogens in the Little Bighorn River, Montana. Int. J. Environ. Res. Public Health. 2019, 16, 1097. [Google Scholar] [CrossRef]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.J.; Lee, S.H.; Cho, J.C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome. 2020, 8, 75. [Google Scholar] [CrossRef]
- McKinney, C.W.; Dungan, R.S.; Moore, A.; Leytem, A.B. Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- McGarvey, K.M.; Queitsch, K.; Fields, S. Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Appl Environ Microbiol. 2012, 78, 1708–1714. [Google Scholar] [CrossRef]
- Armalytė, J.; Skerniškytė, J.; Bakienė, E.; Krasauskas, R.; Šiugždinienė, R.; Kareivienė, V.; Kerzienė, S.; Klimienė, I.; Sužiedėlienė, E.; Ružauskas, M. Microbial diversity and antimicrobial resistance profile in microbiota from soils of conventional and organic farming systems. Front. Microbiol. 2019, 10, 892. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2016; Available online: https://wellcomecollection.org/works/thvwsuba (accessed on 8 February 2023).
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Danko, D.; Bezdan, D.; Afshin, E.E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; Donnellan, D.; Hecht, J.; Jackson, K.; et al. International MetaSUB Consortium. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell 2021, 184, 3376–3393.e17. [Google Scholar] [CrossRef] [PubMed]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Moayeri, M.; Chen, Z.; Harma, H.; Zhao, J.; Hu, H.; Purcell, R.H.; Leppla, S.H.; Hewlett, I.K. Detection of anthrax toxin by an ultrasensitive immunoassay using europium nanoparticles. Clin. Vaccine Immunol. 2009, 16, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Gang, J.J. Challenges of microarray applications for microbial detection and gene expression profiling in food. J. Microbial. Biochem. Technol. 2011, S2, 2. [Google Scholar] [CrossRef]
- Call, D. Challenges and opportunities for pathogen detection using DNA microarrays. Crit. Rev. Microbiol. 2005, 31, 91–99. [Google Scholar] [CrossRef]
- Stoodley, P.; Conti, S.F.; DeMeo, P.J.; Nistico, L.; Melton-Kreft, R.; Johnson, S.; Darabi, A.; Ehrlich, G.D.; Costerton, J.W.; Kathju, S. Characterization of a mixed MRSA/MRSE biofilm in an explanted total ankle arthroplasty. FEMS Immunol. Med. Microbiol. 2011, 62, 66–74. [Google Scholar] [CrossRef]
- Mosier-Boss, P.A. Review on SERS of bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hu, Z.; Yang, D.; Xie, S.; Jiang, Z.; Niessner, R.; Haisch, C.; Zhou, H.; Sun, P. Bacteria detection: From powerful SERS to its advanced compatible techniques. Adv. Sci. 2020, 7, 2001739. [Google Scholar] [CrossRef]
- Golabi, M.; Kuralay, F.; Jager, E.W.H.; Beni, V.; Turner, A.P.F. Electrochemical bacterial detection using poly(3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017, 93, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Sai-Anand, G.; Sivanesan, A.; Benzigar, M.R.; Singh, G.; Gopalan, A.-I.; Vijay, A.; Hamid, B.; Ramadass, I.K.; Kambala, V.; Vinu, A. Recent progress on the sensing of pathogenic bacteria using advanced nanostructures. Bull. Chem. Soc. Jpn. 2019, 92, 216–244. [Google Scholar] [CrossRef]
- Benoit, P.W.; Donahue, D.W. Methods for rapid separation and concentration of bacteria in food that bypass time-consuming cultural enrichment. J. Food Prot. 2003, 66, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Xue, L.; Zhang, H.; Guo, R.; Li, Y.; Liao, M.; Wang, M.; Lin, J. An enzyme-free biosensor for sensitive detection of Salmonella using curcumin as signal reporter and click chemistry for signal amplification. Theranostics 2018, 8, 6263–6273. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Sohn, B.; Choi, J.; Jeon, S. Recent advances in magnetic nanoparticle-based microfluidic devices for the pretreatment of pathogenic bacteria. Biomed. Eng. Lett. 2021, 11, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Duncan, B.; Wang, Z.; Wang, L.S.; Rotello, V.M.; Nugen, S.R. Bacteriophage-based nanoprobes for rapid bacteria separation. Nanoscale 2015, 7, 16230–16236. [Google Scholar] [CrossRef] [PubMed]
- Tsuchido, Y.; Horiuchi, R.; Hashimoto, T.; Ishihara, K.; Kanzawa, N.; Hayashita, T. Rapid and selective discrimination of Gram-positive and Gram-negative bacteria by boronic acid-modified poly(amidoamine) dendrimer. Anal. Chem. 2019, 91, 3929–3935. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New class of polymers: Starburst-dendritic macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef]
- Kitamura, A.; Kasai, Y.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Design and function of fluorescent silica nanoparticles for bacteria detection. J. Ion Exch. 2018, 29, 121–125. [Google Scholar] [CrossRef]
- Bull, S.D.; Davidson, M.G.; van den Elsen, J.M.H.; Fossey, J.S.; Jenkins, A.T.A.; Jiang, Y.-B.; Kubo, Y.; Marken, F.; Sakurai, K.; Zhao, J.; et al. Exploiting the reversible covalent bonding of boronic acids: Recognition, sensing, and assembly. Acc. Chem. Res. 2013, 46, 312–326. [Google Scholar] [CrossRef]
