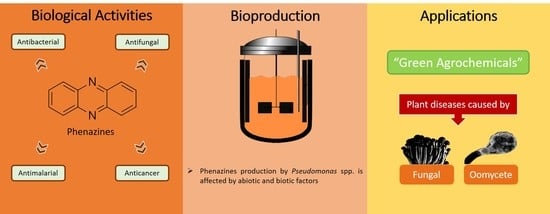
Recent Developments in the Biological Activities, Bioproduction, and Applications of Pseudomonas spp. Phenazines
Abstract
1. Introduction
| Compound | Phenazine Derivative | Bacterial Strain | Reference |
|---|---|---|---|
| 1 | Pyocyanin (PYO) | Pseudomonas aeruginosa | [10] |
| 2 | 1-hydroxyphenazine (1-OH-PHZ) | Pseudomonas aeruginosa | [10] |
| 3 | 2-hydroxyphenazine (2-OH-PHZ) | Pseudomonas chlororaphis | [11] |
| 4 | phenazine-1-carboxylic acid (PCA) | Pseudomonas chlororaphis | [11] |
| 5 | phenazine-1-carboxamide (PCN) | Pseudomonas aeruginosa; Pseudomonas chlororaphis | [11,12] |
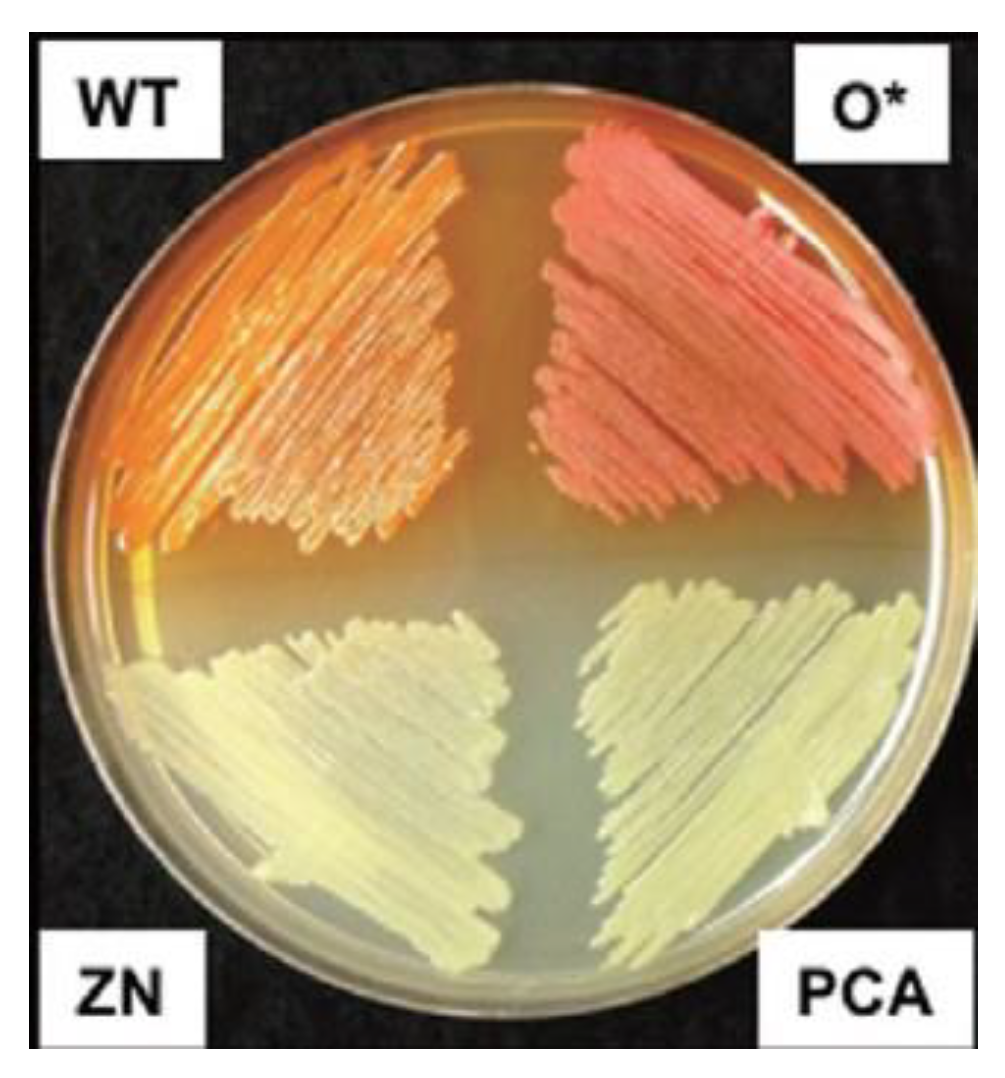
2. Phenazine-Producing Strains
| Strain Name | Strain Characteristics | Phenazine Type | Phenazine Production (mg/L) | Reference |
|---|---|---|---|---|
| Pseudomonas aeruginosa JY21 | Wild type | PYO | 311.1 | [28] |
| Pseudomonas aeruginosa LV | Wild type | PCA | 112.89 | [27] |
| PCN | 177.31 | |||
| Pseudomonas aeruginosa PA1201 | Wild type | PCA | 180 | [24] |
| Pseudomonas aeruginosa PA-IV | Delete phzS, phzM, phzH, pabB/C, trpE, and pchA; overexpression of aroG and phzC1; engineering the promoters of PCA biosynthetic gene clusters and the efflux pump, etc. | PCA | 9882 | [24] |
| Pseudomonas spp. MCC 3145 | Wild type | PYO | 313.94 | [34] |
| Pseudomonas sp. M18 | Wild type | PCA | 48.0 | [35] |
| Pseudomonas sp. M18 G | gacA deficient | PCA | 2597 | [33] |
| Pseudomonas sp. M18 GQ | gacA deficient; overexpression of phz gene cluster | PCA | 6365 | [33] |
| Pseudomonas chlororaphis HT66 | Wild type | PCN | 424.87 | [12] |
| Pseudomonas chlororaphis HT66 P3∆lon | Point mutations in 138 genes; deletion of lon gene | PCN | 9174 | [12] |
| Pseudomonas chlororaphis GP72 | Wild type | PCA | 22.0 | [20] |
| 2-OH-PHZ | 4.5 | |||
| Pseudomonas chlororaphis GP72AN | Inactivation of rpeA gene | PCA | 432/NR | [20,36] |
| 2-OH-PHZ | 170/258 | |||
| Pseudomonas fluorescens 2-79 | Wild type | PCA | 1010 | [37] |
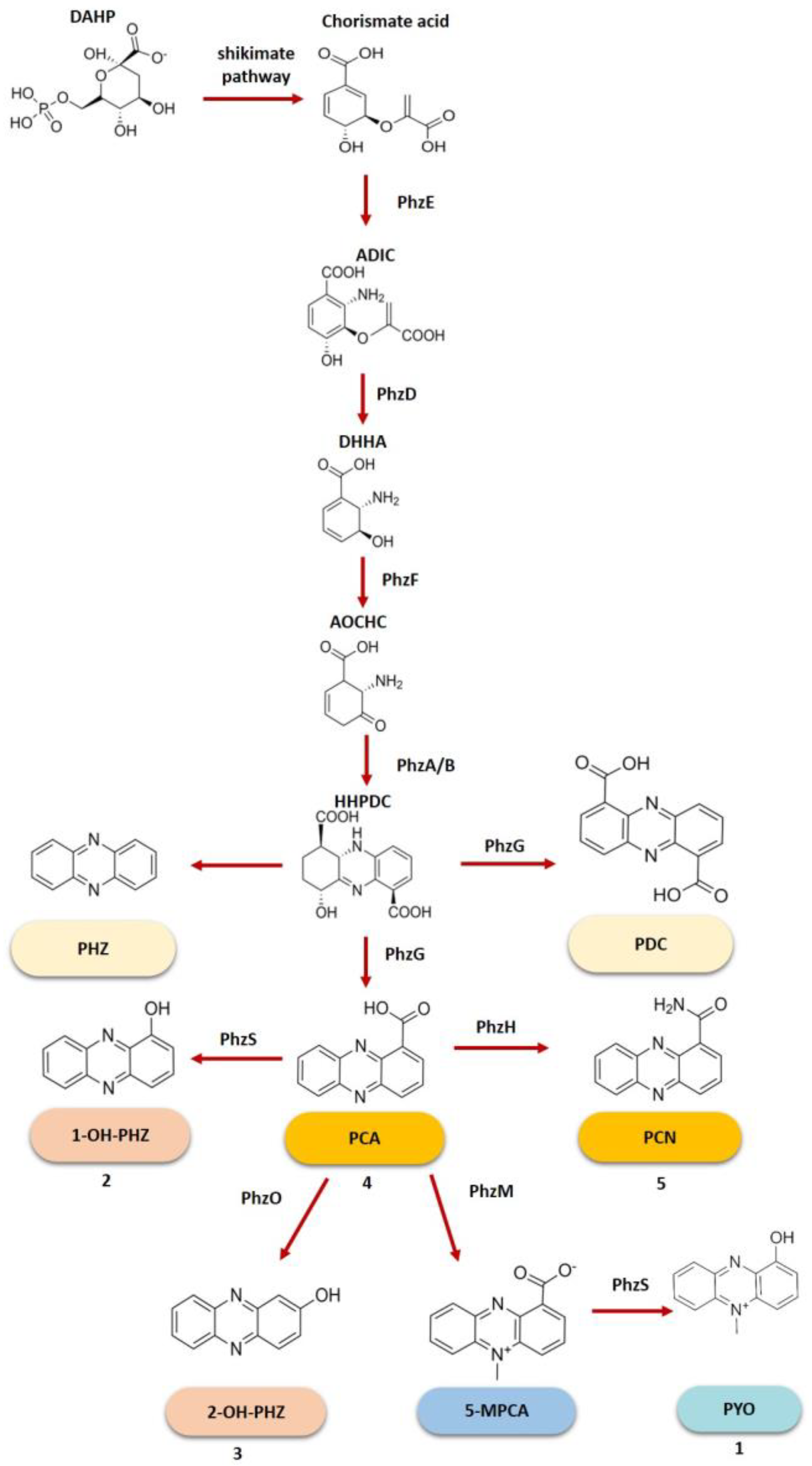
3. Phenazines Biosynthesis
3.1. Metabolic Pathways
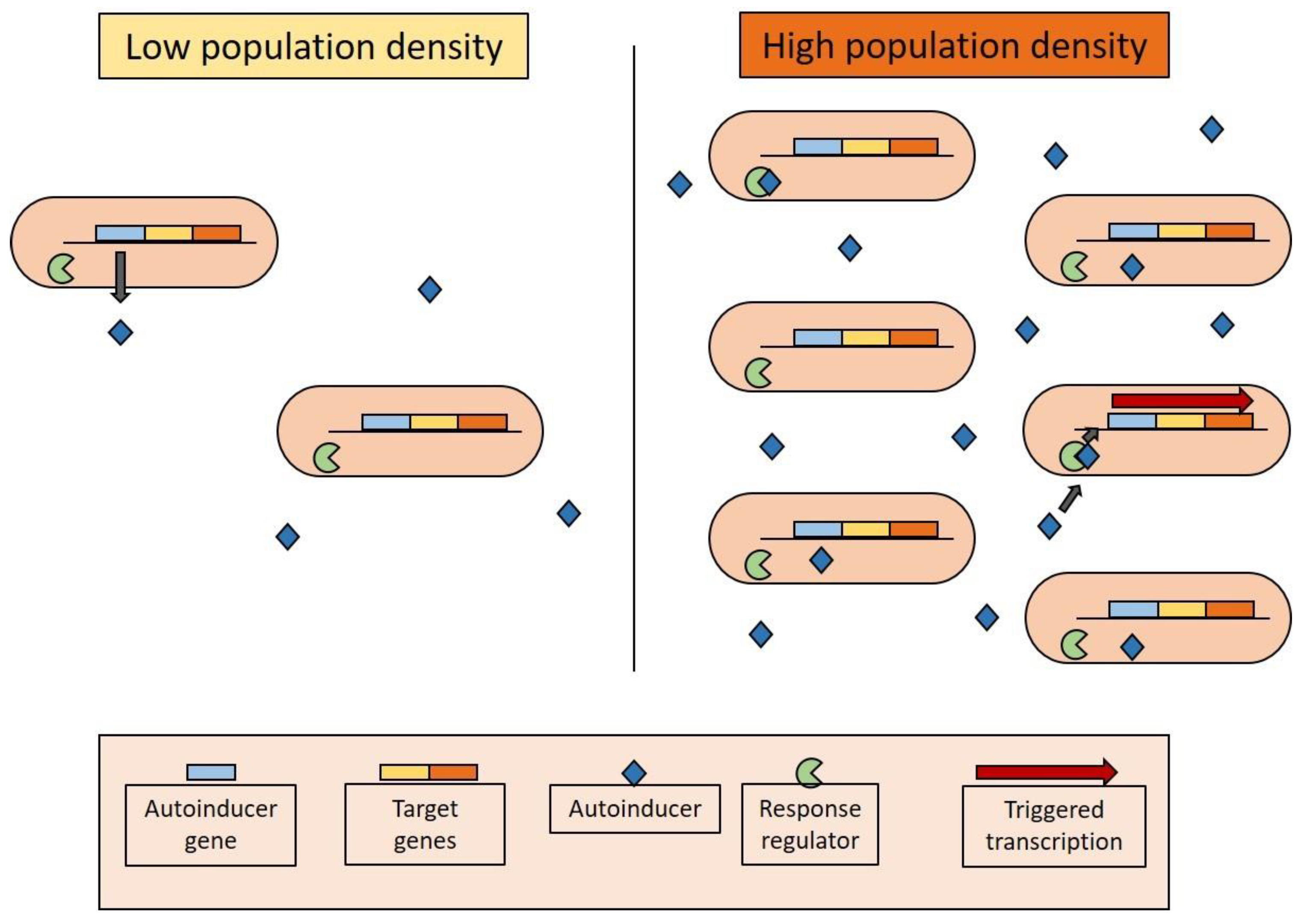
3.2. Biological Regulation
4. Phenazine Production
5. Extraction and Purification of Phenazines
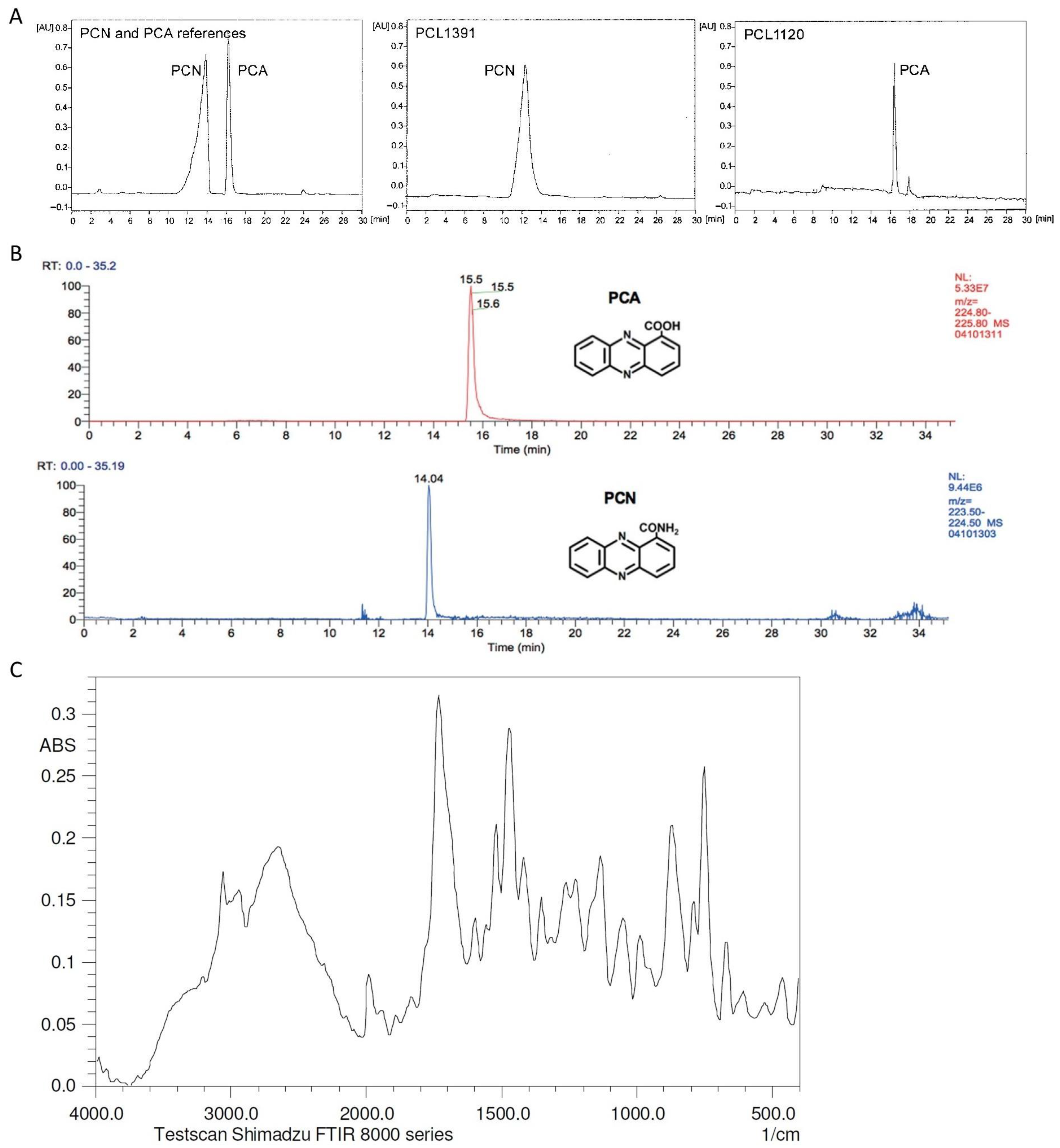
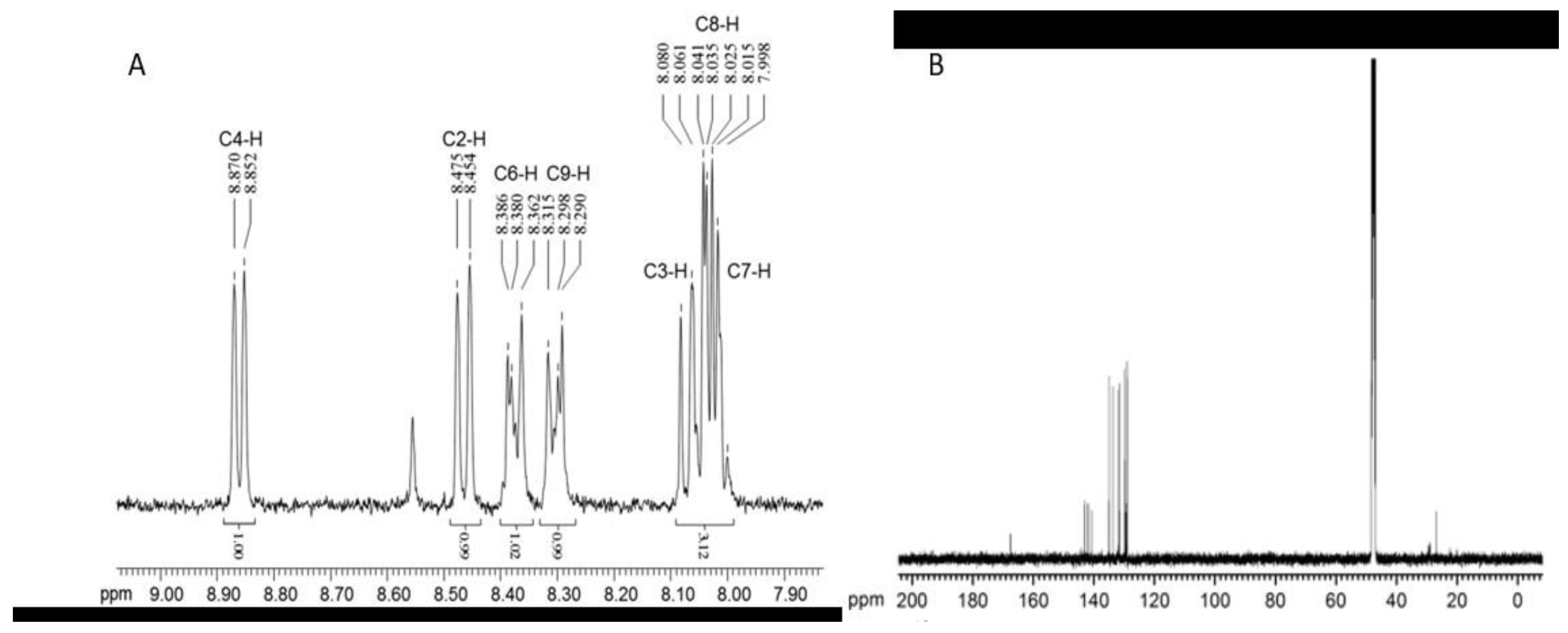
6. Phenazine Characterization
7. Properties, Applications, and Commercial Products
8. Final Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fordos, M.J. Recherches sur la matière colorante des suppurations bleues: Pyocyanine. CR Acad. Sci. 1860, 51, 215–217. [Google Scholar]
