Metabolic Engineering of Microorganisms to Produce Pyruvate and Derived Compounds
Abstract
1. Introduction
2. Metabolic Engineering Strategies to Enhance the Production of Pyruvate
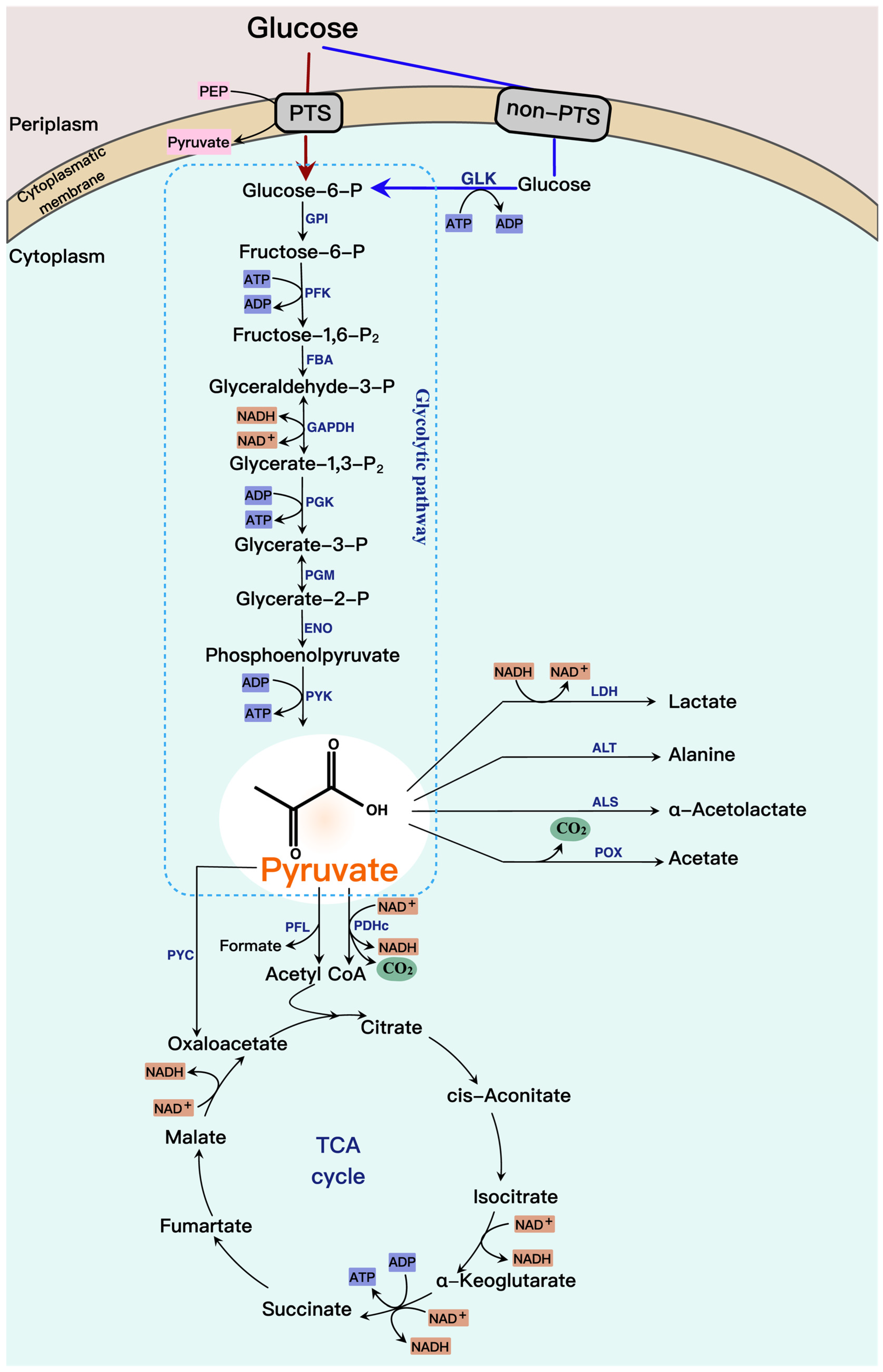
2.1. Pyruvate Pathway and Its Regulation
2.2. Engineering to Expand the Glycolytic Flux for Pyruvate Production
2.3. Engineering to Increase the Carbon Flux for Pyruvate Accumulation
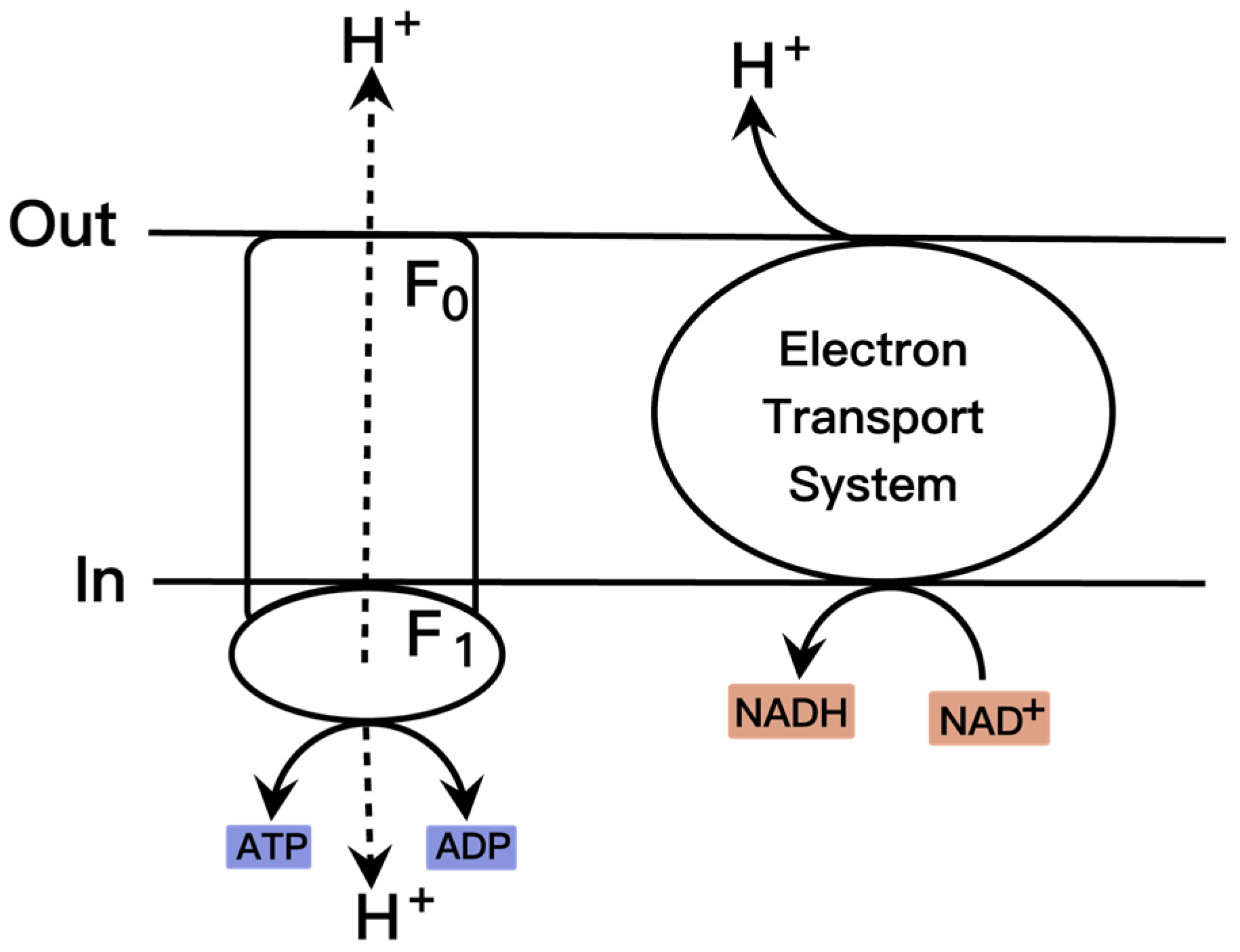
2.4. Cofactor Engineering
2.5. Engineering for Growth and Production Balance
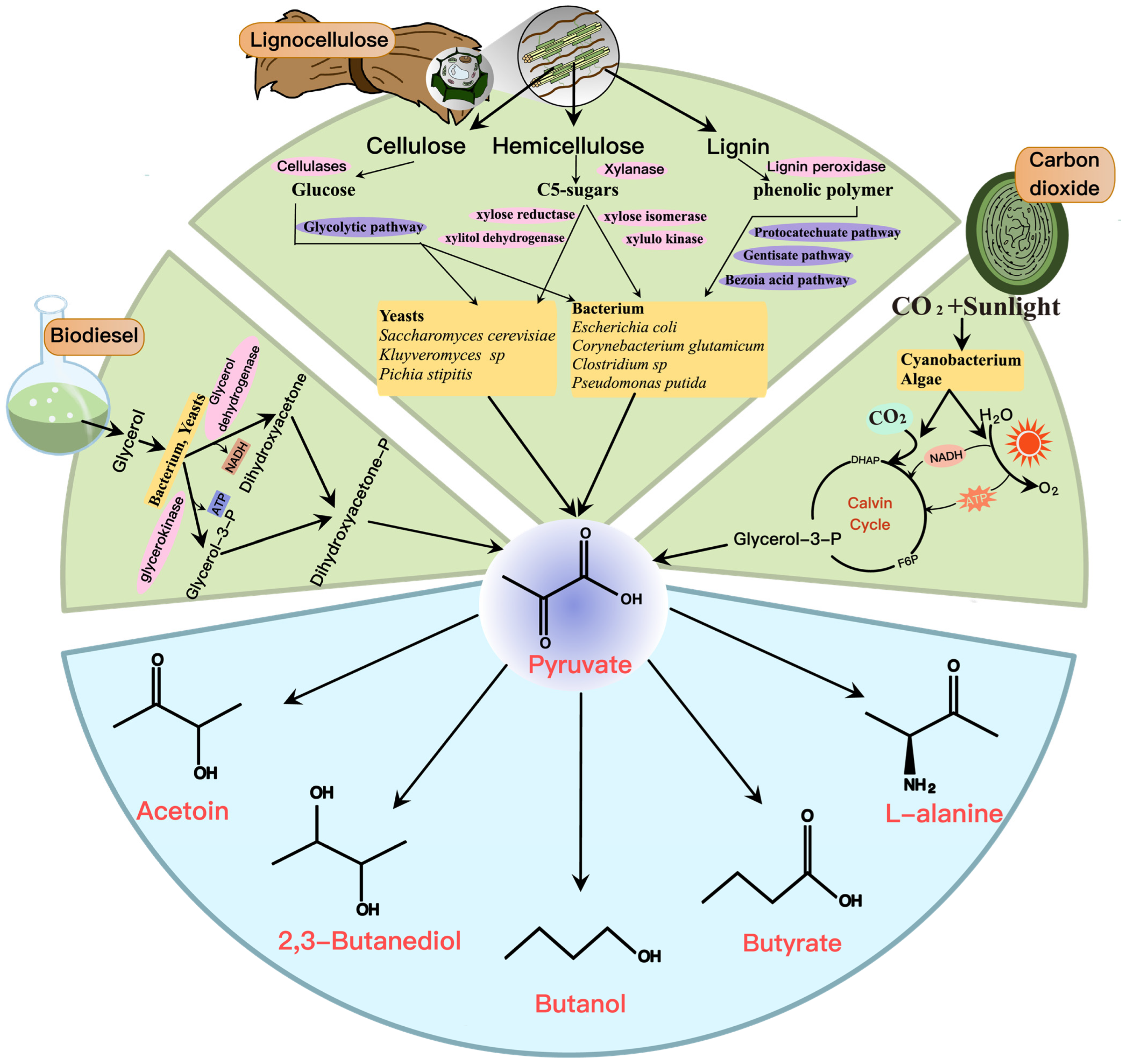
3. Metabolic Engineering for the Production of Pyruvate-Derived Compounds
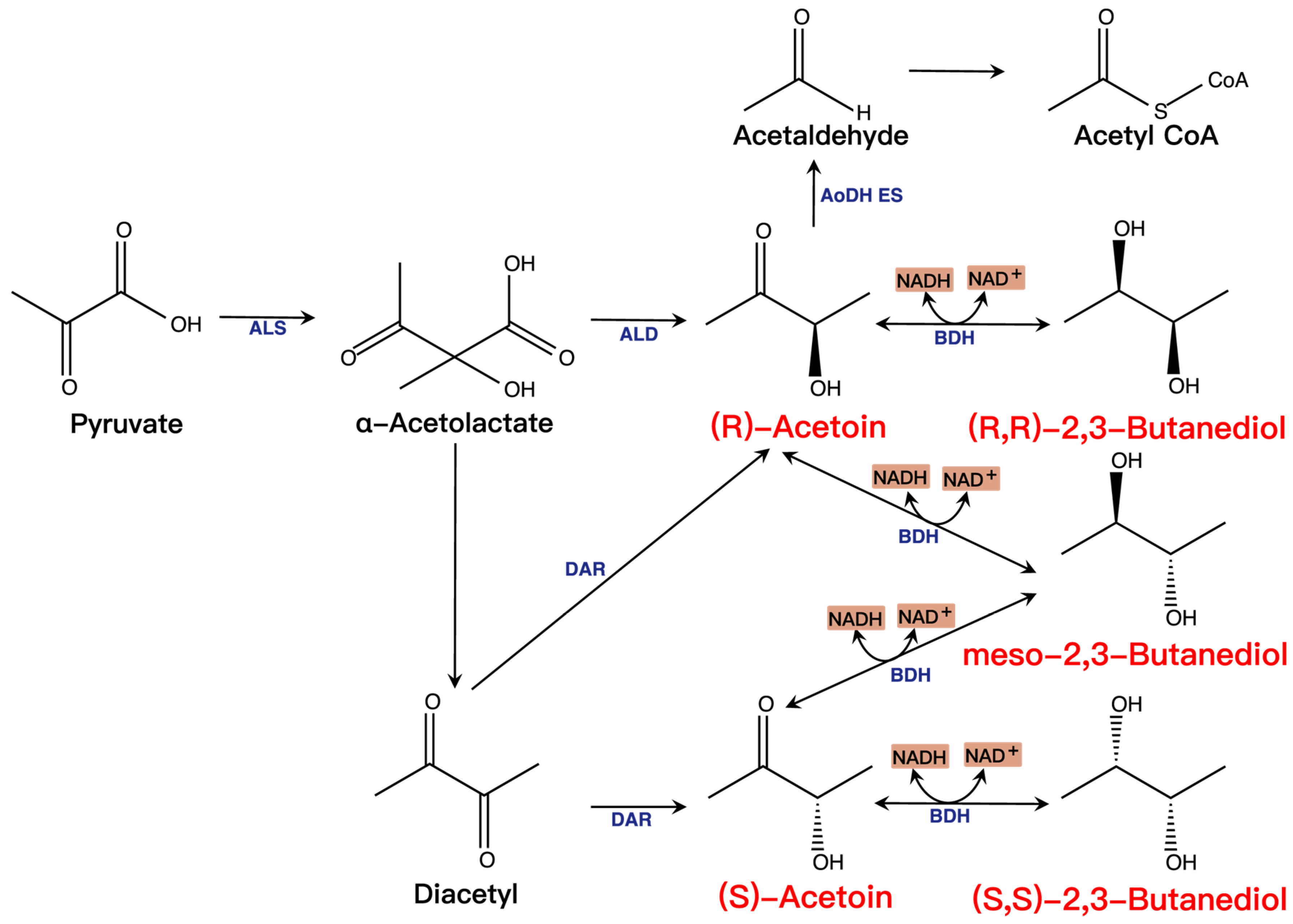
3.1. Acetoin and 2,3-BD
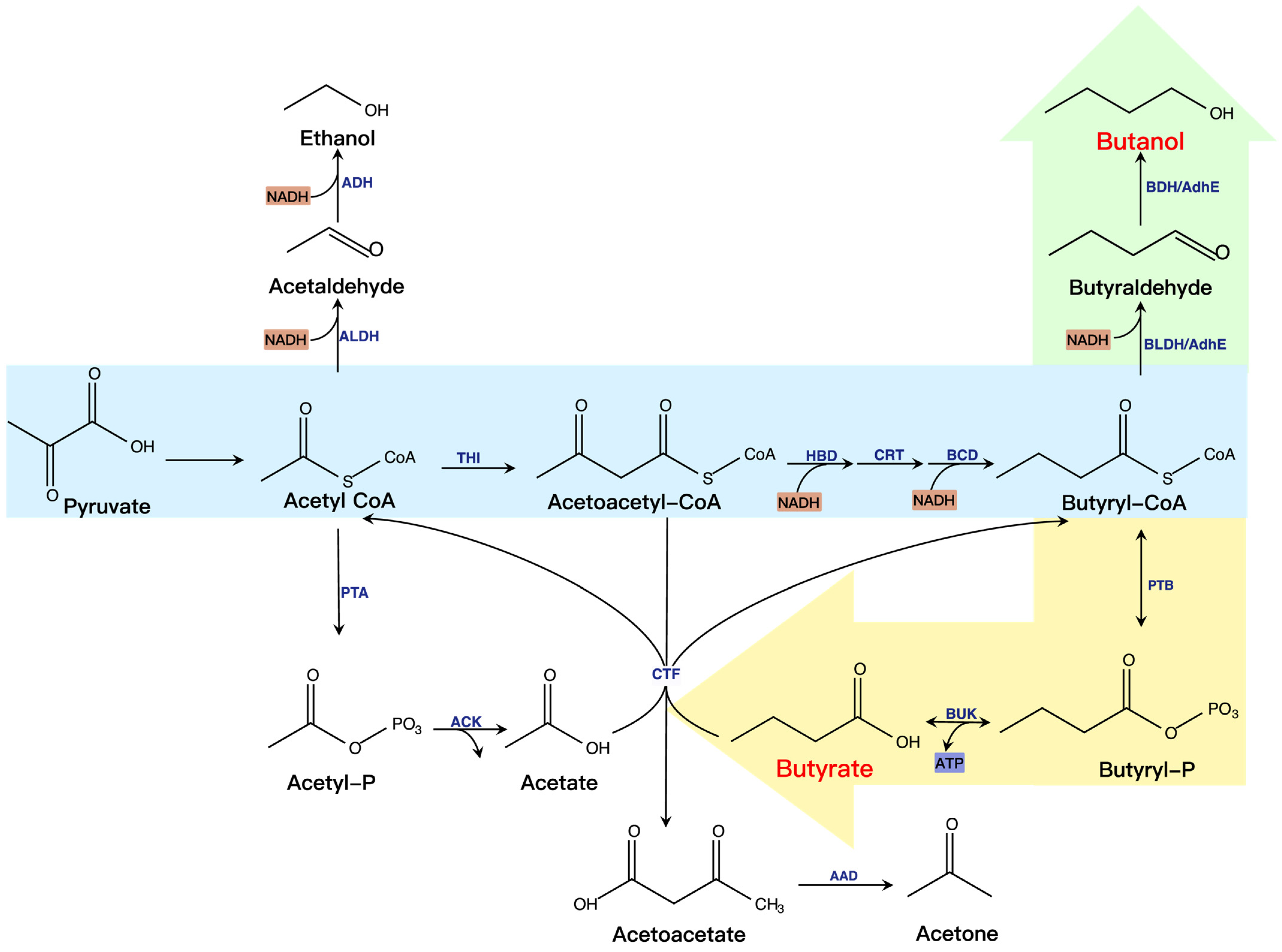
3.2. Butanol and Butyrate
3.3. L-Alanine
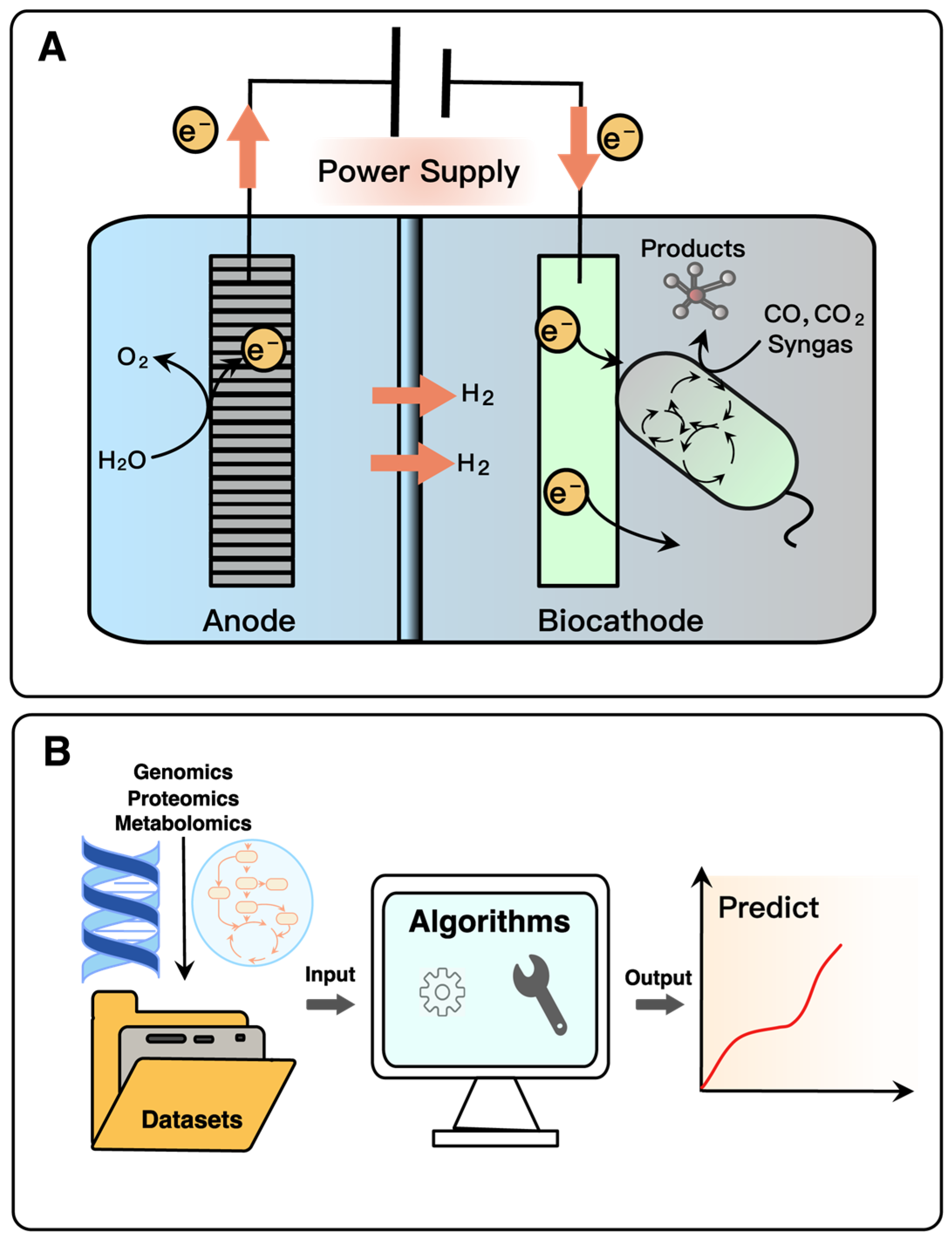
4. Alternative Sustainable Strategies for Pyruvate and Derivatives
4.1. Engineering of Alternative Substrates
4.2. New Tools and Applications in Metabolic Engineering
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stephanopoulos, G.; Vallino, J.J. Network rigidity and metabolic engineering in metabolite overproduction. Science 1991, 252, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Guo, L.; Hu, G.; Luo, Q.; Liu, J.; Nielsen, J.; Chen, J.; Liu, L. DCEO biotechnology: Tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem. Rev. 2018, 118, 4–72. [Google Scholar] [CrossRef] [PubMed]
- Si, T.; Xiao, H.; Zhao, H. Rapid prototyping of microbial cell factories via genome-scale engineering. Biotechnol. Adv. 2015, 33, 1420–1432. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Teo, W.; Chen, B.; Leong, S.S.J.; Chang, M.W. Microbial tolerance engineering toward biochemical production: From lignocellulose to products. Curr. Opin. Biotechnol. 2014, 29, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhou, Y.J.; Krivoruchko, A.; Huang, M.; Liu, L.; Khoomrung, S.; Siewers, V.; Jiang, B.; Nielsen, J. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine. Nat. Commun. 2015, 6, 8224. [Google Scholar] [CrossRef]
- Reyes, L.H.; Gomez, J.M.; Kao, K.C. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng. 2014, 21, 26–33. [Google Scholar] [CrossRef]
- Petrov, K.; Petrova, P. Current Advances in Microbial Production of Acetoin and 2, 3-Butanediol by Bacillus spp. Fermentation 2021, 7, 307. [Google Scholar] [CrossRef]
- Veza, I.; Said, M.F.M.; Latiff, Z.A. Recent advances in butanol production by acetone-butanol-ethanol (ABE) fermentation. Biomass Bioenergy 2021, 144, 105919. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in energy metabolism: There is still more to learn. Trends Endocrinol. Metab. 2021, 32, 159–169. [Google Scholar] [CrossRef]
- Liu, P.; Xu, H.; Zhang, X. Metabolic engineering of microorganisms for L-alanine production. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab057. [Google Scholar] [CrossRef]
- Yuan, W.; Du, Y.; Yu, K.; Xu, S.; Liu, M.; Wang, S.; Yang, Y.; Zhang, Y.; Sun, J. The Production of Pyruvate in Biological Technology: A Critical Review. Microorganisms 2022, 10, 2454. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Terasawa, Y.; Takaoka, N.; Shimizu, H.; Tomita, F. Pyruvic acid production by an F1-ATPase-defective mutant of Escherichia coli W1485lip2. Biosci. Biotechnol. Biochem. 1994, 58, 2164–2167. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Du, G.; Chen, J. Increasing glycolytic flux in Torulopsis glabrata by redirecting ATP production from oxidative phosphorylation to substrate-level phosphorylation. J. Appl. Microbiol. 2006, 100, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Huang, L.; Liu, L.; Chen, J. Enhancement of pyruvate productivity by inducible expression of a F0F1-ATPase inhibitor INH1 in Torulopsis glabrata CCTCC M202019. J. Biotechnol. 2009, 144, 120–126. [Google Scholar] [CrossRef]
- Zhu, Y.; Eiteman, M.A.; Altman, R.; Altman, E. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl. Environ. Microbiol. 2008, 74, 6649–6655. [Google Scholar] [CrossRef]
- Tomar, A.; Eiteman, M.; Altman, E. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 62, 76–82. [Google Scholar] [CrossRef]
- Wang, D.; Wang, L.; Hou, L.; Deng, X.; Gao, Q.; Gao, N. Metabolic engineering of Saccharomyces cerevisiae for accumulating pyruvic acid. Ann. Microbiol. 2015, 65, 2323–2331. [Google Scholar] [CrossRef]
- Causey, T.; Zhou, S.; Shanmugam, K.; Ingram, L. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: Homoacetate production. Proc. Natl. Acad. Sci. USA 2003, 100, 825–832. [Google Scholar] [CrossRef]
