Molecular Modeling and In Vitro Evaluation of Piplartine Analogs against Oral Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
2.1. Chemistry
2.2. Cytotoxicity
2.2.1. Cell Viability (Cytotoxicity) Assay
2.2.2. Selectivity
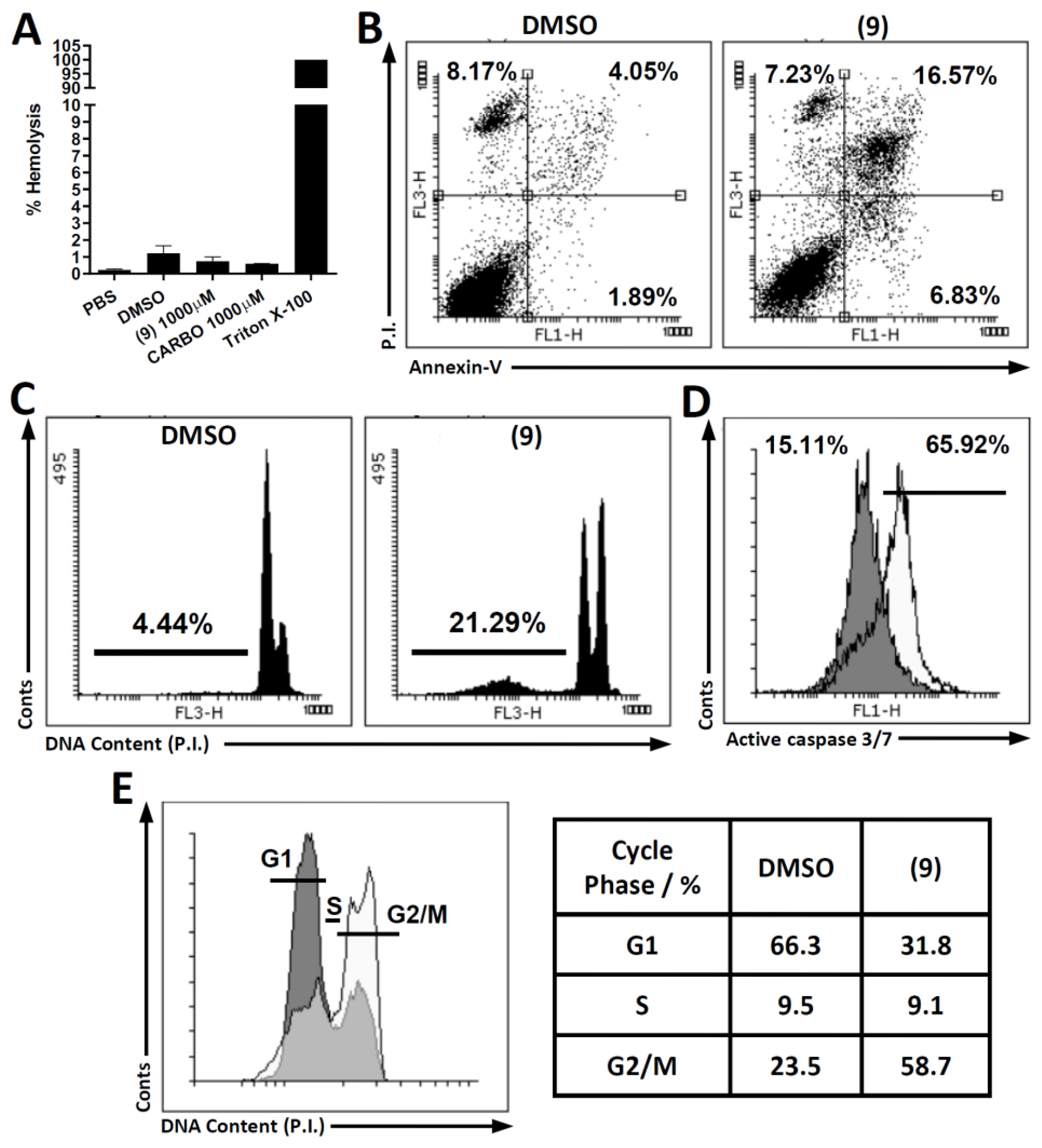
2.2.3. Hemolytic Activity
2.2.4. Investigation of Cell Death Pathway
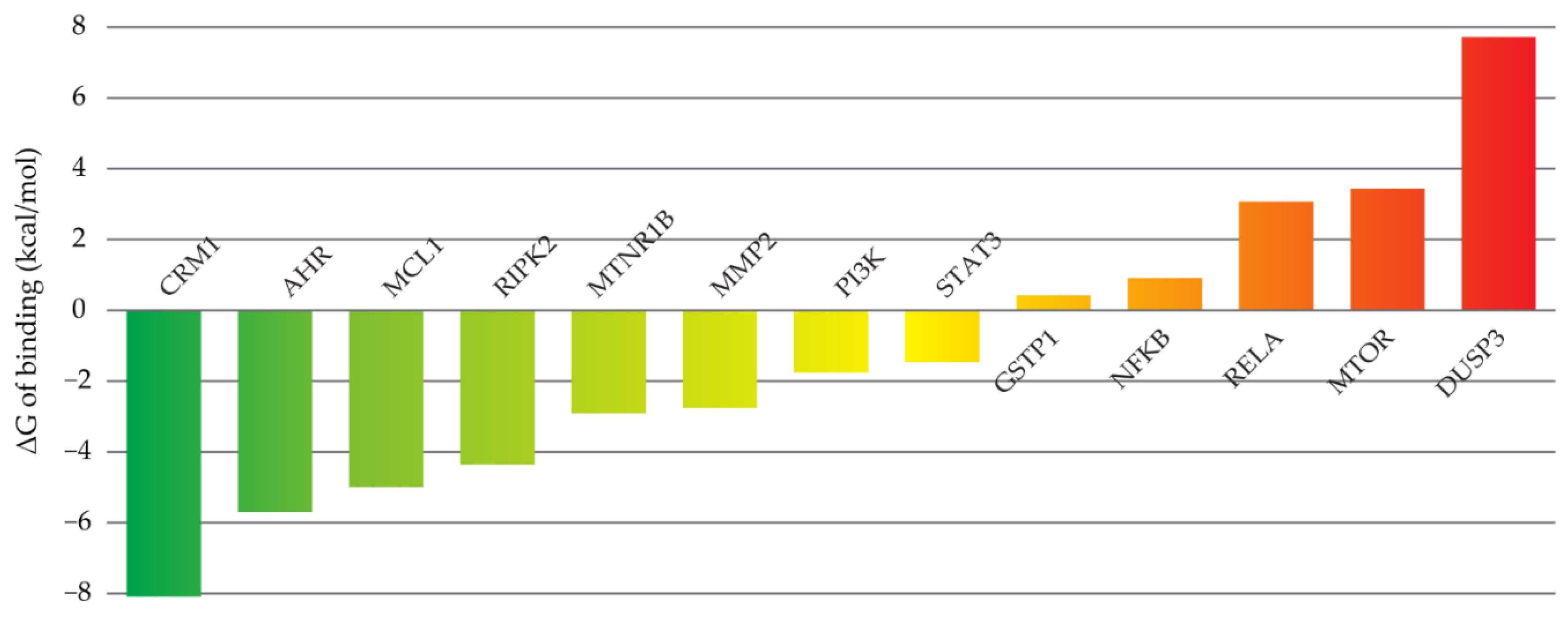
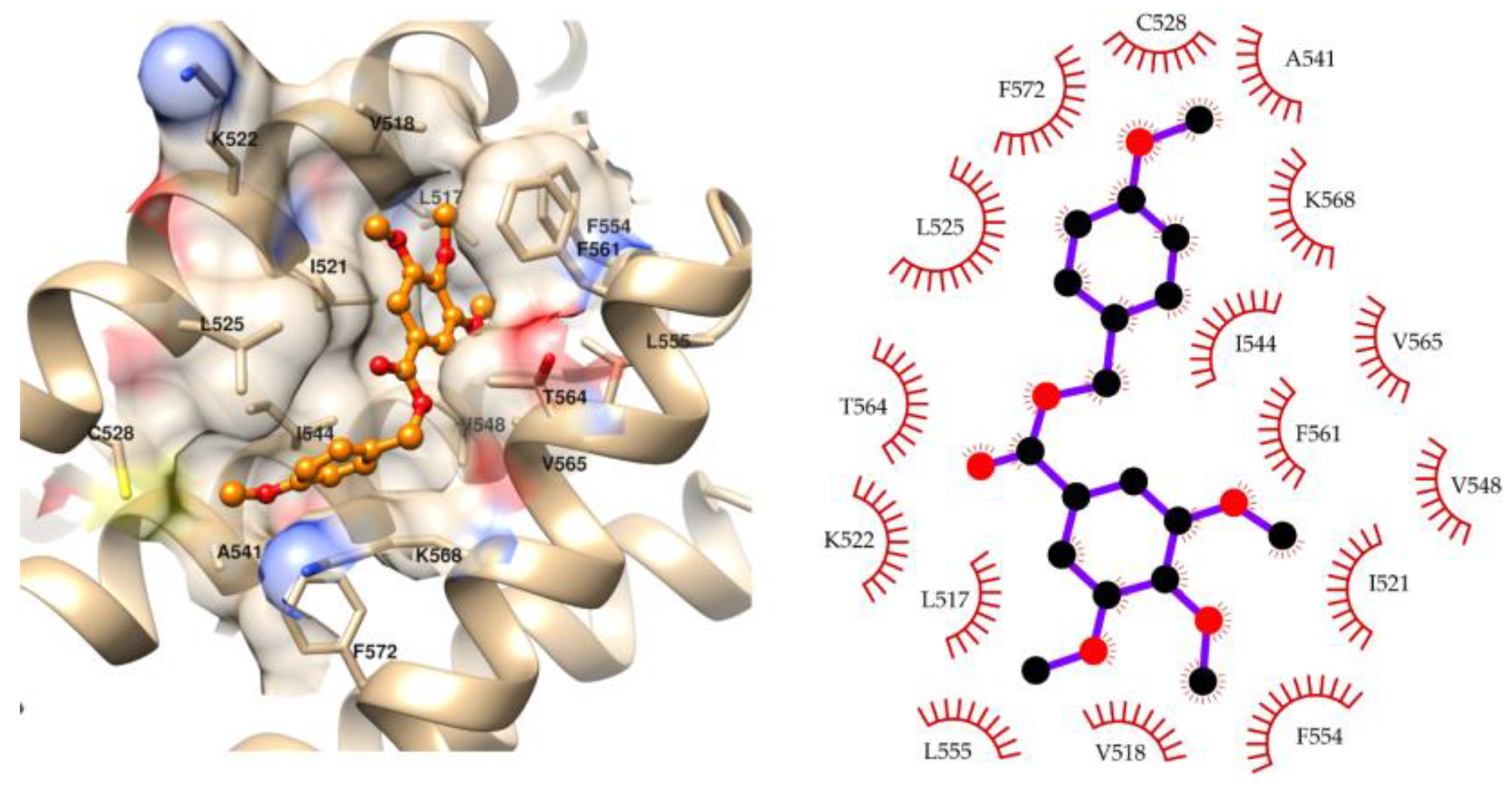
2.3. Molecular Modeling
3. Discussion
4. Materials and Methods
4.1. Preparation of Compounds 1–6
4.2. Preparation of Compounds 7–12
4.3. Preparation of Compounds 13–19
- Methyl 3,4,5–trimethoxybenzoate (1): White crystalline solid; yield 94.9% (101.2 mg; 0.44 mmol); M.P.: 81–83 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.46; IR ʋmax (KBr, cm−1): 3021, 2953; 1716, 1674, 1592 and 1467, 1338 and 1132, 1229 and 992, 863; 1H NMR (400 MHz, CDCl3,): δ 7.29 (s, 2H); 3.89 (s, 12H). 13C NMR (100 MHz, CDCl3): δ 166.59; 152.85; 142.34; 125.26; 106.86; 60.89; 56.23; 52.19 [57].
- Ethyl 3,4,5–trimethoxybenzoate (2): White solid; yield 94.6% (107.1 mg; 0.44 mmol); M.P.: 53–54 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.52; IR ʋmax (KBr, cm−1): 3014, 2964, 1706, 1664, 1591 and 1456, 1332 and 1132, 1228 and 1042, 863; 1H NMR (400 MHz, CDCl3,): δ 7.29 (s, 2H); 4.39 (q, J = 7.1 Hz, 2H); 3.90 (s, 6H); 3.89 (s, 3H); 1.39 (t, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 166.2); 152.91; 142.09; 125.54; 106.77; 61.13 ; 60.91; 56.24; 14.42 [53].
- Propyl 3,4,5–trimethoxybenzoate (3): White crystalline solid; yield 99.1% (118.8 mg; 0.46 mmol); M.P.: 34–35 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.56; IR ʋmax (KBr, cm−1): 3114, 2964, 1706, 1664, 1590 and 1459, 1333 and 1124, 1227 and 1008, 858; 1H NMR (400 MHz, CDCl3): δ 7.30 (s, 2H); 4.27 (t, J = 6.7 Hz, 2H); 3.93 (s, 9H); 1.79 (sext, J = 7.4 Hz, 2H); 1.02 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, CDCl3): δ 166.14; 152.66; 142.06; 125.46; 106.67; 66.64; 60.95; 56.18; 22.05; 10.44 [57].