- Guttenberger, N.; Blankenfeldt, W.; Breinbauer, R. Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorg. Med. Chem. 2017, 25, 6149–6166. [Google Scholar] [CrossRef] [PubMed]
- Wrede, F.; Strack, E. Über das pyocyanin, Den blauen Farbstoff des Bacillus pyocyaneus. Hoppe-Seyler´s Z. Physiol. Chem. 1924, 140, 1–15. [Google Scholar] [CrossRef]
- Gonçalves, T.; Vasconcelos, U. Colour me blue: The history and the Biotechnological potential of pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef]
- Yan, J.; Liu, W.; Cai, J.; Wang, Y.; Li, D.; Hua, H.; Cao, H. Advances in phenazines over the past decade: Review of their pharmacological activities, mechanisms of action, biosynthetic pathways and synthetic strategies. Mar. Drugs 2021, 19, 610. [Google Scholar] [CrossRef]
- Schmitz, S.; Rosenbaum, M.A. Controlling the production of Pseudomonas phenazines by modulating the genetic repertoire. ACS Chem. Biol. 2020, 15, 3244–3252. [Google Scholar] [CrossRef]
- Chincholkar, S.; Thomashow, L. Microbial Phenazines, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Wang, S.-Y.; Shi, X.-C.; Chen, X.; Laborda, P.; Zhao, Y.-Y.; Liu, F.-Q.; Laborda, P. Biocontrol ability of phenazine-producing strains for the management of Fungal Plant Pathogens: A Review. Biol. Control 2021, 155, 104548. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, H.; Xian, M.; Huang, W. Biosynthetic pathway construction and production enhancement of 1-hydroxyphenazine derivatives in Pseudomonas chlororaphis H18. J. Agric. Food Chem. 2022, 70, 1223–1231. [Google Scholar] [CrossRef]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; HOiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocianin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S. Phenazine compounds in fluorescent Pseudomonas spp. Biosynthesis and regulation. Annu. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, P.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2018, 17, 1–12. [Google Scholar] [CrossRef]
- Blankenfeldt, W.; Parsons, J.F. The structural biology of phenazines biosynthesis. Curr. Opin. Struct. Biol. 2014, 29, 26–33. [Google Scholar] [CrossRef]
- Pierson, L.S.; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behaviour of bacteria in the environment and Biotechnological Processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef]
- Bilal, M.; Guo, S.; Iqbal, H.M.; Hu, H.; Wang, W.; Zhang, X. Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: A Review. World J. Microbiol. Biotechnol. 2017, 33, 1–11. [Google Scholar] [CrossRef]