- Nakashima, N.; Ohno, S.; Yoshikawa, K.; Shimizu, H.; Tamura, T. A vector library for silencing central carbon metabolism genes with antisense RNAs in Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 564–573. [Google Scholar] [CrossRef]
- Ziegler, M.; Hägele, L.; Gäbele, T.; Takors, R. CRISPRi enables fast growth followed by stable aerobic pyruvate formation in Escherichia coli without auxotrophy. Eng. Life Sci. 2022, 22, 70–84. [Google Scholar] [CrossRef]
- Akita, H.; Nakashima, N.; Hoshino, T. Pyruvate production using engineered Escherichia coli. Amb Express 2016, 6, 94. [Google Scholar] [CrossRef]
- Moxley, W.C.; Eiteman, M.A. Pyruvate production by Escherichia coli by use of pyruvate dehydrogenase variants. Appl. Environ. Microbiol. 2021, 87, e00487-00421. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cao, Z. Regulation of NADH oxidase expression via a thermo-regulated genetic switch for pyruvate production in Escherichia coli. Biotechnol. Bioprocess Eng. 2018, 23, 93–99. [Google Scholar] [CrossRef]
- Luo, Z.; Zeng, W.; Du, G.; Chen, J.; Zhou, J. Enhancement of pyruvic acid production in Candida glabrata by engineering hypoxia-inducible factor 1. Bioresour. Technol. 2020, 295, 122248. [Google Scholar] [CrossRef] [PubMed]
- Soma, Y.; Yamaji, T.; Hanai, T. Dynamic metabolic engineering of Escherichia coli improves fermentation for the production of pyruvate and its derivatives. J. Biosci. Bioeng. 2022, 133, 56–63. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Bao, T.; Yang, T.; Xu, M.; Li, H.; Xu, Z.; Rao, Z. Moderate expression of the transcriptional regulator ALsR enhances acetoin production by Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2013, 40, 1067–1076. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Bao, T.; Rao, Z.; Yang, T.; Xu, M.; Xu, Z.; Li, H.; Yang, S. The rebalanced pathway significantly enhances acetoin production by disruption of acetoin reductase gene and moderate-expression of a new water-forming NADH oxidase in Bacillus subtilis. Metab. Eng. 2014, 23, 34–41. [Google Scholar] [CrossRef]
- Wang, D.; Zhou, J.; Chen, C.; Wei, D.; Shi, J.; Jiang, B.; Liu, P.; Hao, J. R-acetoin accumulation and dissimilation in Klebsiella pneumoniae. J. Ind. Microbiol. Biotechnol. 2015, 42, 1105–1115. [Google Scholar] [CrossRef]
- Sun, J.-A.; Zhang, L.-Y.; Rao, B.; Shen, Y.-L.; Wei, D.-Z. Enhanced acetoin production by Serratia marcescens H32 with expression of a water-forming NADH oxidase. Bioresour. Technol. 2012, 119, 94–98. [Google Scholar] [CrossRef]
- Li, S.; Xu, N.; Liu, L.; Chen, J. Engineering of carboligase activity reaction in Candida glabrata for acetoin production. Metab. Eng. 2014, 22, 32–39. [Google Scholar] [CrossRef]
- Jang, J.-W.; Jung, H.-M.; Im, D.-K.; Jung, M.-Y.; Oh, M.-K. Pathway engineering of Enterobacter aerogenes to improve acetoin production by reducing by-products formation. Enzym. Microb. Technol. 2017, 106, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, C.; Yu, X.; Liu, S.; Wang, Z.; Tang, Y.J.; Chen, T. Highly efficient hemicellulose utilization for acetoin production by an engineered Bacillus subtilis. J. Chem. Technol. Biotechnol. 2018, 93, 3428–3435. [Google Scholar] [CrossRef]
- Ishii, J.; Morita, K.; Ida, K.; Kato, H.; Kinoshita, S.; Hataya, S.; Shimizu, H.; Kondo, A.; Matsuda, F. A pyruvate carbon flux tugging strategy for increasing 2, 3-butanediol production and reducing ethanol subgeneration in the yeast Saccharomyces cerevisiae. Biotechnol. Biofuels 2018, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, S.; Osire, T.; Zhang, X.; Xu, M.; Yang, S.-T.; Yang, T.; Rao, Z. Engineering the 2, 3-BD pathway in Bacillus subtilis by shifting the carbon flux in favor of 2, 3-BD synthesis. Biochem. Eng. J. 2021, 169, 107969. [Google Scholar] [CrossRef]
- Ji, X.-J.; Huang, H.; Zhu, J.-G.; Ren, L.-J.; Nie, Z.-K.; Du, J.; Li, S. Engineering Klebsiella oxytoca for efficient 2, 3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl. Microbiol. Biotechnol. 2010, 85, 1751–1758. [Google Scholar] [CrossRef]
- Liang, K.; Shen, C.R. Selection of an endogenous 2, 3-butanediol pathway in Escherichia coli by fermentative redox balance. Metab. Eng. 2017, 39, 181–191. [Google Scholar] [CrossRef]
- Park, J.M.; Song, H.; Lee, H.J.; Seung, D. In silico aided metabolic engineering of Klebsiella oxytoca and fermentation optimization for enhanced 2, 3-butanediol production. J. Ind. Microbiol. Biotechnol. 2013, 40, 1057–1066. [Google Scholar] [CrossRef]
