Leaves of Moringa oleifera Are Potential Source of Bioactive Compound β-Carotene: Evidence from In Silico and Quantitative Gene Expression Analysis
Abstract
1. Introduction
2. Results
2.1. Retrieval of Zeta-Carotene Desaturase (ZDS) and Phytoene Synthase (PSY) Genes
2.2. Sequencing of ZDS and PSY Genes of M. oleifera and Submission to NCBI Database
2.3. Multiple Sequence Alignments and Phylogenetic Analysis
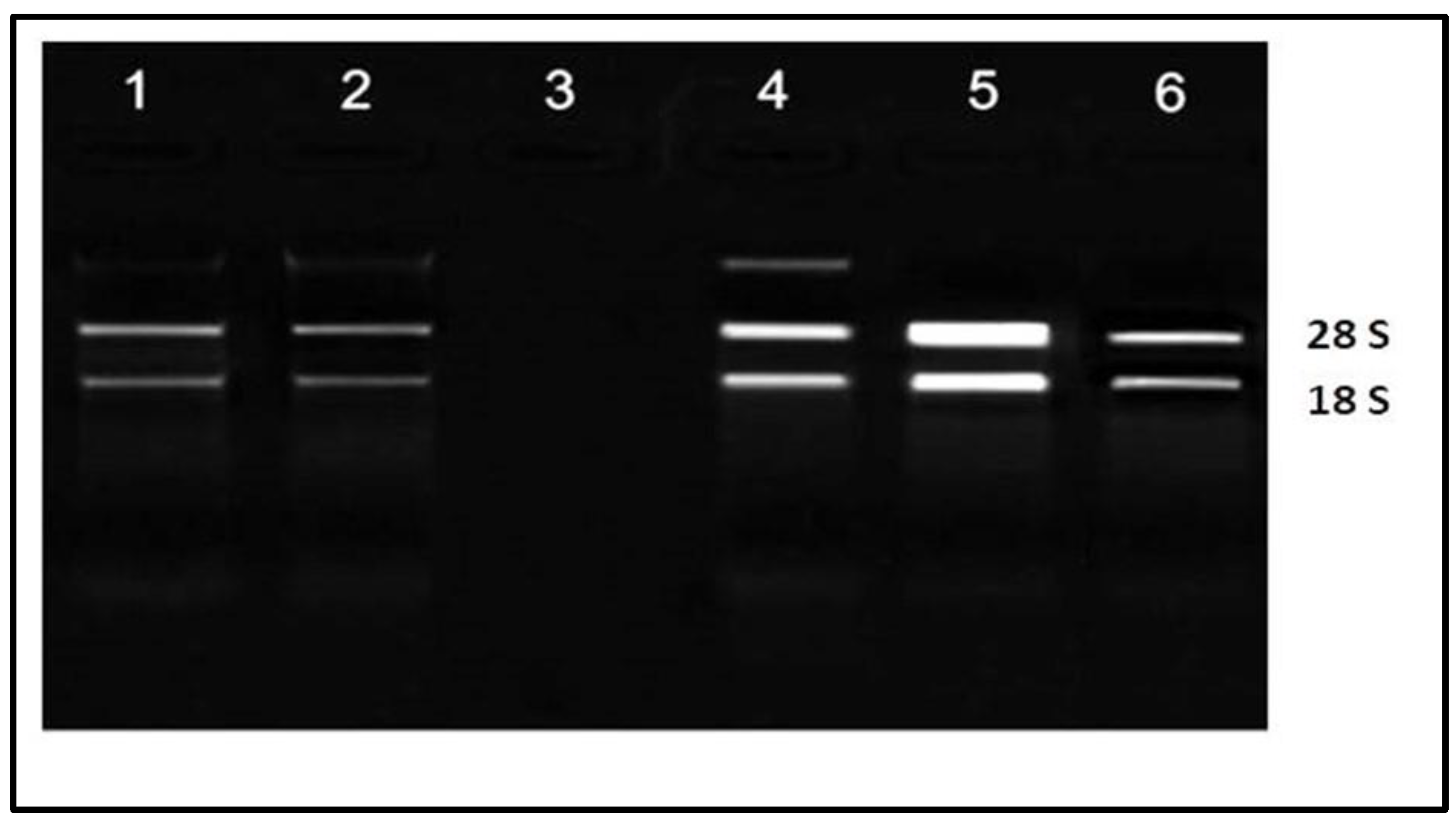
2.4. RNA Extraction and cDNA Preparation
2.5. qRT-PCR Data Interpretation
3. Discussion
4. Materials and Methods
4.1. Assembling the Genes Data Set
4.2. DNA Extraction, PCR and Sequencing of ZDS and PSY Genes
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. Protein Profiling and Analysis
4.5. Primer Design
4.6. Sample Collection and RNA Extraction
4.7. cDNA Preparation
4.8. qRT-PCR Analysis
4.9. Quantitative Gene Expression Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kamboj, V.P. Herbal medicine. Curr. Sci. 2000, 78, 35–39. Available online: https://www.jstor.org/stable/24103844 (accessed on 9 November 2022).