- Laursen, J.B.; Nielsen, J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 2004, 104, 1663–1686. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, X.; Peng, H.; Hu, H.; Wang, W.; Zhang, X. Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting Rhizobacterium. Genom. Data 2015, 4, 33–42. [Google Scholar] [CrossRef]
- Yu, J.M.; Wang, D.; Pierson, L.S.; Pierson, E.A. Effect of producing different phenazines on bacterial fitness and biological control in Pseudomonas chlororaphis 30-84. Plant Pathol. J. 2018, 34, 44–58. [Google Scholar] [CrossRef]
- Yao, R.; Pan, K.; Peng, H.; Feng, L.; Hu, H.; Zhang, X. Engineering and systems-level analysis of Pseudomonas chlororaphis for production of phenazine-1-carboxamide using glycerol as the cost-effective carbon source. Biotechnol. Biofuels 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Huang, L.; Chen, M.-M.; Wang, W.; Hu, H.-B.; Peng, H.-S.; Xu, Y.-Q.; Zhang, X.-H. Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. App. Microbiol. Biotechnol. 2010, 89, 169–177. [Google Scholar] [CrossRef]
- Liu, K.; Hu, H.; Wang, W.; Zhang, X. Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb. Cell Fact. 2016, 15, 1–12. [Google Scholar] [CrossRef]
- Van Rij, E.T.; Wesselink, M.; Chin-A-Woeng, T.F.; Bloemberg, G.V.; Lugtenberg, B.J. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant Microbe Interact. 2004, 17, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Zhou, L.; Jiang, H.; Sun, S.; Fang, Y.; Liu, J.; Zhang, X.; He, Y.-W. Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab. Eng. 2015, 32, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa pao1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Ma, G.; Deng, L.; Li, R. A Kinetic Model for Phenazine-1-Carboxylic Acid Production by Pseudomonas sp. M18G. Int. J. Clin. Exp. Med. 2017, 10, 6468–6474. [Google Scholar]
- Li, Y.; Jiang, H.; Xu, Y.; Zhang, X. Optimization of nutrient components for enhanced phenazine-1-carboxylic acid production by GACA-inactivated Pseudomonas sp. M18G using response surface method. App. Microbiol. Biotechnol. 2008, 77, 1207–1217. [Google Scholar] [CrossRef]
- Bedoya, J.C.; Dealis, M.L.; Silva, C.S.; Niekawa, E.T.G.; Navarro, M.O.P.; Simionato, A.S.; Modolon, F.; Chryssafidis, A.L.; Andrade, G. Enhanced Production of Target Bioactive Metabolites Produced by Pseudomonas aeruginosa LV Strain. Biocatal. Agric. Biotechnol. 2019, 17, 545–556. [Google Scholar] [CrossRef]
- Abo-Zaid, G.A.; Wagih, E.E.; Matar, S.M.; Ashmawy, N.A.; Hafez, E.E. Optimization of Pyocyanin Production from Pseudomonas aeruginosa JY21 Using Statistical Experimental Designs. Int. J. ChemTech Res. 2015, 8, 137–148. [Google Scholar]