- Jung, M.-Y.; Mazumdar, S.; Shin, S.H.; Yang, K.-S.; Lee, J.; Oh, M.-K. Improvement of 2, 3-butanediol yield in Klebsiella pneumoniae by deletion of the pyruvate formate-lyase gene. Appl. Environ. Microbiol. 2014, 80, 6195–6203. [Google Scholar] [CrossRef]
- Erian, A.M.; Gibisch, M.; Pflügl, S. Engineered E. coli W enables efficient 2, 3-butanediol production from glucose and sugar beet molasses using defined minimal medium as economic basis. Microb. Cell Factories 2018, 17, 190. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Zhang, X.; Xu, M.; Xu, Z.; Yang, S.-T. Enhanced 2, 3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb. Cell Factories 2015, 14, 122. [Google Scholar] [CrossRef]
- Bao, T.; Hou, W.; Wu, X.; Lu, L.; Zhang, X.; Yang, S.T. Engineering Clostridium cellulovorans for highly selective n-butanol production from cellulose in consolidated bioprocessing. Biotechnol. Bioeng. 2021, 118, 2703–2718. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dong, S.; Wang, Y. Enhancement of solvent production by overexpressing key genes of the acetone-butanol-ethanol fermentation pathway in Clostridium saccharoperbutylacetonicum N1-4. Bioresour. Technol. 2017, 245, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Seong, W.; Han, G.H.; Lee, D.-H.; Lee, S.-G. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb. Cell Factories 2017, 16, 188. [Google Scholar] [CrossRef] [PubMed]
- Nitta, K.; Laviña, W.A.; Pontrelli, S.; Liao, J.C.; Putri, S.P.; Fukusaki, E. Metabolome analysis revealed the knockout of glyoxylate shunt as an effective strategy for improvement of 1-butanol production in transgenic Escherichia coli. J. Biosci. Bioeng. 2019, 127, 301–308. [Google Scholar] [CrossRef]
- Jiang, Y.; Xu, C.; Dong, F.; Yang, Y.; Jiang, W.; Yang, S. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 2009, 11, 284–291. [Google Scholar] [CrossRef]
- Suo, Y.; Ren, M.; Yang, X.; Liao, Z.; Fu, H.; Wang, J. Metabolic engineering of Clostridium tyrobutyricum for enhanced butyric acid production with high butyrate/acetate ratio. Appl. Microbiol. Biotechnol. 2018, 102, 4511–4522. [Google Scholar] [CrossRef]
- Wang, L.; Chauliac, D.; Moritz, B.E.; Zhang, G.; Ingram, L.O.; Shanmugam, K. Metabolic engineering of Escherichia coli for the production of butyric acid at high titer and productivity. Biotechnol. Biofuels 2019, 12, 62. [Google Scholar] [CrossRef]
- Kataoka, N.; Vangnai, A.S.; Pongtharangkul, T.; Yakushi, T.; Matsushita, K. Butyrate production under aerobic growth conditions by engineered Escherichia coli. J. Biosci. Bioeng. 2017, 123, 562–568. [Google Scholar] [CrossRef]
- Fu, H.; Yang, S.-T.; Wang, M.; Wang, J.; Tang, I.-C. Butyric acid production from lignocellulosic biomass hydrolysates by engineered Clostridium tyrobutyricum overexpressing xylose catabolism genes for glucose and xylose co-utilization. Bioresour. Technol. 2017, 234, 389–396. [Google Scholar] [CrossRef]
- Suo, Y.; Fu, H.; Ren, M.; Yang, X.; Liao, Z.; Wang, J. Butyric acid production from lignocellulosic biomass hydrolysates by engineered Clostridium tyrobutyricum overexpressing Class I heat shock protein GroESL. Bioresour. Technol. 2018, 250, 691–698. [Google Scholar] [CrossRef]
- Uhlenbusch, I.; Sahm, H.; Sprenger, G.A. Expression of an L-alanine dehydrogenase gene in Zymomonas mobilis and excretion of L-alanine. Appl. Environ. Microbiol. 1991, 57, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Deng, C.; Cui, W.-J.; Liu, Z.-M.; Zhou, Z.-M. Efficient L-alanine production by a thermo-regulated switch in Escherichia coli. Appl. Biochem. Biotechnol. 2016, 178, 324–337. [Google Scholar] [CrossRef]
- Lee, M.; Smith, G.; Eiteman, M.; Altman, E. Aerobic production of alanine by Escherichia coli aceF ldhA mutants expressing the Bacillus sphaericus alaD gene. Appl. Microbiol. Biotechnol. 2004, 65, 56–60. [Google Scholar] [CrossRef]
- Smith, G.M.; Lee, S.A.; Reilly, K.C.; Eiteman, M.A.; Altman, E. Fed-batch two-phase production of alanine by a metabolically engineered Escherichia coli. Biotechnol. Lett. 2006, 28, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Jojima, T.; Fujii, M.; Mori, E.; Inui, M.; Yukawa, H. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid L-alanine under oxygen deprivation. Appl. Microbiol. Biotechnol. 2010, 87, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Deng, C.; Cui, W.-J.; Liu, Z.-M.; Zhou, Z.-M. L-Alanine Production through Glycerol Fermentation by Recombinant Escherichia coli. Mod. Food Sci. Technol. 2016, 32, 163–169. [Google Scholar] [CrossRef]