- Isopropyl 3,4,5–trimethoxybenzoate (4): Light brown oil; yield 57.3% (68.6 mg; 0.26 mmol); TLC (8:2 Hexane/AcOEt), Rf = 0.56; IR ʋmax (KBr, cm−1): 3024, 2981, 1711, 1679, 1590 and 1461, 1327 and 1129, 1229 and 1007, 865; 1H NMR (400 MHz, CDCl3): δ 7.28 (s, 2H); 5.24 (sept, J = 2.2 Hz, 1H); 3.90 (s, 6H); 3.89 (s, 3H); 1.36 (d, J = 6.3 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ 165.74; 152.84; 142.03; 125.91; 106.76; 68.59; 60.91; 56.23; 21.9 [57].
- Butyl 3,4,5–trimethoxybenzoate (5): Colorless oil; yield 99.6% (125.9 mg; 0.46 mmol); TLC (8:2 Hexane/AcOEt), Rf = 0.64; IR ʋmax (KBr, cm−1): 3006, 2961, 1716, 1655, 1590 and 1459, 1335 and 1129, 1225 and 1006, 865; 1H NMR (500 MHz, CDCl3): δ 7.29 (s, 2H); 4.31 (t, J = 6.7 Hz, 2H); 3.90 (s, 6H); 3.90 (s, 3H); 1.76 (quint, J = 6.7 Hz, 2H); 1.47 (sex, J = 7.4 Hz, 2H); 0.98 (t, J = 7.3 Hz, 3H); 13C NMR (125 MHz, CDCl3): δ 166.4); 153.04; 142.26; 125.69; 106.96; 65.19; 61.04; 56.40; 30.97; 19.47; 13.91 [53].
- Isopentyl 3,4,5–trimethoxybenzoate (6): Brown oil, yield 45.3% (60.3 mg; 0.21 mmol); TLC (8:2 Hexane/AcOEt), Rf = 0.60; IR ʋmax (KBr, cm−1): 3002, 2959, 1717, 1655, 1590 and 1459, 1334 and 1129, 1225 and 1006, 865; 1H NMR (400 MHz, CDCl3): δ 7.29 (s, 2H); 4.34 (t, J = 6.8 Hz, 2H); 3.90 (s, 9H); 1.81–1.71 (m, 1H); 1.66 (q, J = 6.8 Hz, 2H); 0.98 (d, J = 6.6 Hz, 6H); 13C NMR (100 MHz, CDCl3): δ 166.57; 153.03; 142.29; 125.75; 107.00; 64.13; 61.05; 56.41; 37.62; 25.49; 22.65. HRMS (FT-ICR) analyze: C15H22O5 calculated theoretical value [M+H]+: 283.1540. Found = 283.1538.