- Haggag, W.M.; Abo El Soud, M. Production and optimization of Pseudomonas fluorescens biomass and metabolites for biocontrol of Strawberry Grey Mould. Am. J. Plant Sci. 2012, 03, 836–845. [Google Scholar] [CrossRef]
- Cui, J.; Wang, W.; Hu, H.; Zhang, H.; Zhang, X. Enhanced phenazine-1-carboxamide production in Pseudomonas chlororaphis h5△fleq△rela through fermentation optimization. Fermentation 2022, 8, 188. [Google Scholar] [CrossRef]
- Zhou, Q.; Su, J.; Jiang, H.; Huang, X.; Xu, Y. Optimization of Phenazine-1-Carboxylic Acid Production by a GacA/QscR-Inactivated Pseudomonas sp. M18GQ Harboring PME6032Phz Using Response Surface Methodology. App. Microbiol. Biotechnol. 2010, 86, 1761–1773. [Google Scholar] [CrossRef]
- Song, C.; Yue, S.-J.; Liu, W.-H.; Zheng, Y.-F.; Zhang, C.-H.; Feng, T.-T.; Hu, H.-B.; Wang, W.; Zhang, X.-H. Engineering of glycerol utilization in Pseudomonas chlororaphis GP72 for enhancing phenazine-1-carboxylic acid production. World J. Microbiol. Biotechnol. 2020, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, H.; Du, X.; Huang, X.; Zhang, X.; Xu, Y.; Xu, Y. Enhancement of phenazine-1-carboxylic acid production using batch and fed-batch culture of GacA inactivated Pseudomonas sp. M18G. Biores. Technol. 2010, 101, 3649–3656. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Nikam, M.; Patil, H.; Anokhina, T.; Kochetkov, V.; Chaudhari, A. Bioactive pigment production by Pseudomonas spp. MCC 3145: Statistical media optimization, biochemical characterization, fungicidal and DNA intercalation-based cytostatic activity. Proc. Biochem. 2017, 58, 298–305. [Google Scholar] [CrossRef]
- Ge, Y.; Huang, X.; Wang, S.; Zhang, X.; Xu, Y. Phenazine-1-carboxylic acid is negatively regulated and pyoluteorin positively regulated by GacA in Pseudomona ssp. M18. FEMS Microbiol. Lett. 2004, 237, 41–47. [Google Scholar] [CrossRef]
- Yue, S.-J.; Huang, P.; Li, S.; Jan, M.; Hu, H.-B.; Wang, W.; Zhang, X.-H. Enhanced production of 2-hydroxyphenazine from glycerol by a two-stage fermentation strategy in Pseudomonas chlororaphis GP72AN. J. Agric. Food Chem. 2019, 68, 561–566. [Google Scholar] [CrossRef]
- Slininger, P.J.; Shea-Wilbur, M.A. Liquid-culture pH, temperature, and carbon (not nitrogen) source regulate phenazine productivity of the take-all biocontrol agent Pseudomonas fluorescens 2-79. App. Microbiol. Biotechnol. 1995, 43, 794–800. [Google Scholar] [CrossRef]
- McDonald, M.; Mavrodi, D.V.; Thomashow, L.S.; Floss, H.G. Phenazine Biosynthesis in Pseudomonas fluorescens: Branchpoint from the Primary Shikimate Biosynthetic Pathway and Role of Phenazine-1,6-Dicarboxylic Acid. J. Am. Chem. Soc. 2001, 123, 9459–9460. [Google Scholar] [CrossRef]