- Maleki, N.; Eiteman, M.A. Recent progress in the microbial production of pyruvic acid. Fermentation 2017, 3, 8. [Google Scholar] [CrossRef]
- Oliver, S. Demand management in cells. Nature 2002, 418, 33–34. [Google Scholar] [CrossRef]
- Mesecar, A.D.; Nowak, T. Metal-ion-mediated allosteric triggering of yeast pyruvate kinase. 1. A multidimensional kinetic linked-function analysis. Biochemistry 1997, 36, 6792–6802. [Google Scholar] [CrossRef]
- Solem, C.; Koebmann, B.J.; Jensen, P.R. Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J. Bacteriol. 2003, 185, 1564–1571. [Google Scholar] [CrossRef]
- Jensen, P.R.; Michelsen, O. Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 1992, 174, 7635–7641. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Liu, S.; Du, G.; Xu, S.; Zhou, J.; Chen, J. Enhanced pyruvate production in Candida glabrata by carrier engineering. Biotechnol. Bioeng. 2018, 115, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.T.; Chen, A.Y.; Lan, E.I. Metabolic engineering design strategies for increasing acetyl-CoA flux. Metabolites 2020, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, J.; Schwentner, A.; Brunnenkan, B.; Gabris, C.; Grimm, S.; Gerstmeir, R.; Takors, R.; Eikmanns, B.J.; Blombach, B. Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L-lysine, L-valine, and 2-ketoisovalerate. Appl. Environ. Microbiol. 2013, 79, 5566–5575. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.; Henning, U. Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Et Biophys. Acta (BBA)-Enzymol. Biol. Oxid. 1966, 122, 355–358. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Shi, Z.; Du, G.; Chen, J. Enhancement of pyruvate productivity in Torulopsis glabrata: Increase of NAD+ availability. J. Biotechnol. 2006, 126, 173–185. [Google Scholar] [CrossRef]
- Yang, M.; Xing, J. Improvement of pyruvate production based on regulation of intracellular redox state in engineered Escherichia coli. Biotechnol. Bioprocess Eng. 2017, 22, 376–381. [Google Scholar] [CrossRef]
- Hou, J.; Gao, C.; Guo, L.; Nielsen, J.; Ding, Q.; Tang, W.; Hu, G.; Chen, X.; Liu, L. Rewiring carbon flux in Escherichia coli using a bifunctional molecular switch. Metab. Eng. 2020, 61, 47–57. [Google Scholar] [CrossRef]
- Kierans, S.; Taylor, C. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Liu, Y.; Zhu, Y.; Li, J.; Du, G.; Chen, J.; Ledesma-Amaro, R.; Liu, L. Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat. Chem. Biol. 2020, 16, 1261–1268. [Google Scholar] [CrossRef]
- Doong, S.J.; Gupta, A.; Prather, K.L. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, 2964–2969. [Google Scholar] [CrossRef] [PubMed]
- Soma, Y.; Takahashi, M.; Fujiwara, Y.; Shinohara, T.; Izumi, Y.; Hanai, T.; Bamba, T. Design of synthetic quorum sensing achieving induction timing-independent signal stabilization for dynamic metabolic engineering of E. coli. ACS Synth. Biol. 2021, 10, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Tsau, J.-L.; Guffanti, A.A.; Montville, T.J. Conversion of pyruvate to acetoin helps to maintain pH homeostasis in Lactobacillus plantarum. Appl. Environ. Microbiol. 1992, 58, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Magee, R.J.; Kosaric, N. The microbial production of 2, 3-butanediol. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 32, pp. 89–161. [Google Scholar]
- Xiao, Z.; Xu, P. Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 2007, 33, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Lu, J.R. Strategies for enhancing fermentative production of acetoin: A review. Biotechnol. Adv. 2014, 32, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Gao, T.; Bai, R.; Yang, J.; Xu, Y.; Chu, W.; Jiang, K.; Zhang, J.; Xu, F.; Zhao, H. Regulation of carbon flux and NADH/NAD+ supply to enhance 2, 3-butanediol production in Enterobacter aerogenes. J. Biotechnol. 2022, 358, 67–75. [Google Scholar] [CrossRef]
- Ji, X.-J.; Xia, Z.-F.; Fu, N.-H.; Nie, Z.-K.; Shen, M.-Q.; Tian, Q.-Q.; Huang, H. Cofactor engineering through heterologous expression of an NADH oxidase and its impact on metabolic flux redistribution in Klebsiella pneumoniae. Biotechnol. Biofuels 2013, 6, 7. [Google Scholar] [CrossRef]
- Dong, H.; Zhao, C.; Zhang, T.; Lin, Z.; Li, Y.; Zhang, Y. Engineering Escherichia coli cell factories for n-butanol production. Bioreact. Eng. Res. Ind. Appl. I 2015, 155, 141–163. [Google Scholar]