- Pentyl 3,4,5–trimethoxybenzoate (7): White oil; yield 75.2% (100.1 mg; 0.35 mmol); TLC (8:2 Hexane/AcOEt), Rf = 0.60; IR ʋmax (KBr, cm−1): 3012, 2957, 1714, 1657, 1590 and 1461, 1335 and 1129, 1226 and 1006, 865; 1H NMR (400 MHz, CDCl3): δ 7.29 (s, 2H); 4.29 (t, J = 6.8 Hz, 2H); 3.89 (s, 9H); 1.75 (quint, J = 7.2 Hz, 2H); 1.44–1.34 (m, 4H); 0.92 (t, J = 7.0 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 166.27 (C=O); 152.93 (C-3, C-5); 142.13 (C-4); 125.57 (C-1); 106.80; 65.30; 60.96; 56.23; 28.42; 28.18; 22.36; 13.99 [53].
- Decyl 3,4,5–trimethoxybenzoate (8): Amorphous solid, yield 40.6% (67.4 mg; 0.19 mmol); M.P.: 49–50 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.64; IR ʋmax (KBr, cm−1): 3015, 2956, 1709, 1672, 1590 and 1465, 1336 and 1131, 1226 and 990, 864; 1H NMR (400 MHz, CDCl3): δ 7.29 (s, 2H); 4.30 (t, J = 6.8 Hz, 2H); 3.90 (s, 9H); 1.80 (quint, J = 6.8 Hz, 2H); 1.42–1.26 (m, 14H); 0.87 (t, J = 7.0 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 166.23; 152.59; 142.05; 125.43; 106.46; 65.33; 60.81; 56.23; 31.89; 29.54; 29.31; 29.28; 28.75; 26.07; 22.71; 14.10 [58].
- 4-Methoxy-benzyl 3,4,5–trimethoxybenzoate (9): White cristalline solid, yield 67.5% (105.7 mg; 0.32 mmol); M.P.: 83–84 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.36; IR ʋmax (KBr, cm−1): 3008, 2944, 1711, 1670, 1589 and 1465, 1332 and 1126, 1228 and 1005, 864; 1H NMR (400 MHz, CDCl3): δ 7.38 (d, J = 9.5 Hz, 2H); 7.31 (s, 2H, H-2); 6.91 (d, J = 8.8 Hz, 2H); 5.29 (s, 2H,); 3.89 (s, 9H); 3.81 (s, 3H); 13C NMR (100 MHz, CDCl3): δ 166.25; 159.66; 152.82, 142.12; 130.01; 128.19; 125.24; 113.89 ; 106.90; 66.69; 60.94; 56.14; 55.27. HRMS (FT-ICR) analyze: C18H20O6 calculated theoretical value [M+H]+: 333.1332. Found = 333.1331.
- 4-Bromo-benzyl 3,4,5–trimethoxybenzoate (10): White cristalline solid, yield 55.0% (197.0 mg; 0.51 mmol); M.P.: 104–105 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.62; IR ʋmax (KBr, cm−1): 3034, 2958, 1712, 1664, 1594 and 1454, 1334 and 1133, 1228 and 1010, 1070, 801; 1H NMR (500 MHz, CDCl3): δ 7.51 (d, J = 8.4 Hz, 2H); 7.32 (d, J = 10 Hz, 2H); 7.31 (s, 2H); 5.30 (s, 2H); 3.90 (s, 9H,); 13C NMR (125 MHz, CDCl3): δ 166.10; 153.12; 142.68; 135.26; 131.93; 130.04; 125.00; 122.50; 107.17; 66.16; 61.10; 56.43. HRMS (FT-ICR) analyze: C17H17O5Br calculated theoretical value [M+H]+: 381.0332. Found = 381.0330.
- 2-Methylnaphthalene 3,4,5–trimethoxybenzoate (11): White solid; yield 48.4% (160.9 mg; 0.45 mmol); M.P.: 84–85 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.58; IR ʋmax (KBr, cm−1): 3002, 2936, 1714, 1670, 1590 and 1463, 1329 and 1129, 1225 and 1008, 864; 1H NMR (500 MHz, CDCl3): δ 7.91–7.85 (m, 4H); 7.55 (dd, J = 8.4 Hz; 1.6 Hz, 1H), 7.53–7.45 (m, 2H); 7.36 (s, 2H); 5,53 (s, 2H); 3.90 (s, 9H); 13C NMR (125 MHz, CDCl3): δ 166.26; 153.14; 142.55; 133.64; 133.34; 133.29; 128.57; 128.12; 127.85; 127.57; 126.47; 126.44; 126.05; 125.25; 107.18; 67.14; 61.04; 56.41. HRMS (FT-ICR) analyze: C21H20O5 calculated theoretical value [M+H]+: 353.1383. Found = 353.1381.
- Diphenyl 3,4,5–trimethoxybenzoate (12): Yellow solid; yield 41.5% (148.2 mg; 0.39 mmol); M.P.: 57–58 °C; TLC (8:2 Hexane/AcOEt), Rf = 0.64; IR ʋmax (KBr, cm−1): 3026, 2939, 1727, 1655, 1587 and 1456, 1338 and 1172, 1226 and 1127, 855; 1H NMR (500 MHz, CDCl3,): δ 7.59–7.47 (m, 12H); 7.44 (s, 1H); 4.06 (s, 9H); 13C NMR (125 MHz, CDCl3,): δ 165.29; 152.96; 142.55; 140.19; 128.58*; 128.47*; 127.99#; 127.53 #; 127.15; 126.53; 125.17; 107.15; 77.63; 60.93; 56.31. HRMS (FT-ICR) analyze: C23H22O5 calculated theoretical value [M+H]+: 379.1540. Found= 379.1512.
- N-Butyl-3,4,5–trimethoxybenzamide (13): White solid; yield 89.1% (112.2 mg; 0.41 mmol M.P.: 115–116 °C; TLC (6:4 Hexane/AcOEt), Rf = 0.36; IR ʋmax (KBr, cm−1): 3294, 3017, 2932, 1681, 1634, 1583 and 1459, 1541 and 1506, 1236 and 1131, 843; 1H NMR (400 MHz, CDCl3): δ 6.98 (s, 2H); 6.22 (s, 1H); 3.87 (s, 6H); 3.86 (s, 3H); 3.43 (q, J = 5.8 Hz, 2H); 1.62 (quint, J = 7.2 Hz, 2H); 1.38 (sex, J = 7.5 Hz, 2H); 0.94 (t, J = 7.3 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 167.32; 153.21; 140.87; 130.53; 104.46; 60.97; 56.28; 40.03; 31.91; 20.19; 13.91 [59].
- N-Isobutyl-3,4,5–trimethoxybenzamide (14): Yellow solid; yield 91.3% (115.0 mg; 0.43 mmol); M.P.: 118–119 °C; TLC (6:4 Hexane/AcOEt), Rf = 0.36 ; IR ʋmax (KBr, cm−1): 3307, 3015, 2955, 1687, 1634, 1583 and 1469, 1543 and 1504, 1237 and 1131, 842; 1H NMR (500 MHz, CDCl3,): δ 6.98 (s, 2H), 6.30 (s, 1H), 3.86 (s, 6H), 3.85 (s, 3H); 3.24 (t, J = 6.5 Hz, 2H), 1.92–1.84 (m, 1H), 0.95 (d, J = 6.7 Hz, 6H). 13C NMR (125 MHz, CDCl3,): δ 167.49; 153.27; 140.75; 130.45; 104.46; 60.94; 56.41; 47.59; 28.74; 20.25 [59].
- N-Cyclohexyl-3,4,5–trimethoxybenzamide (15): White crystalline solid; yield 44.4% (61.4 mg; 0.21 mmol); M.P.: 179–180 °C; TLC (6:4 Hexane/AcOEt), Rf = 2.2; IR ʋmax (KBr, cm−1): 3468, 3077, 2871, 1677, 1621, 1582 and 1463, 1510 and 1417, 1239 and 1004, 844; 1H NMR (500 MHz, DMSO-d6): δ 7.16 (s, 1H); 6.19 (s, 2H); 2.86 (s, 6H); 2.72 (s, 3H); 1.54 (quint J = 1.8 Hz, 1H); 0.88–0.76 (m, 4H); 0.67–0.62 (m, 2H); 0.36-0.32 (m, 4H); 13C NMR (125 MHz, DMSO-d6): δ 164.91; 152.50; 139.87; 130.09; 104.94; 60.22; 56.10; 48.64; 32.63; 25.41; 25.15 [60].