- Wang, S.; Fu, C.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Enhanced biosynthesis of Arbutin by engineering shikimate pathway in Pseudomonas chlororaphis p3. Microb. Cell Fact. 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Wang, S.; Liu, D.; Bilal, M.; Wang, W.; Zhang, X. Uncovering the role of phzc as DAHP synthase in shikimate pathway of Pseudomonas chlororaphis HT66. Biology 2022, 11, 86. [Google Scholar] [CrossRef]
- Blankenfeldt, W.; Kuzin, A.P.; Skarina, T.; Korniyenko, Y.; Tong, L.; Bayer, P.; Janning, P.; Thomashow, L.S.; Mavrodi, D.V. Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc. Natl. Acad. Sci. USA 2004, 101, 16431–16436. [Google Scholar] [CrossRef]
- Parsons, J.F.; Song, F.; Parsons, L.; Calabrese, K.; Eisenstein, E.; Ladner, J.E. Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79. Biochemistry 2004, 43, 12427–12435. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, E.G.; Janning, P.; Mentel, M.; Graebsch, A.; Breinbauer, R.; Hiller, W.; Costisella, B.; Thomashow, L.S.; Mavrodi, D.V.; Blankenfeldt, W. PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis. J. Am. Chem. Soc. 2008, 130, 17053–17061. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Dai, B.; Wang, W.; Hu, H.; Huang, X.; Zhang, X. PhzA, the shunt switch of phenazine-1,6-dicarboxylic acid biosynthesis in Pseudomonas chlororaphis HT66. App. Microb. Biotechnol. 2017, 101, 7165–7175. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, Y.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Microbial synthesis of antibacterial phenazine-1,6-dicarboxylic acid and the role of PhzG in Pseudomonas chlororaphis GP72AN. J. Agric. Food Chem. 2020, 68, 2373–2380. [Google Scholar] [CrossRef]
- Delaney, S.M.; Mavrodi, D.V.; Bonsall, R.F.; Thomashow, L.S. PhzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30-84. J. Bacteriol. 2001, 183, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Ahuja, E.K.; Janning, P.; Mavrodi, D.V.; Thomashow, L.S.; Blankenfeldt, W. Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis. Acta Crystallog. D Biol. Crystallogr. 2013, 69, 1403–1413. [Google Scholar] [CrossRef]
- Huang, L.; Huang, Y.; Lou, Y.; Qian, H.; Xu, D.; Ma, L.; Jiang, C.; Zhang, D. Pyocyanin-modifying genes phzM and phzS regulated the extracellular electron transfer in microbiologically-influenced corrosion of X80 carbon steel by Pseudomonas aeruginosa. Corros. Sci. 2020, 164, 108355. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.; Thomas-Oates, J.E.; Lugtenberg, B.J.; Bloemberg, G.V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant Microbe Interact. 2001, 14, 1006–1015. [Google Scholar] [CrossRef]