- Sun, L.; Gong, M.; Lv, X.; Huang, Z.; Gu, Y.; Li, J.; Du, G.; Liu, L. Current advance in biological production of short-chain organic acid. Appl. Microbiol. Biotechnol. 2020, 104, 9109–9124. [Google Scholar] [CrossRef]
- Sandoval, N.R.; Papoutsakis, E.T. Engineering membrane and cell-wall programs for tolerance to toxic chemicals: Beyond solo genes. Curr. Opin. Microbiol. 2016, 33, 56–66. [Google Scholar] [CrossRef]
- Murínová, S.; Dercová, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 2014, 873081. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L.H.; Almario, M.P.; Winkler, J.; Orozco, M.M.; Kao, K.C. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab. Eng. 2012, 14, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lang, N.; Yang, G.; Yang, S.; Jiang, W.; Gu, Y. Improving the performance of solventogenic clostridia by reinforcing the biotin synthetic pathway. Metab. Eng. 2016, 35, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, A.S.; Ageez, A.M.; El-Hadi, A.; Abdallah, N.A. Genetic improvement of n-butanol tolerance in Escherichia coli by heterologous overexpression of groESL operon from Clostridium acetobutylicum. 3 Biotech 2015, 5, 401–410. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, Y.; Yang, S.T. Construction and characterization of ack deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid and hydrogen production. Biotechnol. Prog. 2006, 22, 1265–1275. [Google Scholar] [CrossRef]
- Hashimoto, S.-I.; Katsumata, R. L-alanine fermentation by an alanine racemase-deficient mutant of the DL-alanine hyperproducing bacterium Arthrobacter oxydans HAP-1. J. Ferment. Bioeng. 1998, 86, 385–390. [Google Scholar] [CrossRef]
- Zhang, X.; Jantama, K.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Production of L-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007, 77, 355–366. [Google Scholar] [CrossRef]
- Ohashima, T.; Soda, K. Purification and properties of alanine dehydrogenase from Bacillus sphaericus. Eur. J. Biochem. 1979, 100, 29–30. [Google Scholar] [CrossRef]
- Örlygsson, J.; Anderson, R.; Svensson, B.H. Alanine as an end product during fermentation of monosaccharides by Clostridium strain P2. Antonie Van Leeuwenhoek 1995, 68, 273–280. [Google Scholar] [CrossRef]
- Ashokkumar, V.; Venkatkarthick, R.; Jayashree, S.; Chuetor, S.; Dharmaraj, S.; Kumar, G.; Chen, W.-H.; Ngamcharussrivichai, C. Recent advances in lignocellulosic biomass for biofuels and value-added bioproducts-A critical review. Bioresour. Technol. 2022, 344, 126195. [Google Scholar] [CrossRef]
- Chandrakant, P.; Bisaria, V. Simultaneous bioconversion of glucose and xylose to ethanol by Saccharomyces cerevisiae in the presence of xylose isomerase. Appl. Microbiol. Biotechnol. 2000, 53, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.-Y.; Lee, J.-W.; Min, W.-K.; Park, Y.-C.; Seo, J.-H. Simultaneous conversion of glucose and xylose to 3-hydroxypropionic acid in engineered Escherichia coli by modulation of sugar transport and glycerol synthesis. Bioresour. Technol. 2015, 198, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xu, M.; Tang, I.C.; Yang, S.T. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production through co-utilization of glucose and xylose. Biotechnol. Bioeng. 2015, 112, 2134–2141. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, H.; Yoshihara, K.; Hara, K.Y.; Hasunuma, T.; Ogino, C.; Kondo, A. Metabolome analysis-based design and engineering of a metabolic pathway in Corynebacterium glutamicum to match rates of simultaneous utilization of d-glucose and l-arabinose. Microb. Cell Factories 2018, 17, 76. [Google Scholar] [CrossRef] [PubMed]
- Maleki, N.; Safari, M.; Eiteman, M.A. Conversion of glucose-xylose mixtures to pyruvate using a consortium of metabolically engineered Escherichia coli. Eng. Life Sci. 2018, 18, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef]
- Luo, H.; Yang, R.; Zhao, Y.; Wang, Z.; Liu, Z.; Huang, M.; Zeng, Q. Recent advances and strategies in process and strain engineering for the production of butyric acid by microbial fermentation. Bioresour. Technol. 2018, 253, 343–354. [Google Scholar] [CrossRef]
- Johnson, D.T.; Taconi, K.A. The glycerin glut: Options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ. Prog. 2007, 26, 338–348. [Google Scholar] [CrossRef]
- Joung, S.-M.; Kurumbang, N.P.; Sang, B.-I.; Oh, M.-K. Effects of carbon source and metabolic engineering on butyrate production in Escherichia coli. Korean J. Chem. Eng. 2011, 28, 1587–1592. [Google Scholar] [CrossRef]