- N-4-Hydroxybenzyl-3,4,5–trimethoxybenzamide (16): White solid, yield 57.15% (256.3 mg; 0.81 mmol); M.P.: 227–229 °C; TLC (5:5 Hexane/AcOEt), Rf = 0.37; IR ʋmax (KBr, cm−1): 3379 and 3314, 3346, 3019, 2099, 1634, 1611, 1574 and 1449, 1545 and 1499, 1414 and 1231, 1211 and 1122, 823; 1H NMR (400 MHz, DMSO-d6): δ 8,40 (s, 1H); 7.99 (t, J = 5.7 Hz, 1H,); 6.35 (s, 2H); 6.24 (d, J = 8.5 Hz, 2H); 5.82 (d, J = 10.0 Hz, 2H); 3.48 (d, J = 5.7 Hz, 2H); 2.93 (s, 6H); 2.81 (s, 3H); 13C NMR (100 MHz, DMSO-d6): δ 165.44; 156.32; 152.65; 139.90; 129.89; 129.61; 128.68; 115.06; 104.84; 60.10; 56.01; 42.30 [60].
- N-Benzyl-3,4,5–trimethoxybenzamide (17): White crystalline solid, yield 59.6% (84.6 mg; 0.27 mmol); M.P.: 138–139 °C; TLC (6:4 Hexane/AcOEt), Rf = 0.38; IR ʋmax (KBr, cm−1): 3305, 3028, 2942, 1655, 1625, 1580 and 1459, 1528 and 1499, 1237 and 1127, 840; 1H NMR (400 MHz, CDCl3): δ 7.35–7.27 (m, 5H); 7.03 (s, 2H); 6.60 (s, 1H); 4.61 (d, J = 5,8 Hz, 2H); 3.86 (s, 3H); 3.85 (s, 6H); 13C NMR (100 MHz, CDCl3): δ 167.11; 153.34; 141.10; 138.22; 129.87; 128.86; 128.01; 127.70; 104.55; 61.04; 56.34; 44.30 [53].
- N-(4-Fluorobenzyl)-3,4,5–trimethoxybenzamide (18): White crystalline solid, yield 62.5% (90 mg; 0.28 mmol); M.P.: 131–132 °C; TLC (6:4 Hexane/AcOEt), Rf = 0.34; IR ʋmax (KBr, cm−1): 3288, 3012, 2947, 1672, 1634, 1585 and 1459, 1545 and 1508, 1280 and 1130, 1219 and 1098, 827; 1H NMR (400 MHz, CDCl3): δ 7.29–7.27 (m, 2H); 7.01 (s, 2H); 6.98 (d, J = 8,7 Hz, 2H); 6.69 (s, 1H); 4.55 (d, J = 5,8 Hz, 2H); 3.85 (s, 6H); 3.84 (s, 3H); 13C NMR (100 MHz, CDCl3): δ 167.15; 163.50; 161.04; 153.28; 141.17; 134.05; 129.69; 129.66*; 129.58*; 115.74#; 115.52#; 104.55; 60.90; 56.29; 43.48. HRMS (FT-ICR) analyze: C17H18FNO4 calculated theoretical value [M+H]+: 320.1292. Found = 320.1290.
- *, # Interchangeable.
- N-4-Chlorobenzyl-3,4,5–trimethoxybenzamide (19): White solid; yield 86.6% (137.0 mg; 0.41 mmol); M.P.: 157–158 °C; TLC (6:4 Hexane/AcOEt), Rf = 0.34; IR ʋmax (KBr, cm−1): 3254, 3004, 2946, 1653, 1629, 1582 and 1457, 1538 and 1498, 1235 and 1128, 1070 and 997, 816; 1H NMR (500 MHz, CDCl3): δ 7.31 (d, J = 6.4 Hz, 2H); 7.28 (d, J = 6.4 Hz, 2H); 7.04 (s, 2H); 6.74 (s, 1H); 4.58 (d, J = 5,7 Hz, 2H); 3.88 (s, 3H); 3.87 (s, 6H); 13C NMR (125 MHz, CDCl3): δ 167.30; 153.17; 141.26; 136.97; 133.37; 129.60; 129.27; 128.92; 104.58; 61.01; 56.43; 43.55. HRMS (FT-ICR) analyze: C17H18ClNO4 calculated theoretical value [M+H]+: 336.0997. Found =336.0995.
4.4. Cytotoxic Activity
4.4.1. Cell Viability (Cytotoxicity) Assay
4.4.2. Hemolysis Assay
4.4.3. Cell Cycle and SubG1 Analysis
4.4.4. Phosphatidylserine Exposure Analysis (Apoptosis)
4.4.5. Caspase Analysis
4.4.6. Statistical Analysis, Calculation of IC50, and Selectivity Index (SI)
4.5. Modeling Study
4.5.1. Target Selection
4.5.2. Molecular Docking
4.5.3. Molecular Dynamics Simulations and Free Energies of Binding
4.5.4. ADMET Predictions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
List of Abbreviations
| AcOEt | ethyl acetate |
| ADMET | Absorption, Distribution, Metabolism, Excretion and Toxicity |
| AHR | Aryl hydrocarbon receptor |
| ATCC | American Type Culture Collection |
| B16-F10 | cell lines from origin of melanoma |
| CC | column chromatography |
| CDCl3 | deuterated chloroform |
| CRM1 | Exportin-1 |
| CTD2 | Cancer Target Discovery and Development |
| DMEM | Dulbecco′s Modified Eagle′s Medium |
| DMSO-d6 | Dimethyl sulfoxide-d6 |
| DUSP3 | Receptor-interacting serine/threonine-protein kinase 2 |
| EC | Esophageal Cancer |
| Average normalized score of target i | |
| FITC | Fluorescein isothiocyanate |
| FSi | fluid-structure interaction |
| FTIR | Fourier Transform Infrared Spectroscopy |
| GSTP1 | Glutathione S-transferase P |
| HRMS | High Resolution Mass spectra |
| HEP-G2 | cell lines from origin of hepatocarcinoma |
| HT-29 | cell lines from origin of colon adenocarcinoma |
| Hz | Hertz |
| IC50 | inhibitory concentration 50% |
| 1H NMR | Hydrogen Nuclear Magnetic Resonance |
| 13C NMR | Carbon Thirteen Nuclear Magnetic Resonance |
| INCA | National Cancer Institute |
| i-th | Target |
| KBr | Potassium bromide |
| M | Number of target fishing algorithms |
| MCL1 | Induced myeloid leukemia cell differentiation protein Mcl-1 |
| MD | Molecular Dynamics |
| MD-based | Molecular Dynamics-based |
| MHz | Megahertz |
| MMP2 | 72 kDa type IV collagenase |
| MP | Melting points |
| MTOR | Serine/threonine-protein kinase mTOR |
| MTNR1B | Melatonin receptor type 1B |
| MTT | 3,4,5-dimethiazol-2,5-diphenyltetrazoliumbromide |
| N | Number of methods identifying |
| N.D. | not determined |
| NFKB | Nuclear factor NF-kappa-B p105 subunit |
| NIH | National Institutes of Health |
| NP-40 | Tergitol Type NP-40 |
| OSCC | Oral Squamous Cell Carcinoma |
| PBS | Phosphate buffered saline |
| P.I. | propidium iodide |
| PI3K | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
| PyBOP | (Benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate |
| RELA | Transcription factor p65 |
| Rf | Retardation factor |
| RIPK2 | 72 kDa type IV collagenase |
| SAR | STRUCTURE ACTIVITY RELATION |
| SAS | Tongue squamous cell carcinoma cell line. |
| STAT3 | Signal transducer and activator of transcription 3 |
| SCC | Squamous Cell Carcinoma |
| SCC4 | Squamous Cell Carcinoma—4 |
| SCC9 | Squamous Cell Carcinoma—9 |
| S.I. | selectivity index |
| S.D. | standard deviation |
| TLC | thin layer chromatography |
| V-FITC | ANNEXIN V-FITC |
References
- American Cancer Society (ACS). Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-andstatistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf (accessed on 9 July 2018).