- Peng, H.; Ouyang, Y.; Bilal, M.; Wang, W.; Hu, H.; Zhang, X. Identification, synthesis and regulatory function of the N-acylated homoserine lactone signals produced by Pseudomonas chlororaphis HT66. Microb. Cell Fact. 2018, 17, 1–11. [Google Scholar] [CrossRef]
- Chen, M.; Cao, H.; Peng, H.; Hu, H.; Wang, W.; Zhang, X. Reaction kinetics for the biocatalytic conversion of phenazine-1-carboxylic acid to 2-hydroxyphenazine. PLoS ONE 2014, 9, e98537. [Google Scholar] [CrossRef]
- Dasgupta, D.; Kumar, A.; Mukhopadhyay, B.; Sengupta, T.K. Isolation of phenazine 1,6-di-carboxylic acid from Pseudomonas aeruginosa strain HRW.1-S3 and its role in biofilm-mediated crude oil degradation and cytotoxicity against bacterial and cancer cells. App. Microbiol. Biotechnol. 2015, 99, 8653–8665. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Peever, T.L.; Mavrodi, O.V.; Parejko, J.A.; Raaijmakers, J.M.; Lemanceau, P.; Mazurier, S.; Heide, L.; Blankenfeldt, W.; Weller, D.M.; et al. Diversity and evolution of the phenazine biosynthesis pathway. App. Environ. Microbiol. 2010, 76, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S., III.; Wood, D.W.; Pierson, E.A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 1998, 36, 207–225. [Google Scholar] [CrossRef] [PubMed]
- De Maeyer, K.; D’aes, J.; Hua, G.K.; Perneel, M.; Vanhaecke, L.; Noppe, H.; Höfte, M. N-acylhomoserine lactone quorum-sensing signalling in antagonistic phenazine-producing Pseudomonas isolates from the red cocoyam rhizosphere. Microbiology 2011, 157, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Bourret, R.B.; Silversmith, R.E. Two-component signal transduction. Curr. Opin. Microbiol. 2010, 13, 113–115. [Google Scholar] [CrossRef]
- Bauer, J.S.; Hauck, N.; Christof, L.; Mehnaz, S.; Gust, B.; Gross, H. The systematic investigation of the Quorum Sensing system of the biocontrol strain Pseudomonas chlororaphis subsp. aurantiaca PB-ST2 unveils auri to be a biosynthetic origin for 3-oxo-homoserine lactones. PLoS ONE 2016, 11, e0167002. [Google Scholar] [CrossRef]
- Morohoshi, T.; Yamaguchi, T.; Xie, X.; Wang, W.Z.; Takeuchi, K.; Someya, N. Complete genome sequence of Pseudomonas chlororaphis subsp. aurantiaca reveals a triplicate quorum-sensing mechanism for regulation of phenazine production. Microbes Environ. 2017, 32, 47–53. [Google Scholar] [CrossRef]
- Heeb, S.; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 2001, 14, 1351–1363. [Google Scholar] [CrossRef]