- Shaikh, B.T.; Hatcher, J. Complementary and alternative medicine in Pakistan: Prospects and limitations. Evid. Based Complement. Alternat. Med. 2005, 2, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, J.W.; Pettus, K.; Wheeler, K.E.; Hallam, C.; Bewley, T.; Attaran, A.; Gelb, A.W. Access to controlled medicines for anesthesia and surgical care in low-income countries: A narrative review of international drug control systems and policies. Can. J. Anaesth. 2017, 64, 296–307. [Google Scholar] [CrossRef]
- Saleem, A.; Ahotupa, M.; Pihlaja, K. Total phenolics concentration and antioxidant potential of extracts of medicinal plants of Pakistan. Z. Naturforsch C J. Biosci. 2001, 56, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Adeyemi, O.S.; Elebiyo, T.C. Moringa oleifera Supplemented Diets Prevented Nickel-Induced Nephrotoxicity in Wistar Rats. J. Nutr. Metab. 2014, 2014, 958621. [Google Scholar] [CrossRef]
- Shinwari, Z.K. Medicinal plants research in Pakistan. J. Med. Plants Res. 2010, 4, 161–176. [Google Scholar]
- Pari, L.; Kumar, N.A. Hepatoprotective activity of Moringa oleifera on antitubercular drug-induced liver damage in rats. J. Med. Food 2002, 5, 171–177. [Google Scholar] [CrossRef]
- Mayne, S.T. Beta-carotene, carotenoids, and disease prevention in humans. FASEB J. 1996, 10, 690–701. [Google Scholar] [CrossRef]
- Paiva, S.A.; Russell, R.M. β-carotene and other carotenoids as antioxidants. J. Am. Coll. Nutr. 1999, 18, 426–433. [Google Scholar] [CrossRef]
- Biesalski, H.K.; Dragsted, L.O.; Elmadfa, I.; Grossklaus, R.; Muller, M.; Schrenk, D.; Weber, P. Bioactive compounds: Safety and efficacy. Nutrition 2009, 25, 1206–1211. [Google Scholar] [CrossRef]
- Parsons, A.B.; Brost, R.L.; Ding, H.; Li, Z.; Zhang, C.; Sheikh, B.; Brown, G.W.; Kane, P.M.; Hughes, T.R.; Boone, C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004, 22, 62–69. [Google Scholar] [CrossRef]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Cultivation, genetic, ethnopharmacology, phytochemistry and pharmacology of Moringa oleifera leaves: An overview. Int. J. Mol. Sci. 2015, 16, 12791–12835. [Google Scholar] [CrossRef]
- Masoudzadeh, S.H.; Mohammadabadi, M.R.; Khezri, A.; Kochuk-Yashchenko, O.A.; Kucher, D.M.; Babenko, O.I.; Bushtruk, M.V.; Tkachenko, S.V.; Stavetska, R.V.; Klopenko, N.I.; et al. Dlk1 gene expression in different Tissues of lamb. Iran. J. Appl. Anim. Sci. 2020, 10, 669–677. Available online: https://sid.ir/paper/782944/en (accessed on 31 January 2023).