- Instituto Nacional de Câncer José Alencar Gomes da Silva. Available online: https://www.inca.gov.br/en/node/3776#:~:text=A%20menos%20que%20sejam%20tomadas,ser%20evitadas%20com%20medidas%20adequadas (accessed on 16 September 2022).
- Instituto Nacional de Câncer (INCA). Available online: http://www.inca.gov.br/estimativa/2014/index.asp?ID=2 (accessed on 9 July 2018).
- Organização Mundial da Saúde (OMS). Available online: http://www.who.int/mediacentre/news/releases/2003/pr27/en/ (accessed on 9 July 2018).
- Zhang, Y.; Somtakoune, S.D.; Cheung, C.; Listiawan, M.; Feng, X. Therapeutic Application of Pharmacogenomics in Oncology. AAPS J. 2016, 18, 819–829. [Google Scholar] [CrossRef]
- Brener, S.; Jeunon, F.A.; Barbosa, A.A.; Grandinetti, H.D.A.M. Carcinoma De células Escamosas Bucal: Uma revisão De Literatura Entre O Perfil Do Paciente, Estadiamento clínico E Tratamento Proposto. Rev. Bras. Cancerol. 2007, 53, 63–69. [Google Scholar] [CrossRef]
- Massano, J.; Regateiro, F.S.; Januário, G.; Ferreira, A. Oral squamous cell carcinoma: Review of prognostic and predictive factors. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2006, 102, 67–76. [Google Scholar] [CrossRef]
- Blatt, S.; Ziebart, T.; Krüger, M.; Pabst, A.M. Diagnosing oral squamous cell carcinoma: How much imaging do we really need? A review of the current literature. J. Cranio-Maxillofac. Surg. 2016, 44, 538–549. [Google Scholar] [CrossRef]
- Feller, L.; Lemmer, J. Oral Squamous Cell Carcinoma: Epidemiology, Clinical Presentation and Treatment. J. Cancer Ther. 2012, 03, 263–268. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Morris-Natschke, S.L.; Yang, J.; Niu, H.-M.; Long, C.-L.; Lee, K.-H. Anticancer Principles from Medicinal Piper (胡椒 Hú Jiāo) Plants. J. Tradit. Complement. Med. 2014, 4, 8–16. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Pessoa, C.; de Moraes, M.O.; Saker-Neto, N.; Silveira, E.R.; Costa-Lotufo, L.V. Overview of the therapeutic potential of piplartine (piperlongumine). Eur. J. Pharm. Sci. 2013, 48, 453–463. [Google Scholar] [CrossRef]
- Regasini, L.O.; Cotinguiba, F.; Passerini, G.D.; Bolzani, V.D.S.; Cicarelli, R.M.B.; Kato, M.; Furlan, M. Trypanocidal activity of Piper arboreum and Piper tuberculatum (Piperaceae). Rev. Bras. de Farm. 2009, 19, 199–203. [Google Scholar] [CrossRef]
- Costa-Lotufo, L.V.; Montenegro, R.C.; Alves, A.P.; Madeira, S.V.; Pessoa, C.; Moraes, M.E.; Moraes, M.O. A contribuição dos produtos naturais como fonte de novos fármacos anticâncer: Estudos no Laboratório Nacional de Oncologia Experimental da Universidade Federal do Ceará. Rev. Virtual de Quím 2010, 2, 47–58. [Google Scholar] [CrossRef]
- da Fonseca, A.C.C.; de Queiroz, L.N.; Felisberto, J.S.; Ramos, Y.J.; Marques, A.M.; Wermelinger, G.F.; Pontes, B.; Moreira, D.D.L.; Robbs, B.K. Cytotoxic effect of pure compounds from Piper rivinoides Kunth against oral squamous cell carcinoma. Nat. Prod. Res. 2020, 35, 6163–6167. [Google Scholar] [CrossRef]
- Nóbrega, F.R.; Silva, L.V.; Bezerra Filho, C.D.; Lima, T.C.; Castillo, Y.P.; Bezerra, D.P.; Lima, T.K.; Sousa, D.P. Design, Antileishmanial Activity, and QSAR Studies of a Series of Piplartine Analogues. J. Chem. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Mahavorasirikul, W.; Viyanant, V.; Chaijaroenkul, W.; Itharat, A.; Na-Bangchang, K. Cytotoxic activity of Thai medicinal plants against human cholangiocarcinoma, laryngeal and hepatocarcinoma cells in vitro. BMC Complement. Altern. Med. 2010, 10, 55. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Pessoa, C.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.S.; Elmiro, F.J.M.; Costa-Lotufo, L.V. Antiproliferative Effects of Two Amides, Piperine and Piplartine, from Piper Species. Z. Naturforsch C J. Biosci. 2005, 60, 539–543. [Google Scholar] [CrossRef]
- Bollu, L.R.; Mazumdar, A.; Savage, M.I.; Brown, P.H. Molecular Pathways: Targeting Protein Tyrosine Phosphatases in Cancer. Clin. Cancer Res. 2017, 23, 2136–2142. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Xavier, C.E.; Aurtenetxe, O.; Zaldumbide, L.; López-Almaraz, R.; Erramuzpe, A.; Cortés, J.M.; López, J.I.; Pulido, R. Protein tyrosine phosphatase PTPN1 modulates cell growth and associates with poor outcome in human neuroblastoma. Diagn. Pathol. 2019, 14, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wu, N.; Li, X.; Guo, C.; Li, C.; Jiang, B.; Wang, H.; Shi, D. Inhibition of PTP1B blocks pancreatic cancer progression by targeting the PKM2/AMPK/mTOC1 pathway. Cell Death Dis. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Wang, M.; Yang, K.; Chi, T.; Liao, Z.; Wei, P. PPAR-α Modulators as Current and Potential Cancer Treatments. Front. Oncol. 2021, 11, 9995. [Google Scholar] [CrossRef]
- Heublein, S.; Mayr, R.; Meindl, A.; Angele, M.; Gallwas, J.; Jeschke, U.; Ditsch, N. Thyroid Hormone Receptors Predict Prognosis in BRCA1 Associated Breast Cancer in Opposing Ways. PLoS ONE 2015, 10, e0127072. [Google Scholar] [CrossRef]
- Shinderman-Maman, E.; Cohen, K.; Moskovich, D.; Hercbergs, A.; Werner, H.; Davis, P.J.; Ellis, M.; Ashur-Fabian, O. Thyroid hormones derivatives reduce proliferation and induce cell death and DNA damage in ovarian cancer. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Davidson, C.D.; Gillis, N.E.; Carr, F.E. Thyroid Hormone Receptor Beta as Tumor Suppressor: Untapped Potential in Treatment and Diagnostics in Solid Tumors. Cancers 2021, 13, 4254. [Google Scholar] [CrossRef]
- Xi, H.; Wu, R.; Liu, J.; Zhang, L.; Li, Z. Role of acetylcholinesterase in lung cancer. Thorac. Cancer 2015, 6, 390–398. [Google Scholar] [CrossRef]
- Hakem, R.; Hakem, A.; Duncan, G.S.; Henderson, J.T.; Woo, M.; Soengas, M.S.; Elia, A.; de la Pompa, J.L.; Kagi, D.; Khoo, W.; et al. Differential Requirement for Caspase 9 in Apoptotic Pathways In Vivo. Cell 1998, 94, 339–352. [Google Scholar] [CrossRef]
- Wang, J.; Morin, P.; Wang, W.; Kollman, P.A. Use of MM-PBSA in Reproducing the Binding Free Energies to HIV-1 RT of TIBO Derivatives and Predicting the Binding Mode to HIV-1 RT of Efavirenz by Docking and MM-PBSA. J. Am. Chem. Soc. 2001, 123, 5221–5230. [Google Scholar] [CrossRef]