- Wang, D.; Yu, J.M.; Pierson, L.S.; Pierson, E.A. Differential regulation of phenazine biosynthesis by RPEA and RpeB in Pseudomonas chlororaphis 30-84. Microbiology 2012, 158, 1745–1757. [Google Scholar] [CrossRef]
- Girard, G.; van Rij, E.T.; Lugtenberg, B.J.; Bloemberg, G.V. Regulatory roles of PSRA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology 2006, 152, 43–58. [Google Scholar] [CrossRef]
- Morohoshi, T.; Yabe, N.; Yaguchi, N.; Xie, X.; Someya, N. Regulation of phenazine-1-carboxamide production by quorum sensing in type strains of Pseudomonas chlororaphis subsp. chlororaphis and Pseudomonas chlororaphis subsp. piscium. J. Biosci. Bioeng. 2022, 133, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.-J.; Peng, H.-S.; Hu, H.-B.; Huang, X.-Q.; Wang, W.; Zhang, X.-H. ITRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis p3. Sci. Rep. 2016, 6, 27393. [Google Scholar] [CrossRef]
- Yuan, L.-L.; Li, Y.-Q.; Wang, Y.; Zhang, X.-H.; Xu, Y.-Q. Optimization of critical medium components using response surface methodology for phenazine-1-carboxylic acid production by Pseudomonas sp. M-18Q. J. Biosci. Bioeng. 2008, 105, 232–237. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Xu, Y.-Q.; Zhang, X.-H. Medium factor optimization and fermentation kinetics for phenazine-1-carboxylic acid production by Pseudomonas sp. M18G. Biotechnol. Bioeng. 2008, 100, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Aloui, H.; Khomlaem, C.; Torres, C.A.V.; Freitas, F.; Reis, M.A.M.; Kim, B.S. Enhanced co-production of medium-chain-length polyhydroxyalkanoates and phenazines from crude glycerol by high cell density cultivation of Pseudomonas chlororaphis in membrane bioreactor. Int. J. Biol. Macromol. 2022, 211, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Yue, S.-J.; Hu, H.-B.; Wang, W.; Zhang, X.-H. Adsorption/desorption characteristics, separation and purification of phenazine-1-carboxylic acid from fermentation extract by macroporous adsorbing resins. J. Cheml. Technol. Biotechnol. 2018, 93, 3176–3184. [Google Scholar] [CrossRef]
- Jasim, B.; Anisha, C.; Rohini, S.; Kurian, J.M.; Jyothis, M.; Radhakrishnan, E.K. Phenazine carboxylic acid production and rhizome protective effect of endophytic Pseudomonas aeruginosa isolated from Zingiber officinale. World J. Microbiol. Biotechnol. 2013, 30, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Fan, L.-Y.; Zhang, W.; Guo, C.-G.; Li, S.; Xu, Y.-Q.; Cao, C.-X. Purification of low-concentration phenazine-1-carboxylic acid from fermentation broth of Pseudomonas sp. M18 via free flow electrophoresis with Gratis gravity. Electrophoresis 2010, 31, 3499–3507. [Google Scholar] [CrossRef]