- Mohamadipoor, L.; Mohammadabadi, M.; Amiri, Z.; Babenko, O.; Stavetska, R.; Kalashnik, O.; Kucher, D.; Kochuk-Yashchenko, O.; Asadollahpour, H. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. BMC Vet. Res. 2021, 17, 369. [Google Scholar] [CrossRef]
- Mohammadinejad, F.; Mohammadabadi, M.; Roudbari, Z.; Sadkowski, T. Identification of Key Genes and Biological Pathways Associated with Skeletal Muscle Maturation and Hypertrophy in Bos taurus, Ovis aries, and Sus scrofa. Animals 2022, 12, 3471. [Google Scholar] [CrossRef]
- Shahsavari, M.; Mohammadabadi, M.; Khezri, A.; Asadi Fozi, M.; Babenko, O.; Kalashnyk, O.; Oleshko, V.; Tkachenko, S. Correlation between insulin-like growth factor 1 gene expression and fennel (Foeniculum vulgare) seed powder consumption in muscle of sheep. Anim. Biotechnol. 2021, 1–11. [Google Scholar] [CrossRef]
- Malik, F.; Hussain, S.; Mirza, T.; Hameed, A.; Ahmad, S.; Riaz, H.; Usmanghani, K. Screening for antimicrobial activity of thirty-three medicinal plants used in the traditional system of medicine in Pakistan. J. Med. Plant Res. 2011, 5, 3052–3060. [Google Scholar] [CrossRef]