- Katsila, T.; Spyroulias, G.A.; Patrinos, G.P.; Matsoukas, M.-T. Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 2016, 14, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Guterres, H.; Im, W. Improving Protein-Ligand Docking Results with High-Throughput Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020, 60, 2189–2198. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA Methods in Virtual Screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef] [PubMed]
- Shaikhqasem, A.; Dickmanns, A.; Neumann, P.; Ficner, R. Characterization of Inhibition Reveals Distinctive Properties for Human and Saccharomyces cerevisiae CRM1. J. Med. Chem. 2020, 63, 7545–7558. [Google Scholar] [CrossRef]
- Etchin, J.; Sun, Q.; Kentsis, A.; Farmer, A.; Zhang, Z.C.; Sanda, T.; Mansour, M.; Barceló, C.; McCauley, D.; Kauffman, M.; et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 2012, 27, 66–74. [Google Scholar] [CrossRef]
- Lei, Y.; An, Q.; Shen, X.-F.; Sui, M.; Li, C.; Jia, D.; Luo, Y.; Sun, Q. Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1. J. Med. Chem. 2021, 64, 6596–6607. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Ferreira, B.I.; Cautain, B.; Grenho, I.; Link, W. Small Molecule Inhibitors of CRM1. Front. Pharmacol. 2020, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Dickmanns, A.; Monecke, T.; Ficner, R. Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells 2015, 4, 538–568. [Google Scholar] [CrossRef]
- Azizian, N.G.; Li, Y. XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol. 2020, 13, 61. [Google Scholar] [CrossRef]
- Niu, M.; Xu, X.; Shen, Y.; Yao, Y.; Qiao, J.; Zhu, F.; Zeng, L.; Liu, X.; Xu, K. Piperlongumine is a novel nuclear export inhibitor with potent anticancer activity. Chem. Interact. 2015, 237, 66–72. [Google Scholar] [CrossRef]
- Garces, H.I.; Mora, P.A.R.; Alves, F.V.G.; Carmo, C.C.D.; Grazziotin, R.; Fernandes, A.C.F.M.; Nogueira-Rodrigues, A.; de Melo, A.C. First-Line Paclitaxel and Carboplatin in Persistent/Recurrent or Advanced Cervical Cancer. Int. J. Gynecol. Cancer 2013, 23, 743–748. [Google Scholar] [CrossRef]
- Avelino, C.U.R.; Cardoso, R.M.; De Aguiar, S.S.; Da Silva, M.J.S. Assessment of quality of life in patients with advanced non-small cell lung carcinoma treated with a combination of carboplatin and paclitaxel. J. Bras. Pneumol. 2015, 41, 133–142. [Google Scholar] [CrossRef]
- Díaz-Guardamino, I.E. Identificación de Predictores de Respuesta a Quimioterapia Neodayuvante con Carboplatino-docetaxel de Pacientes con Cáncer de Mama Triple Negative. Ph.D. Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2017. [Google Scholar]
- Fan, X.; Song, J.; Zhao, Z.; Chen, M.; Tu, J.; Lu, C.; Wu, F.; Zhang, D.; Weng, Q.; Zheng, L.; et al. Piplartine suppresses proliferation and invasion of hepatocellular carcinoma by LINC01391-modulated Wnt/β-catenin pathway inactivation through ICAT. Cancer Lett. 2019, 460, 119–127. [Google Scholar] [CrossRef]
- Turkez, H.; da Nóbrega, F.R.; Ozdemir, O.; Filho, C.D.S.M.B.; de Almeida, R.N.; Tejera, E.; Perez-Castillo, Y.; de Sousa, D.P. NFBTA: A Potent Cytotoxic Agent against Glioblastoma. Molecules 2019, 24, 2411. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef]
- Zorzanelli, B.C.; de Queiroz, L.N.; Santos, R.M.; Menezes, L.M.; Gomes, F.C.; Ferreira, V.F.; Silva, F.D.C.D.; Robbs, B.K. Potential cytotoxic and selective effect of new benzo[b]xanthenes against oral squamous cell carcinoma. Futur. Med. Chem. 2018, 10, 1141–1157. [Google Scholar] [CrossRef]
- Chipoline, I.C.; da Fonseca, A.C.C.; da Costa, G.R.M.; de Souza, M.P.; Rabelo, V.W.-H.; de Queiroz, L.N.; de Souza, T.L.F.; de Almeida, E.C.P.; Abreu, P.A.; Pontes, B.; et al. Molecular mechanism of action of new 1,4-naphthoquinones tethered to 1,2,3-1H-triazoles with cytotoxic and selective effect against oral squamous cell carcinoma. Bioorganic Chem. 2020, 101, 103984. [Google Scholar] [CrossRef] [PubMed]
- Machado, T.Q.; Felisberto, J.R.S.; Guimarães, E.F.; de Queiroz, G.A.; da Fonseca, A.C.C.; Ramos, Y.J.; Marques, A.M.; Moreira, D.D.L.; Robbs, B.K. Apoptotic effect of β-pinene on oral squamous cell carcinoma as one of the major compounds from essential oil of medicinal plant Piper rivinoides Kunth. Nat. Prod. Res. 2021, 36, 1636–1640. [Google Scholar] [CrossRef] [PubMed]
- Zorzanelli, B.C.; Ouverney, G.; Pauli, F.P.; da Fonseca, A.C.C.; de Almeida, E.C.P.; de Carvalho, D.G.; Possik, P.A.; Rabelo, V.W.-H.; Abreu, P.A.; Pontes, B.; et al. Pro-Apoptotic Antitumoral Effect of Novel Acridine-Core Naphthoquinone Compounds against Oral Squamous Cell Carcinoma. Molecules 2022, 27, 5148. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Militão, G.C.G.; de Castro, F.O.; Pessoa, C.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.S.; Elmiro, F.J.M.; Costa-Lotufo, L.V. Piplartine induces inhibition of leukemia cell proliferation triggering both apoptosis and necrosis pathways. Toxicol. In Vitro 2007, 21, 1–8. [Google Scholar] [CrossRef]
- Jyothi, D.; Vanathi, P.; Gowri, P.M.; Rao, V.R.S.; Rao, J.M.; Sreedhar, A. Diferuloylmethane augments the cytotoxic effects of piplartine isolated from Piper chaba. Toxicol. In Vitro 2009, 23, 1085–1091. [Google Scholar] [CrossRef]
- Da Nóbrega, F.R.; Ozdemir, O.; Sousa, S.C.S.N.; Barboza, J.N.; Turkez, H.; De Sousa, D.P. Piplartine Analogues and Cytotoxic Evaluation against Glioblastoma. Molecules 2018, 23, 1382. [Google Scholar] [CrossRef]
- Ahn, S.-C.; Kong, E.; Kim, Y.-J.; Cho, H.-J.; Yu, S.-N.; Kim, K.-Y.; Chang, J.-H.; Ahn, T. Piplartine induces caspase-mediated apoptosis in PC-3 human prostate cancer cells. Oncol. Rep. 1994, 20, 785–792. [Google Scholar] [CrossRef]
- Huang, H.-W.; Tang, J.-Y.; Ou-Yang, F.; Wang, H.-R.; Guan, P.-Y.; Huang, C.-Y.; Chen, C.-Y.; Hou, M.-F.; Sheu, J.-H.; Chang, H.-W. Sinularin Selectively Kills Breast Cancer Cells Showing G2/M Arrest, Apoptosis, and Oxidative DNA Damage. Molecules 2018, 23, 849. [Google Scholar] [CrossRef]
- Bezerra, D.P.; Moura, D.J.; Rosa, R.M.; de Vasconcellos, M.C.; e Silva, A.C.R.; de Moraes, M.O.; Silveira, E.R.; Lima, M.A.S.; Henriques, J.A.P.; Costa-Lotufo, L.V.; et al. Evaluation of the genotoxicity of piplartine, an alkamide of Piper tuberculatum, in yeast and mammalian V79 cells. Mutat. Res. Toxicol. Environ. Mutagen. 2008, 652, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Green, R.A.; Pletcher, D.; Leach, S.G.; Brown, R.C.D. N-Heterocyclic Carbene-Mediated Oxidative Electrosynthesis of Esters in a Microflow Cell. Org. Lett. 2015, 17, 3290–3293. [Google Scholar] [CrossRef]