- Varrone, C.; Floriotis, G.; Heggeset, T.M.; Le, S.B.; Markussen, S.; Skiadas, I.V.; Gavala, H.N. Continuous fermentation and kinetic experiments for the conversion of crude glycerol derived from second-generation biodiesel into 1, 3 propanediol and butyric acid. Biochem. Eng. J. 2017, 128, 149–161. [Google Scholar] [CrossRef]
- Andrews, F.; Faulkner, M.; Toogood, H.S.; Scrutton, N.S. Combinatorial use of environmental stresses and genetic engineering to increase ethanol titres in cyanobacteria. Biotechnol. Biofuels 2021, 14, 240. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Liao, J.C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011, 13, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Liao, J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 6018–6023. [Google Scholar] [CrossRef] [PubMed]
- Nozzi, N.E.; Atsumi, S. Genome engineering of the 2, 3-butanediol biosynthetic pathway for tight regulation in cyanobacteria. ACS Synth. Biol. 2015, 4, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- McEwen, J.T.; Kanno, M.; Atsumi, S. 2, 3 Butanediol production in an obligate photoautotrophic cyanobacterium in dark conditions via diverse sugar consumption. Metab. Eng. 2016, 36, 28–36. [Google Scholar] [CrossRef]
- Lai, M.J.; Lan, E.I. Photoautotrophic synthesis of butyrate by metabolically engineered cyanobacteria. Biotechnol. Bioeng. 2019, 116, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Rabaey, K.; Rozendal, R.A. Microbial electrosynthesis—Revisiting the electrical route for microbial production. Nat. Rev. Microbiol. 2010, 8, 706–716. [Google Scholar] [CrossRef]
- Kerzenmacher, S. Engineering of microbial electrodes. Bioelectrosynthesis 2017, 167, 135–180. [Google Scholar]
- Ganigué, R.; Puig, S.; Batlle-Vilanova, P.; Balaguer, M.D.; Colprim, J. Microbial electrosynthesis of butyrate from carbon dioxide. Chem. Commun. 2015, 51, 3235–3238. [Google Scholar] [CrossRef]
- Batlle-Vilanova, P.; Ganigue, R.; Ramió-Pujol, S.; Baneras, L.; Jiménez, G.; Hidalgo, M.; Balaguer, M.D.; Colprim, J.; Puig, S. Microbial electrosynthesis of butyrate from carbon dioxide: Production and extraction. Bioelectrochemistry 2017, 117, 57–64. [Google Scholar] [CrossRef]
- Vassilev, I.; Hernandez, P.A.; Batlle-Vilanova, P.; Freguia, S.; Krömer, J.O.; Keller, J.r.; Ledezma, P.; Virdis, B. Microbial electrosynthesis of isobutyric, butyric, caproic acids, and corresponding alcohols from carbon dioxide. ACS Sustain. Chem. Eng. 2018, 6, 8485–8493. [Google Scholar] [CrossRef]
- Bao, Z.; HamediRad, M.; Xue, P.; Xiao, H.; Tasan, I.; Chao, R.; Liang, J.; Zhao, H. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat. Biotechnol. 2018, 36, 505–508. [Google Scholar] [CrossRef]
- Garst, A.D.; Bassalo, M.C.; Pines, G.; Lynch, S.A.; Halweg-Edwards, A.L.; Liu, R.; Liang, L.; Wang, Z.; Zeitoun, R.; Alexander, W.G. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 2017, 35, 48–55. [Google Scholar] [CrossRef]
- Costello, Z.; Martin, H.G. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. NPJ Syst. Biol. Appl. 2018, 4, 19. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C. Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 2017, 7, 2087. [Google Scholar] [CrossRef]
- Lin, G.-M.; Warden-Rothman, R.; Voigt, C.A. Retrosynthetic design of metabolic pathways to chemicals not found in nature. Curr. Opin. Syst. Biol. 2019, 14, 82–107. [Google Scholar] [CrossRef]
- Carbonell, P.; Jervis, A.J.; Robinson, C.J.; Yan, C.; Dunstan, M.; Swainston, N.; Vinaixa, M.; Hollywood, K.A.; Currin, A.; Rattray, N.J. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals. Commun. Biol. 2018, 1, 66. [Google Scholar] [CrossRef]
- Roell, G.W.; Sathish, A.; Wan, N.; Cheng, Q.; Wen, Z.; Tang, Y.J.; Bao, F.S. A comparative evaluation of machine learning algorithms for predicting syngas fermentation outcomes. Biochem. Eng. J. 2022, 186, 108578. [Google Scholar] [CrossRef]
- Zhuang, K.H.; Herrgård, M.J. Multi-scale exploration of the technical, economic, and environmental dimensions of bio-based chemical production. Metab. Eng. 2015, 31, 1–12. [Google Scholar] [CrossRef]
- Yu, Z.; Xiaolin, S.; Sun, X.; Wang, J.; Yuan, Q. Application of dynamic regulation strategies in metabolic engineering. Synth. Biol. J. 2020, 1, 440. [Google Scholar]
- Cui, S.; Lv, X.; Xu, X.; Chen, T.; Zhang, H.; Liu, Y.; Li, J.; Du, G.; Ledesma-Amaro, R.; Liu, L. Multilayer Genetic Circuits for Dynamic Regulation of Metabolic Pathways. ACS Synth. Biol. 2021, 10, 1587–1597. [Google Scholar] [CrossRef]