- Segwatibe, M.K.; Cosa, S.; Bassey, K. Antioxidant and Antimicrobial Evaluations of Moringa oleifera Lam Leaves Extract and Isolated Compounds. Molecules 2023, 28, 899. [Google Scholar] [CrossRef]
- Khouj, W.W.; Gull, M.; Rahimuddin, S.A.; Alshamrani, R.; Najjar, A.A.; Al-Hejin, A.M.; Zaher, G.F. Screening of Antimicrobial and Antioxidant Activities of Moringa oleifera Lam Leaf Extracts Against Multidrug Resistant Pathogenic Bacteria. Biosc. Biotech. Res. Comm. 2020, 13. Available online: https://bit.ly/31rhX1u (accessed on 31 January 2023). [CrossRef]
- Gardner, T.S.; di Bernardo, D.; Lorenz, D.; Collins, J.J. Inferring genetic networks and identifying compound mode of action via expression profiling. Science 2003, 301, 102–105. [Google Scholar] [CrossRef]
- Nugrahedi, P.Y.; Verkerk, R.; Widianarko, B.; Dekker, M. A Mechanistic Perspective on Process-Induced Changes in Glucosinolate Content in Brassica Vegetables: A Review. Crit. Rev. Food Sci. Nutr. 2015, 55, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.Á.; Rodríguez-Concepción, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arab. Book 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.D.; Gajardo, H.A.; Huebert, T.; Parkin, I.A.; Iniguez-Luy, F.L.; Federico, M.L. Retention of triplicated phytoene synthase (PSY) genes in Brassica napus L. and its diploid progenitors during the evolution of the Brassiceae. Theor. Appl. Genet. 