- Iranpoor, N.; Firouzabadi, H.; Riazi, A.; Pedrood, K. Regioselective hydrocarbonylation of phenylacetylene to α,β-unsaturated esters and thioesters with Fe(CO)5 and Mo(CO)6. J. Organomet. Chem. 2016, 822, 67–73. [Google Scholar] [CrossRef]
- Zinchenko, A.N.; Muzychka, L.V.; Smolii, O.; Bdzhola, V.G.; Protopopov, M.V.; Yarmoluk, S.M. Synthesis and biological evaluation of novel amino-substituted derivatives of pyrido[2,3-d]pyrimidine as inhibitors of protein kinase CK2. Biopolym. Cell 2017, 33, 367–378. [Google Scholar] [CrossRef]
- Obrecht, D.; Bohdal, U.; Broger, C.; Bur, D.; Lehmann, C.; Ruffieux, R.; Schönholzer, P.; Spiegler, C.; Müller, K. L-Phenylalanine Cyclohexylamide: A simple and convenient auxiliary for the synthesis of optically pure α,α-disubstituted (R)- and (S)-amino acids. Helvetica Chim. Acta 1995, 78, 563–580. [Google Scholar] [CrossRef]
- Faget, D.V.; Lucena, P.I.; Robbs, B.K.; Viola, J.P.B. NFAT1 C-Terminal Domains Are Necessary but Not Sufficient for Inducing Cell Death. PLoS ONE 2012, 7, e47868. [Google Scholar] [CrossRef]
- Yao, Y.; Sun, Y.; Shi, M.; Xia, D.; Zhao, K.; Zeng, L.; Yao, R.; Zhang, Y.; Li, Z.; Niu, M.; et al. Piperlongumine induces apoptosis and reduces bortezomib resistance by inhibiting STAT3 in multiple myeloma cells. Oncotarget 2016, 7, 73497–73508. [Google Scholar] [CrossRef]
- Zheng, J.; Son, D.J.; Gu, S.M.; Woo, J.R.; Ham, Y.W.; Lee, H.P.; Kim, W.J.; Jung, J.K.; Hong, J.T. Piperlongumine inhibits lung tumor growth via inhibition of nuclear factor kappa B signaling pathway. Sci. Rep. 2016, 6, 26357. [Google Scholar] [CrossRef]
- Shrivastava, S.; Kulkarni, P.; Thummuri, D.; Jeengar, M.K.; Naidu, V.G.M.; Alvala, M.; Redddy, G.B.; Ramakrishna, S. Piperlongumine, an alkaloid causes inhibition of PI3 K/Akt/mTOR signaling axis to induce caspase-dependent apoptosis in human triple-negative breast cancer cells. Apoptosis 2014, 19, 1148–1164. [Google Scholar] [CrossRef]
- Harshbarger, W.; Gondi, S.; Ficarro, S.B.; Hunter, J.; Udayakumar, D.; Gurbani, D.; Singer, W.D.; Liu, Y.; Li, L.; Marto, J.A.; et al. Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine. J. Biol. Chem. 2017, 292, 112–120. [Google Scholar] [CrossRef]
- Tejera, E.; Pérez-Castillo, Y.; Toscano, G.; Noboa, A.L.; Ochoa-Herrera, V.; Giampieri, F.; Álvarez-Suarez, J.M. Computational modeling predicts potential effects of the herbal infusion “horchata” against COVID-19. Food Chem. 2021, 366, 130589. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Noboa, A.; Proaño-Ojeda, J.; Guevara, M.; Gallo, B.; Berrueta, L.A.; Giampieri, F.; Perez-Castillo, Y.; Battino, M.; Álvarez-Suarez, J.M.; Tejera, E. Metabolomic profile and computational analysis for the identification of the potential anti-inflammatory mechanisms of action of the traditional medicinal plants Ocimum basilicum and Ocimum tenuiflorum. Food Chem. Toxicol. 2022, 164, 3039. [Google Scholar] [CrossRef] [PubMed]
- Peón, A.; Naulaerts, S.; Ballester, P.J. Predicting the Reliability of Drug-target Interaction Predictions with Maximum Coverage of Target Space. Sci. Rep. 2017, 7, 3820. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Yao, Z.-J.; Dong, J.; Che, Y.-J.; Zhu, M.-F.; Wen, M.; Wang, N.-N.; Wang, S.; Lu, A.-P.; Cao, D.-S. TargetNet: A web service for predicting potential drug–target interaction profiling via multi-target SAR models. J. Comput. Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef]
- Lee, K.; Lee, M.; Kim, D. Utilizing random Forest QSAR models with optimized parameters for target identification and its application to target-fishing server. BMC Bioinform. 2017, 18, 567. [Google Scholar] [CrossRef]
- Awale, M.; Reymond, J.-L. Polypharmacology Browser PPB2: Target Prediction Combining Nearest Neighbors with Machine Learning. J. Chem. Inf. Model. 2018, 59, 10–17. [Google Scholar] [CrossRef]
- Liu, S.-H.; Shen, P.-C.; Chen, C.-Y.; Hsu, A.-N.; Cho, Y.-C.; Lai, Y.-L.; Chen, F.-H.; Li, C.-Y.; Wang, S.-C.; Chen, M.; et al. DriverDBv3: A multi-omics database for cancer driver gene research. Nucleic Acids Res. 2019, 48, D863–D870. [Google Scholar] [CrossRef]
- Aksoy, B.A.; Dančík, V.; Smith, K.; Mazerik, J.N.; Ji, Z.; Gross, B.; Nikolova, O.; Jaber, N.; Califano, A.; Schreiber, S.L.; et al. CTD2 Dashboard: A searchable web interface to connect validated results from the Cancer Target Discovery and Development Network. Database 2017, 2017, 54. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. TheBioGRIDdatabase: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2020, 30, 187–200. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Perez-Castillo, Y.; Lima, T.C.; Ferreira, A.R.; Silva, C.R.; Campos, R.S.; Neto, J.B.A.; Magalhães, H.I.F.; Cavalcanti, B.C.; Júnior, H.V.N.; de Sousa, D.P. Bioactivity and Molecular Docking Studies of Derivatives from Cinnamic and Benzoic Acids. BioMed. Res. Int. 2020, 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lopes, S.P.; Castillo, Y.P.; Monteiro, M.L.; de Menezes, R.R.; Almeida, R.N.; Martins, A.; Sousa, D.P. Trypanocidal Mechanism of Action and in silico Studies of p-Coumaric Acid Derivatives. Int. J. Mol. Sci. 2019, 20, 5916. [Google Scholar] [CrossRef] [PubMed]
- OMEGA. OpenEye Scientific Software, Santa Fe, NM. Available online: https://www.eyesopen.com/search?term=OMEGA (accessed on 20 October 2022).
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- QUACPAC. OpenEye Scientific Software, Santa Fe, NM. Available online: https://www.eyesopen.com/search?term=QUACPAC (accessed on 20 October 2022).
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Case, D.; Aktulga, H.; Belfon, K.; Ben-Shalom, I.; Berryman, J.; Brozell, S.; Cerutti, D.; Cheatham, T., III; Cis-neros, G.; Cruzeiro, V.; et al. AMBER 2022; University of California: San Francisco, CA, USA, 2022. [Google Scholar]
- Araújo, M.O.; Pérez-Castillo, Y.; Oliveira, L.H.G.; Nunes, F.C.; De Sousa, D.P. Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control. Molecules 2020, 26, 61. [Google Scholar] [CrossRef]
- Mayo Clinic. Zinc Protein Simulations Using the Cationic Dummy Atom (CaDA) Method. Available online: https://www.mayo.edu/research/labs/computer-aided-molecular-design/projects/zinc-protein-simulations-using-cationic-dummy-atom-cada-approach (accessed on 21 January 2021).