- Brockman, I.M.; Prather, K.L. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol. J. 2015, 10, 1360–1369. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Yang, L.; Lv, L.; Zhang, Z.; Ren, B.; Dong, L.; Li, N. A novel riboregulator switch system of gene expression for enhanced microbial production of succinic acid. J. Ind. Microbiol. Biotechnol. 2018, 45, 253–269. [Google Scholar] [CrossRef]
- Pandey, N.; Davison, S.A.; Krishnamurthy, M.; Trettel, D.S.; Lo, C.-C.; Starkenburg, S.; Wozniak, K.L.; Kern, T.L.; Reardon, S.D.; Unkefer, C.J.; et al. Precise Genomic Riboregulator Control of Metabolic Flux in Microbial Systems. ACS Synth. Biol. 2022, 11, 3216–3227. [Google Scholar] [CrossRef]
- Kirst, H.; Kerfeld, C.A. Bacterial microcompartments: Catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biol. 2019, 17, 79. [Google Scholar] [CrossRef]
- Müller, V. A synthetic bacterial microcompartment as production platform for pyruvate from formate and acetate. Proc. Natl. Acad. Sci. USA 2022, 119, e2201330119. [Google Scholar] [CrossRef]
- Axen, S.D.; Erbilgin, O.; Kerfeld, C.A. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 2014, 10, e1003898. [Google Scholar] [CrossRef]
- Tang, S.; Liao, D.; Li, X.; Lin, Y.; Han, S.; Zheng, S. Cell-free biosynthesis system: Methodology and perspective of in vitro efficient platform for pyruvate biosynthesis and transformation. ACS Synth. Biol. 2021, 10, 2417–2433. [Google Scholar] [CrossRef]
- Shiming, T.; Jiyuan, H.; Suiping ZHENG, S.H.; Ying, L. Designing, building and rapid prototyping of biosynthesis module based on cell-free system. Synth. Biol. J. 2022, 1, 90–96. [Google Scholar]
- Li, Y.; Chen, J.; Lun, S.-Y. Biotechnological production of pyruvic acid. Appl. Microbiol. Biotechnol. 2001, 57, 451–459. [Google Scholar]
- Rahmati, S.; Doherty, W.; Dubal, D.; Atanda, L.; Moghaddam, L.; Sonar, P.; Hessel, V.; Ostrikov, K.K. Pretreatment and fermentation of lignocellulosic biomass: Reaction mechanisms and process engineering. React. Chem. Eng. 2020, 5, 2017–2047. [Google Scholar] [CrossRef]
- Liu, X.; Xie, H.; Roussou, S.; Lindblad, P. Current advances in engineering cyanobacteria and their applications for photosynthetic butanol production. Curr. Opin. Biotechnol. 2022, 73, 143–150. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.; Eppink, M.H.; Wijffels, R.H. Integrated product recovery will boost industrial cyanobacterial processes. Trends Biotechnol. 2019, 37, 454–463. [Google Scholar] [CrossRef] [PubMed]

| Strain | Engineering Strategy | Substrate | Culture Method | Titer (g/L) | Yield (g/g) | Productivity (g/L/h) | References |
|---|---|---|---|---|---|---|---|
| Pyruvate | |||||||
| E. coli TBLA-1 | atpA mutation | Glucose | Batch | 30 | 0.64 | 1.2 | [12] |
| T.glabrata N07 | Reduced F0F1-ATPase activity | Glucose | Shake flask | 49.8 | 0.52 | 1.25 | [13] |
| T. glabrata INH1 | Expression of INH1 from S. cerevisiae | Glucose | Batch | 67.4 | ns | 1.69 | [14] |
| E. coli ALS929 | ΔaceEF, Δpfl, ΔpoxB, Δpps, ΔldhA, ΔatpFH, ΔarcA | Glucose | Fed-batch | 90 | 0.7 | 2.1 | [15] |
| E. coli CGSC6162 | ΔaceF Δppc | Glucose, acetate | Shake flask | 35 | 0.78 | 1.2 | [16] |
| S. cerevisiae Y2-15 | Δ PDC1, ΔPDC5 | Glucose | Shake flask | 24.65 | ns | 0.26 | [17] |
| E. coli W3110 | ΔpflB ΔpoxB ΔackA ΔldhA ΔadhE ΔfrdBC ΔsucA ΔatpFH | Glucose | Fed-batch | 52 | 0.76 | ns | [18] |
| E. coli MG1655 | ↓aceE, ↓r accA, ↓ppc, ↓gltA, Δ cra | Glucose | Batch | 26 | ns | ns | [19] |
| E. coli MG1655 | ↓aceE, ↓pdhR | Glucose | Shake flask | 11.28 | 0.33 | ns | [20] |
| E. coli LAFCPCPt | tetracycline-regulated promoter regulates aceE,ΔackA-pta, ΔadhE, Δcra, ΔldhA, ΔpflB, ΔpoxB | Glucose | Batch | 26.1 | 0.54 | ns | [21] |
| E. coli ATCC 8739 | Δ ldhA, ΔpoxB, ΔppsA, aceE point mutation | Glucose | Fed-batch | 18.8 | 0.66 | 1.28 | [22] |
| E. coli MP-XB010CN | ΔldhA, ΔpflB, ΔpoxB, ΔackA | Glucose | Two-phase fermentation | 93 | 0.71 | 2.02 | [23] |
| C. glabrata | Engineering HIF1 | Glucose | Batch | 53.1 | ns | ns | [24] |
| E. coli TA3052 | ΔdhA, ΔpoxB, Δpta, ΔadhE, harboring gltA-OFF switch | Glucose | Shake flask | 14.35 | ns | ns | [25] |
| Acetoin | |||||||
| B. subtilis PAR | ↑alsR | Glucose | Shake flask | 41.5 | 0.35 | 0.43 | [26] |
| B. subtilis JNA 3-10 BMN | ΔbdhA, ΔyodC | Glucose | Batch | 56.7 | 0.38 | 0.64 | [27] |
| K. Pneumoniae | ΔacoABCD, ΔbudC | Glucose | Fed-batch | 62.3 | 0.29 | 1.09 | [28] |