2012, 124, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Liu, N.; Zhao, Y.; Yan, H.; Wang, Q. Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem. 2011, 124, 941–947. [Google Scholar] [CrossRef]
- Ratshilivha, N.; Awouafack, M.D.; du Toit, E.S.; Eloff, J.N. The variation in antimicrobial and antioxidant activities of acetone leaf extracts of 12 Moringa oleifera (Moringaceae) trees enables the selection of trees with additional uses. S. Afr. J. Bot. 2014, 92, 59–64. [Google Scholar] [CrossRef]
- Ferreira, P.M.P.; Farias, D.; Oliveira, J.T.A.; Carvalho, A.F.U. Moringa oleifera: Bioactive compounds and nutritional potential Moringa oleifera. Rev. Nutr. 2008, 21, 431–437. [Google Scholar] [CrossRef]
- Prakash, S.; Singh, S. Species Specific Protocol for Extraction of Genomic Dna in Moringa oleifera. J. Biosci. Inform. 2013, 4, 1–17. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.-R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Alam, M.W.; Pandey, P.; Khan, F.; Souayeh, B.; Farhan, M. Study to investigate the potential of combined extract of leaves and seeds of Moringa oleifera in groundwater purification. Int. J. Environ. Res. Public Health 2020, 17, 7468. [Google Scholar] [CrossRef]