- Wu, X.; Li, Z.; Chen, X.X.; Fossey, J.S.; James, T.D.; Jiang, Y.-B. Selective sensing of saccharides using simple boronic acids and their aggregates. Chem. Soc. Rev. 2013, 42, 8032–8048. [Google Scholar] [CrossRef] [PubMed]
- Lorand, J.P.; Edwards, J.O. Polyol complexes and structure of the benzeneboronate ion. J. Org. Chem. 1959, 24, 769–774. [Google Scholar] [CrossRef]
- Mikagi, A.; Manita, K.; Tsuchido, Y.; Kanzawa, N.; Hashimoto, T.; Hayashita, T. Boronic acid-based dendrimers with various surface properties for bacterial recognition with adjustable selectivity. ACS Appl. Bio Mater. 2022, 5, 5255–5263. [Google Scholar] [CrossRef] [PubMed]
- Dumitriu, S. Polysaccharides: Structural Diversity and Functional Versatility, 2nd ed.; CRC Press: Boca Raton, CA, USA, 2014; Chapter 1. [Google Scholar]
- Rahman, M.M.; Hunter, H.N.; Prova, S.; Verma, V.; Qamar, A.; Golemi-Kotra, D. The Staphylococcus aureus methicillin resistance factor FmtA is a D-amino esterase that acts on teichoic acids. MBio 2016, 7, e02070-15. [Google Scholar] [CrossRef] [PubMed]
- Mikagi, A.; Manita, K.; Yoyasu, A.; Tsuchido, Y.; Kanzawa, N.; Hashimoto, T.; Hayashita, T. Rapid bacterial recognition over a wide pH range by boronic acid-based ditopic dendrimer probes for Gram-positive bacteria. Molecules 2021, 27, 256. [Google Scholar] [CrossRef] [PubMed]
- Priem, C.; Geyer, A. Reversible covalent end-capping of collagen model peptides. Chemistry 2019, 25, 14278–14283. [Google Scholar] [CrossRef] [PubMed]
- Nishiyabu, R.; Kubo, Y.; James, T.D.; Fossey, J.S. Boronic acid building blocks: Tools for sensing and separation. Chem. Commun. 2011, 47, 1106–1123. [Google Scholar] [CrossRef]
- Brooks, W.L.A.; Deng, C.C.; Sumerlin, B.S. Structure-Reactivity Relationships in Boronic Acid-Diol Complexation. ACS Omega 2018, 3, 17863–17870. [Google Scholar] [CrossRef]
- Medveďová, A.; Valík, Ľ. Staphylococcus aureus: Characterisation and quantitative growth description in milk and artisanal raw milk cheese production. In Structure and function of food engineering.; Eissa, A.A., Ed.; IntechOpen Limited: London, UK, 2012. [Google Scholar]
- Wilks, J.C.; Slonczewski, J.L. pH of the Cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. Bacteriol. 2007, 189, 5601–5607. [Google Scholar] [CrossRef]
- Hua, X.; Bao, Y.; Wang, H.; Chen, Z.; Wu, F. Bacteria-derived fluorescent carbon dots for microbial live/dead differentiation. Nanoscale 2017, 9, 2150–2161. [Google Scholar] [CrossRef]
- Quan, K.; Jiang, G.; Liu, J.; Zhang, Z.; Ren, Y.; Busscher, H.J.; Van der Mei, H.C.; Peterson, B.W. Influence of interaction between surface-modified magnetic nanoparticles with infectious biofilm components in artificial channel digging and biofilm eradication by antibiotics in vitro and in vivo. Nanoscale 2021, 13, 4644–4653. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, J.; Yu, Q.; Zhang, J.; Niu, X.; Hao, L.; Yang, L.; Zhao, Y. Effects of salts on the self-assembly behavior and antibacterial activity of a surfactant-like peptide. Soft Matter 2018, 16, 9758–9768. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yuan, X.; Song, Z.; Xu, S.; Yang, Y.; Yang, X. Gram-negative Escherichia coli promotes deposition of polymer-capped silver nanoparticles in saturated porous media. Environ. Sci.: Nano 2018, 5, 1495–1505. [Google Scholar] [CrossRef]
- Mikagi, A.; Tsurufusa, R.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Fast and sensitive bacteria detection by boronic acid modified fluorescent dendrimer. Sensors 2021, 21, 3115. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Naylor, A.M.; Goddard III, W.A. Starburst dendrimers: Molecular-level control of size, shape, surface chemistry, topology, and flexibility from atoms to macroscopic matter. Angew. Chem. Int. Ed. 1990, 29, 138–175. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikagi, A.; Takahashi, Y.; Kanzawa, N.; Suzuki, Y.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Aggregation-Based Bacterial Separation with Gram-Positive Selectivity by Using a Benzoxaborole-Modified Dendrimer. Molecules 2023, 28, 1704. https://doi.org/10.3390/molecules28041704
Mikagi A, Takahashi Y, Kanzawa N, Suzuki Y, Tsuchido Y, Hashimoto T, Hayashita T. Aggregation-Based Bacterial Separation with Gram-Positive Selectivity by Using a Benzoxaborole-Modified Dendrimer. Molecules. 2023; 28(4):1704. https://doi.org/10.3390/molecules28041704
Chicago/Turabian StyleMikagi, Ayame, Yotaro Takahashi, Nobuyuki Kanzawa, Yota Suzuki, Yuji Tsuchido, Takeshi Hashimoto, and Takashi Hayashita. 2023. "Aggregation-Based Bacterial Separation with Gram-Positive Selectivity by Using a Benzoxaborole-Modified Dendrimer" Molecules 28, no. 4: 1704. https://doi.org/10.3390/molecules28041704
APA StyleMikagi, A., Takahashi, Y., Kanzawa, N., Suzuki, Y., Tsuchido, Y., Hashimoto, T., & Hayashita, T. (2023). Aggregation-Based Bacterial Separation with Gram-Positive Selectivity by Using a Benzoxaborole-Modified Dendrimer. Molecules, 28(4), 1704. https://doi.org/10.3390/molecules28041704