| S. marcescens H32 | Introduction of the L.brevis nox | Glucose | Fed-batch | 75.2 | 0.36 | 1.88 | [29] |
| C. glabrata | Introduction of the L. lactis nox; ↑PDC1, ↑GPD1, Δadh, Δald, Δbdh | Glucose | Shake flask | 7.33 | ns | ns | [30] |
| E. aerogenes EJW-03 | ΔbudC, ΔldhA, ΔdhaD, Δgcd | Glucose | Fed-batch | 71.1 | 0.32 | 2.87 | [31] |
| B.subtilis BSL24 | Introduction of the Selenomonas ruminantium xsa; introduction of the Clostridium stercorarium xyn10B | Xylose, xylan | Shake flask | 15 | 0.3 | 0.11 | [32] |
| 2,3-BD | |||||||
| S. cerevisiae YHI030 | ΔPDC,↑alsLpOp, ↑aldcLlOp, | Glucose | Fed-batch | 81 | 0.27 | ns | [33] |
| B. subtilis | ALsR regulates the expression of ALS and ALDC; expression of tdh from Clostridium beijerinckii; ΔldhA | Glucose | Three-stage fermentation | 102.6 | ns | 0.93 | [34] |
| Klebsiella oxytoca ME-UD-3 | ΔaldA | Glucose | Fed-batch | 130 | 0.48 | 1.63 | [35] |
| E. coli BW25113 | ΔldhA, ΔadhE, Δfrd, ↑Ec-IlvBN, ↑Ec-GldA | Glucose | Shake flask | 3 | ns | ns | [36] |
| K. oxytoca | ΔldhA, ΔpflB | Glucose | Fed-batch | 113 | 0.45 | 2.1 | [37] |
| K. pneumoniae KMK-05 | ΔwabG, ΔldhA, ΔpflB | Glucose | Shake flask | 3.11 | 0.46 | ns | [38] |
| E. coli W | Expression of budA, budB and budC from Enterobacteriaceae | Glucose | Fed-batch | 68 | 0.4 | 4.5 | [39] |
| B. amyloliquefaciens GAR | pMA5-acr-HapII-dhaD-PbdhA-alsR | glycerol | Fed-batch | 102.3 | 0.44 | 1.16 | [40] |
| Butanol | |||||||
| C. cellulovorans adh E2 | ↑fnrCA, ↑thlACA, ↑hbdCT | Cellulose | Shake flask | 5.6 | 0.34 | ns | [41] |
| C. saccharoperbutylacetonicum N1-4 | ↑thl, ↑hbd, ↑crt, ↑bcd, ↑thl, ↑hbd, ↑crt, ↑bcd, ↑ adhE1, ↑adhE1D485G, ↑thl, ↑thlA1V5A, ↑thlAV5A | Glucose | Bach | 17.4 | ns | ns | [42] |
| E.coli BW25113 | ↓pta, ↓frdA, ↓dhA, ↓adhE | Glucose | Bach | 30 | ns | ns | [43] |
| E. coli JCL299FT | ΔldhA, ΔadhE, ΔfrdBC,Δpta, ΔaceA | Glucose | Shake flask | 25.44 | ns | ns | [44] |
| C. acetobutylicum EA2018 | Δadc | Glucose | Shake flask | 12.2 | 0.203 | ns | [45] |
| Butyrate | |||||||
| C. tyrobutyricum ATCC 25755 | ↑cat1, ↑crt | Glucose | Fed-batch | 46.8 | 13.22 | 0.83 | [46] |
| E.coli LW393 | ΔldhA, ΔfrdABCD, ΔackA, ΔadhE; expression of hbd, crt, ptb, buk, from C. acetobutylicum; expression of ter from T. denticola | Glucose | Batch | 33 | 0.37 | 0.89 | [47] |
| E. coli BW lacIq | Expression of phaA, phaB from Ralstonia eutropha; phaJ from Aeromonas caviae; ter from Treponema denticola | Glucose | Fed-batch | 12.34 | 0.313 | 0.23 | [48] |
| C. tyrobutyricum Ct-pTBA | ↑xylT, ↑xylA,↑xylB | glucose, xylose | Batch | 42.6 | 0.36 | 0.56 | [49] |
| C. tyrobutyricum ATCC 25755 | ↑groESL | corn/rice straw | Fed-batch | 29.6/30.1 | 5.11/5.16 | 0.31/0.31 | [50] |
| L-alanine | |||||||
| Z. mobilis CP4thi | Expression of the B. sphaericus alaD | Glucose | Batch | 8 | 0.16 | ns | [51] |
| E. coli B0016-060BC | △ldhA, △ackA-pta, △pflB, △adhE, △frdA, △dadX::clts857-pR-pL-alaD-FRT | Glucose | Batch | 120.8 | 5.03 | 4.18 | [52] |
| E. coli AL887 | Expression of the B. sphaericus alaD; △ldhA, △aceF | Glucose | Batch | 32 | 0.63 | ns | [53] |
| E. coli ALS929 | Expression of the B. sphaericus alaD; △pfl, △pps, △aceEF, △poxB, △ldhA | Glucose | Fed-batch | 88 | 4 | 1 | [54] |
| C. glutamicum | Expression of the L. sphaericus alaD; ↑gapA, △ldhA, △ppc, △alr | Glucose | Fed-batch | 98 | ns | 0.83 | [55] |
| E. coli B0016-060BC | △ldhA, △ackA-pta, △pflB, △adhE, △frdA, △dadX::clts857-pR-pL-alaD-FRT | glycerol | Two-phase fermentation | 63.64 | 0.63 | 1.91 | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Q.; Ding, N.; Liu, Y.; Zhang, H.; Fang, Y.; Yin, L. Metabolic Engineering of Microorganisms to Produce Pyruvate and Derived Compounds. Molecules 2023, 28, 1418. https://doi.org/10.3390/molecules28031418
Luo Q, Ding N, Liu Y, Zhang H, Fang Y, Yin L. Metabolic Engineering of Microorganisms to Produce Pyruvate and Derived Compounds. Molecules. 2023; 28(3):1418. https://doi.org/10.3390/molecules28031418
Chicago/Turabian StyleLuo, Qian, Nana Ding, Yunfeng Liu, Hailing Zhang, Yu Fang, and Lianghong Yin. 2023. "Metabolic Engineering of Microorganisms to Produce Pyruvate and Derived Compounds" Molecules 28, no. 3: 1418. https://doi.org/10.3390/molecules28031418
APA StyleLuo, Q., Ding, N., Liu, Y., Zhang, H., Fang, Y., & Yin, L. (2023). Metabolic Engineering of Microorganisms to Produce Pyruvate and Derived Compounds. Molecules, 28(3), 1418. https://doi.org/10.3390/molecules28031418