- Simms, D.; Cizdziel, P.; Chomczynski, P. TRIzol: A new reagent for optimal single-step isolation of RNA. Focus 1993, 15, 532–535. [Google Scholar]
- Poyraz, I.; Sozen, E.; Arslanyolu, M. Isolation of Quality Total RNA from the Aromatic Plant Origanum onites. J. Nat. Sci. 2010, 65, 266–270. [Google Scholar] [CrossRef]
- Chum, P.Y.; Haas, A.; Kelley, M.; Lafayette, C.O. Solution for RT-qPCR: Relative gene expression analysis using Thermo Scientific PikoReal real-time PCR system and Solaris gene expression reagents. Target 2012, 1, 33. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 2010, 463, 662–665. [Google Scholar] [CrossRef]
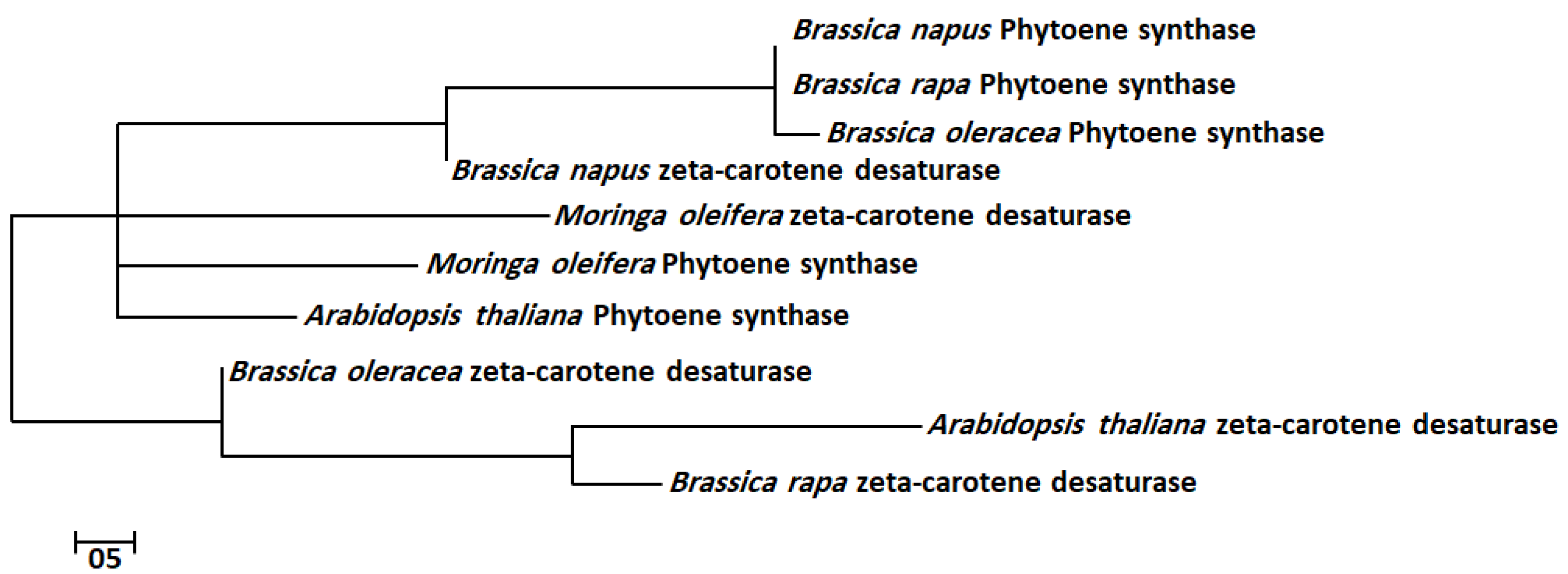
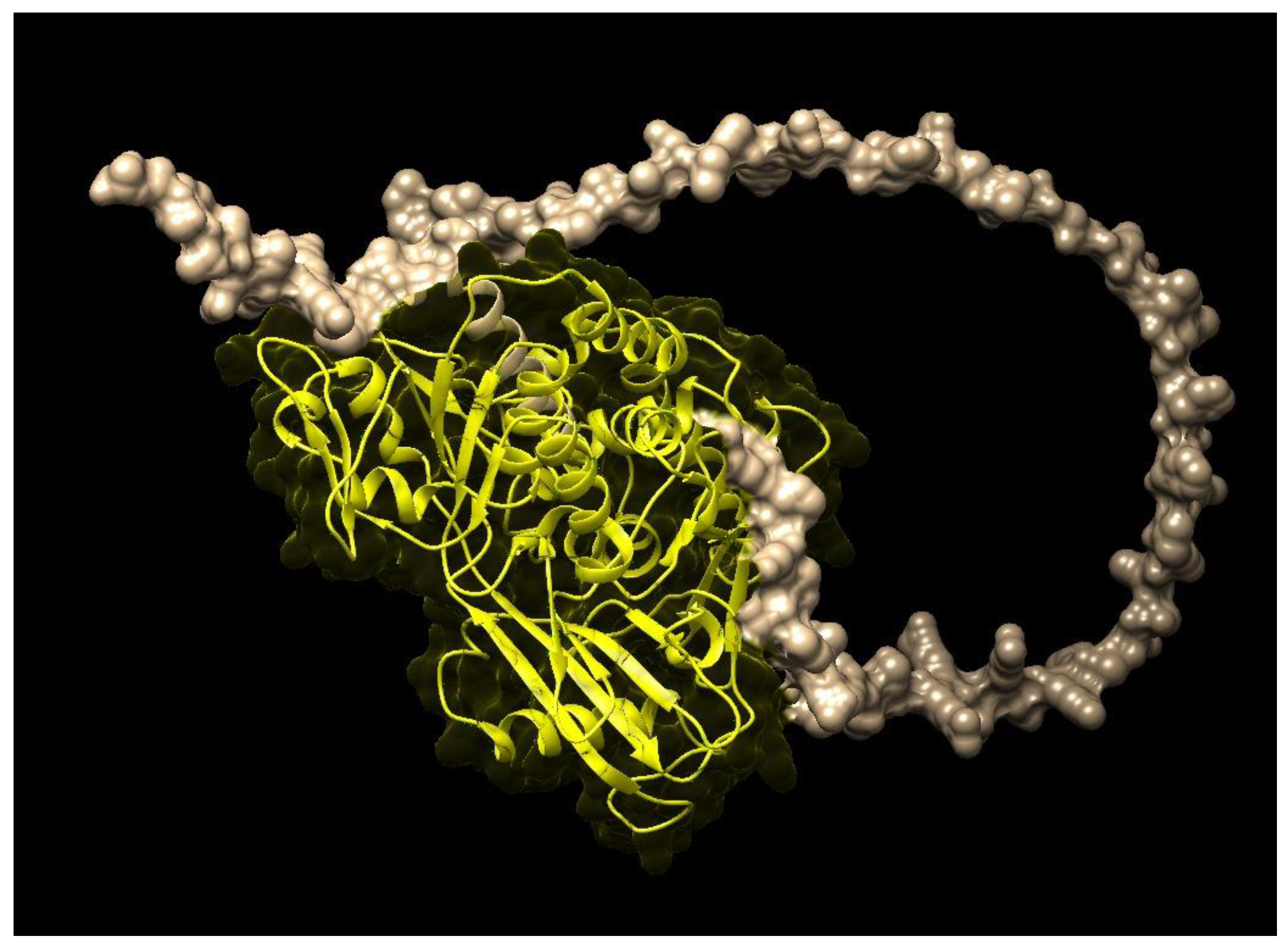
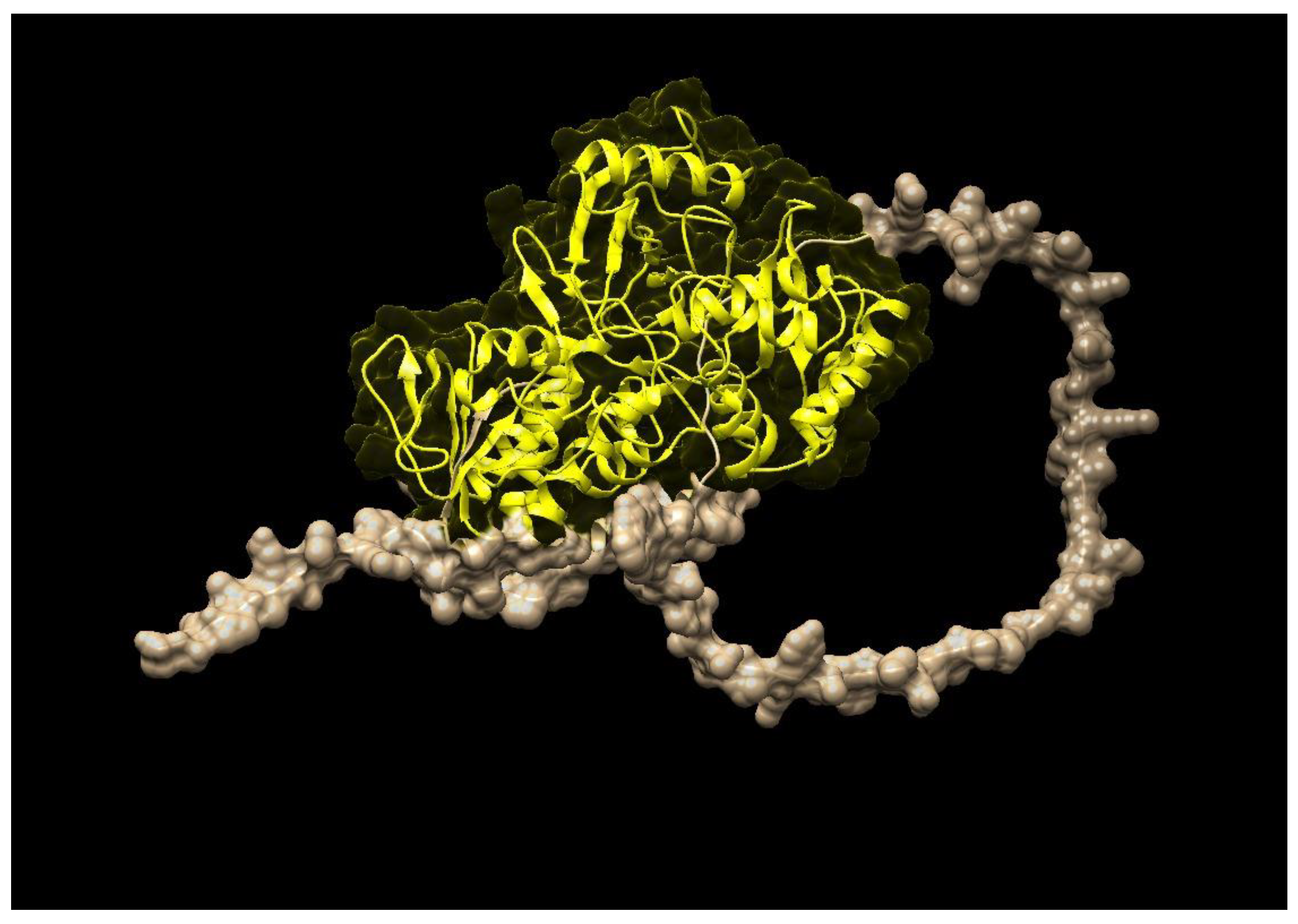
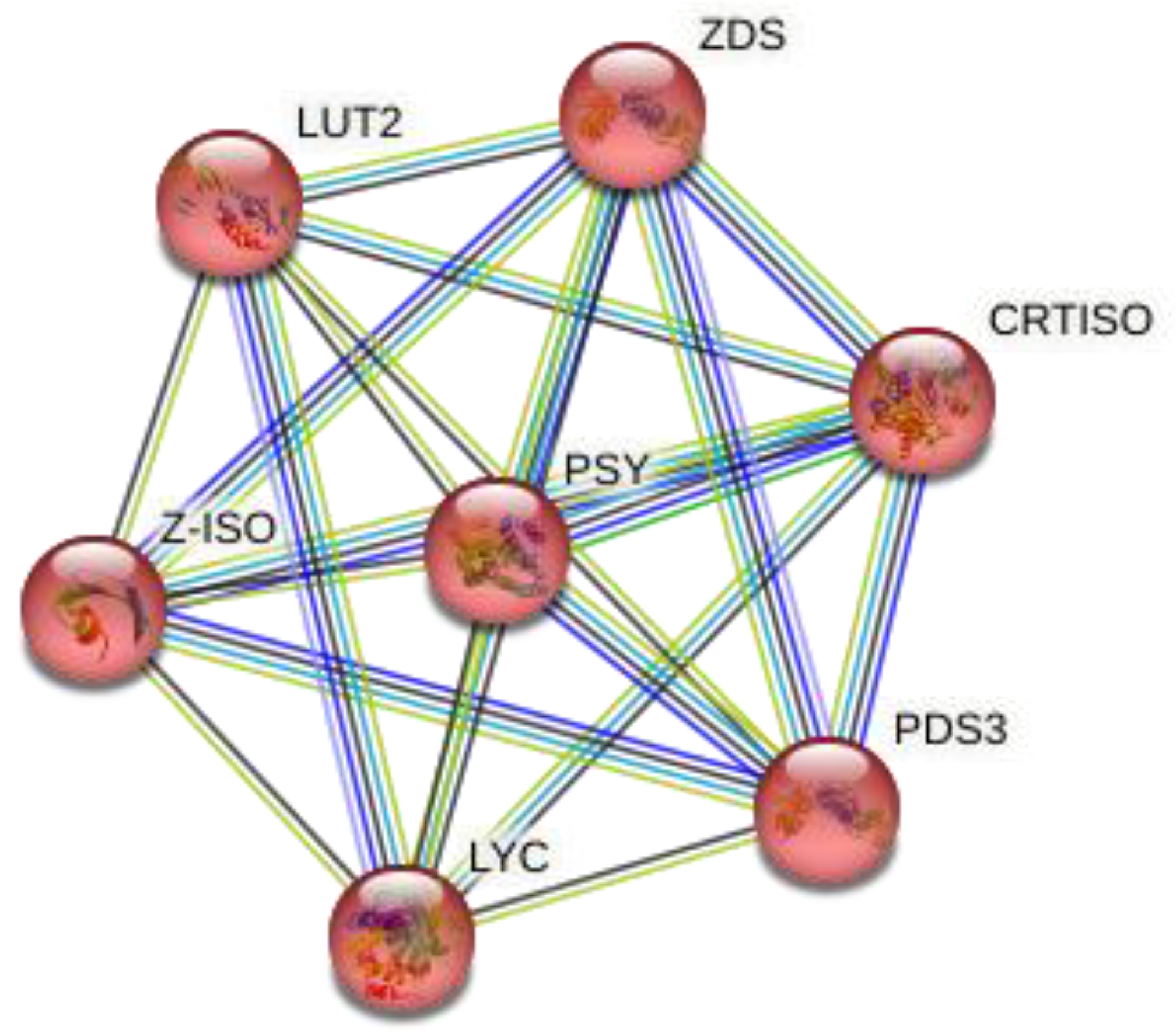
| Species Name | Gene Name | E-Value | Identity (%) | Domain | Family |
|---|---|---|---|---|---|
| Arabidopsis thaliana | ZDS | 0.0 | 98.0 | PLN02487 | Amino oxidase |
| PSY | 0.0 | 97.5 | PLN02632 | Squalene Synthase (SQS) | |