- Pang, Y.P.; Xu, K.; Yazal, J.E.; Prendergas, F.G. Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci. 2000, 9, 1857–1865. [Google Scholar]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]

| COMPOUND | R | SCC9 | Primary Gingival Fibroblast | S.I. | ||
|---|---|---|---|---|---|---|
| IC50 μM (µg/mL) | S.D. μM (µg/mL) | IC50 μM (µg/mL) | S.D. μM (µg/mL) | |||
| ESTERS | ||||||
| 1 | -CH3 | 858.5 (194.21) | 0.10 (0.022) | - | - | - |
| 2 | -CH2CH3 | N.D. | - | - | - | - |
| 3 | -CH2CH2CH3 | 299 (76.03) | 0.06 (0.015) | 1256 (319.37) | 0.04 (0.010) | 4.20 |
| 4 | -CH(CH3)2 | 373.1 (94.87) | 0.05 (0.013) | 1795 (456.43) | 0.07 (0.018) | 4.81 |
| 5 | -CH2CH2CH2CH3 | 204.2 (54.79) | 0.07 (0.019) | 510.4 (136.94) | 0.04 (0.012) | 2.50 |
| 6 | -CH2CH2CH(CH3)2 | 160.2 (45.23) | 0.07 (0.2) | 569.8 (160.87) | 0.07 (0.2) | 3.56 |
| 7 | CH2CH2CH2CH2CH3 | 257.6 (72.73) | 0.08 (0.02) | 417.6 (117.9) | 0.02 (0.0056) | 1.62 |
| 8 | -CH2(CH2)7CH2CH3 | 954.9 (336.57) | 0.18 (0.063) | - | - | - |
| 9 |  | 46.21 (15.36) | 0.16 (0.053) | 740.6 (246.14) | 0.03 (0.009) | 16.02 |
| 10 | 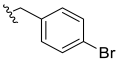 | N.D. | - | - | - | |
| 11 | 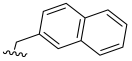 | 309.9 (109.2) | 0.04 (0.014) | 3406 (1200.2) | 0.06 (0.021) | 3.78 |
| 12 | 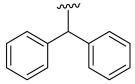 | 829.8 (314.01) | 0.26 (0.098) | - | - | - |
| AMIDES | ||||||
| 13 | -CH2CH2CH2CH3 | 468.8 (125.32) | 0.07 (0.019) | 2359 (630.61) | 0.05 (0.013) | 5.03 |
| 14 | -CH2CH(CH3)2 | N.D. | - | - | - | - |
| 15 | 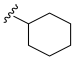 | N.D. | - | - | - | - |
| 16 | 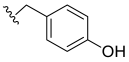 | N.D. | - | - | - | |
| 17 | 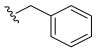 | N.D. | - | - | - | - |
| 18 | 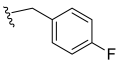 | N.D. | - | - | - | - |
| 19 | 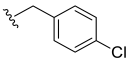 | N.D. | - | - | - | - |
| 3,4,5-Trimethoxybenzoic acid | H | N.D. | - | - | - | - |
| Carboplatin | - | 208.4 (77.37) | 0.05 (0.018) | 512.3 (190.19) | 0.02 (0.0074) | 2.46 |
| Oral Tumor Cells | Primary Gingival Fibroblast | Average S.I. | ||||||
|---|---|---|---|---|---|---|---|---|
| COMPOUND | SCC9 | SCC4 | Average | |||||
| IC50 | S.D. | IC50 | S.D. | IC50 | S.D. | |||
| 3 | 299.0 | 0.06 | N.D. | N.D. | N.D. | 1256 | 0.04 | N.D. |
| 4 | 373.1 | 0.05 | 454.3 | 0.04 | 375.6 | 1795 | 0.07 | 4.79 |
| 9 | 46.21 | 0.16 | 49.81 | 0.34 | 48.01 | 740.6 | 0.03 | 15.42 |
| 13 | 468.8 | 0.07 | N.D. | N.D. | N.D. | 2359 | 0.05 | N.D. |
| CARBOPLATIN | 208.4 | 0.05 | 175.2 | 0.15 | 191.8 | 512.3 | 0.02 | 2.67 |
| Tumor Cells | COMPOUND 9 | ||
|---|---|---|---|
| IC50 | S.D. | S.I. | |
| B16-F10 | 73.29 | 0.06 | 10.10 |
| HEP G2 | 6.63 | 0.15 | 111.7 |
| HT-29 | 7.31 | 0.23 | 101.3 |
| UniProt Accession | Target ID (a) | Description | Source (b) |
|---|---|---|---|
| P40763 (c) | STAT3 | Signal transducer and activator of transcription 3 | AlphaFold |
| P19838 (c) | NFKB | Nuclear factor NF-kappa-B p105 subunit | PDB (1SVC) |
| O14980 (c) | CRM1 | Exportin-1 | PDB (6TVO) |
| P42336 (c) | PI3K | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | PDB (4JPS) |
| P42345 (c) | MTOR | Serine/threonine-protein kinase mTOR | PDB (4JSP) |
| P09211 (c) | GSTP1 | Glutathione S-transferase P | PDB (5J41) |
| P35869 (d) | AHR | Aryl hydrocarbon receptor | SwissModel |
| P49286 (d) | MTNR1B | Melatonin receptor type 1B | PDB (6ME6) |
| Q04206 (d) | RELA | Transcription factor p65 | PDB (3GUT) |
| P08253 (d) | MMP2 | 72 kDa type IV collagenase | PDB (1HOV) |
| O43353 (d) | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | PDB (5J79) |
| O43353 (d) | DUSP3 | Receptor-interacting serine/threonine-protein kinase 2 | PDB (3F81) |
| Q07820 (d) | MCL1 | Induced myeloid leukemia cell differentiation protein Mcl-1 | PDB (6UDV) |
| Parameter | Compound 9 |
|---|---|
| Physicochemical properties | |
| Molecular weight (g/mol) | 332.35 |
| Rotatable bonds | 8 |
| H-bond acceptors | 6 |
| H-bond donors | 0 |
| Fraction Csp3 | 0.28 |
| TPSA (A3) | 63.22 |
| Lipophilicity (Log Po/w) | |
| iLOGP | 3.64 |
| XLOGP3 | 3.15 |
| MLOGP | 2.1 |
| Consensus | 3.06 |
| Absorption | |
| Water solubility (log mol/L) | −4.88 |
| GI (%) | 97.58 |
| Log Kp (skin permeation) cm/s | −2.74 |
| P-gp substrate | No |
| Distribution | |
| BBB permeability (log BB) | −0.124 |
| CNS permeation (Log PS) | −3.02 |
| VD (human) (log L/kg) | −0.425 |
| Metabolism | |
| CYP1A2 inhibitor | Yes |
| CYP2C9 inhibitor | No |
| CYP2C19 inhibitor | Yes |
| CYP3A4 inhibitor | Yes |
| CYP2D6 inhibitor | No |
| Excretion | |
| Total Clearance (log mL/min/kg) | 0.59 |
| Renal OCT2 substrate | No |
| Toxicity | |
| AMES toxicity | Yes |
| Max. tolerated dose (human) (log mg/kg/day) | 1.44 |
| hERG I inhibitor | No |
| hERG II inhibitor | No |
| Oral rat acute toxicity (LD50) (mol/kg) | 2.34 |
| Oral rat chronic toxicity (LOAEL) (log mg/kg_bw/day) | 1.53 |
| Hepatotoxicity | No |
| Skin Sensitisation | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, R.H.N.; Machado, T.Q.; da Fonseca, A.C.C.; Tejera, E.; Perez-Castillo, Y.; Robbs, B.K.; de Sousa, D.P. Molecular Modeling and In Vitro Evaluation of Piplartine Analogs against Oral Squamous Cell Carcinoma. Molecules 2023, 28, 1675. https://doi.org/10.3390/molecules28041675
Silva RHN, Machado TQ, da Fonseca ACC, Tejera E, Perez-Castillo Y, Robbs BK, de Sousa DP. Molecular Modeling and In Vitro Evaluation of Piplartine Analogs against Oral Squamous Cell Carcinoma. Molecules. 2023; 28(4):1675. https://doi.org/10.3390/molecules28041675
Chicago/Turabian StyleSilva, Rayanne H. N., Thaíssa Q. Machado, Anna Carolina C. da Fonseca, Eduardo Tejera, Yunierkis Perez-Castillo, Bruno K. Robbs, and Damião P. de Sousa. 2023. "Molecular Modeling and In Vitro Evaluation of Piplartine Analogs against Oral Squamous Cell Carcinoma" Molecules 28, no. 4: 1675. https://doi.org/10.3390/molecules28041675
APA StyleSilva, R. H. N., Machado, T. Q., da Fonseca, A. C. C., Tejera, E., Perez-Castillo, Y., Robbs, B. K., & de Sousa, D. P. (2023). Molecular Modeling and In Vitro Evaluation of Piplartine Analogs against Oral Squamous Cell Carcinoma. Molecules, 28(4), 1675. https://doi.org/10.3390/molecules28041675