| Brassica napus | ZDS | 1 × 10−121 | 83.4 | PLN02487 | Amino oxidase |
| PSY | 1 × 10−140 | 83.5 | PLN02632 | Squalene Synthase (SQS) | |
| Brassica rapa | ZDS | 1 × 10−11 | 76.4 | PLN02487 | Amino oxidase |
| PSY | 1 × 10−148 | 82.3 | PLN02632 | Squalene Synthase (SQS) | |
| Brassica oleracea | ZDS | 1 × 10−13 | 86.0 | PLN02487 | Amino oxidase |
| PSY | 1 × 10−148 | 83.5 | PLN02632 | Squalene Synthase (SQS) |
| Genes | Expression Changes Relative to That of A. thaliana | ||||
|---|---|---|---|---|---|
| Arabidopsis thaliana | Brassica napus | Brassica rapa | Bixa orellana | Moringa oleifera | |
| ZDS | ↑651.3 * | ↑412.8 * | ↑615.0 * | ↓140.8 * | ↓140.8 * |
| PSY | ↓412.3 * | ↑536.1 * | ↓341.8 | ↓514.1 * | ↓514.1 * |
| Actin | ↑120.9 * | ↑102.6 * | ↑7.4 * | ↓27.0 * | ↓17.0 * |
| S. No. | Genes | Primers | Length | TM (°C) | GC (%) |
|---|---|---|---|---|---|
| 1. | PSY | Forward. TGCAGCTTAAACGAGCAAGA | 20 | 59.90 | 45 |
| Reverse. AGCAATGAAGCCCATACCTG | 20 | 60.10 | 50 | ||
| 2. | ZDS | Forward. GACTCCGATGTTTCCGACAT | 20 | 59.93 | 50 |
| Reverse. CACTTTGCCACCAATGAATG | 20 | 59.96 | 45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muteeb, G.; Aatif, M.; Farhan, M.; Alsultan, A.; Alshoaibi, A.; Alam, M.W. Leaves of Moringa oleifera Are Potential Source of Bioactive Compound β-Carotene: Evidence from In Silico and Quantitative Gene Expression Analysis. Molecules 2023, 28, 1578. https://doi.org/10.3390/molecules28041578
Muteeb G, Aatif M, Farhan M, Alsultan A, Alshoaibi A, Alam MW. Leaves of Moringa oleifera Are Potential Source of Bioactive Compound β-Carotene: Evidence from In Silico and Quantitative Gene Expression Analysis. Molecules. 2023; 28(4):1578. https://doi.org/10.3390/molecules28041578
Chicago/Turabian StyleMuteeb, Ghazala, Mohammad Aatif, Mohd Farhan, Abdulrahman Alsultan, Adil Alshoaibi, and Mir Waqas Alam. 2023. "Leaves of Moringa oleifera Are Potential Source of Bioactive Compound β-Carotene: Evidence from In Silico and Quantitative Gene Expression Analysis" Molecules 28, no. 4: 1578. https://doi.org/10.3390/molecules28041578
APA StyleMuteeb, G., Aatif, M., Farhan, M., Alsultan, A., Alshoaibi, A., & Alam, M. W. (2023). Leaves of Moringa oleifera Are Potential Source of Bioactive Compound β-Carotene: Evidence from In Silico and Quantitative Gene Expression Analysis. Molecules, 28(4), 1578. https://doi.org/10.3390/molecules28041578