- Mehnaz, S. Characterization of a phenazine and hexanoyl homoserine lactone producing Pseudomonas aurantiaca strain PB-st2, isolated from sugarcane stem. J. Microbiol. Biotechnol. 2009, 19, 1688–1694. [Google Scholar] [CrossRef]
- Shanmugaiah, V.; Mathivanan, N.; Varghese, B. Purification, crystal structure and antimicrobial activity of phenazine-1-carboxamide produced by a growth-promoting biocontrol bacterium, Pseudomonas aeruginosa MML2212. J. App. Microbiol. 2010, 108, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Aziz, L.M.; Hamza, S.J. Isolation and characterization of phenazine produced from mutant Pseudomonas aeruginosa. Al-Anbar J. Vet. Sci. 2012, 5, 42–53. [Google Scholar]
- Gorantla, J.N.; Nishanth Kumar, S.; Nisha, G.V.; Sumandu, A.S.; Dileep, C.; Sudaresan, A.; Sree Kumar, M.M.; Lankalapalli, R.S.; Kumar, B.S. Purification and characterization of antifungal phenazines from a fluorescent Pseudomonas strain FPO4 against medically important fungi. J. Mycol. Médicale 2014, 24, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.R.; Adhikari, N. An overview on common organic solvents and their toxicity. J. Pharm. Res. Int. 2019, 28, 1–18. [Google Scholar] [CrossRef]
- Rane, M.R.; Sarode, P.D.; Chaudhari, B.L.; Chincholkar, S.B. Detection, isolation and identification of phenazine-1-carboxylic acid produced by biocontrol strains of Pseudomonas Aeruginosa. J. Sci. Ind. Res. 2007, 66, 627–631. [Google Scholar]
- Chen, K.; Hu, H.; Wang, W.; Zhang, X.; Xu, Y. Metabolic degradation of phenazine-1-carboxylic acid by the strain Sphingomonas sp. DP58: The identification of two metabolites. Biodegradation 2007, 19, 659–667. [Google Scholar] [CrossRef]
- Liu, H.; He, Y.; Jiang, H.; Peng, H.; Huang, X.; Zhang, X.; Thomashow, L.S.; Xu, Y. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr. Microbiol. 2007, 54, 302–306. [Google Scholar] [CrossRef]
- Thacharodi, A.; Priyadharshini, R.; Karthikeyan, G.; Jeganathan, C.; Reghu, A.P.; Pugazhendhi, A. Extraction, purification and characterization of phenazine from Pseudomonas aeruginosa isolate of wastewater sources: A panacea towards clinical pathogens. App. Nanosci. 2021. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, X.; Huang, X.; Cao, C.; Xu, Y. Rapid quantitative analysis of phenazine-1-carboxylic acid and 2-hydroxyphenazine from fermentation culture of Pseudomonas chlororaphis GP72 by capillary zone electrophoresis. Talanta 2008, 76, 276–281. [Google Scholar] [CrossRef]
- Zhou, K.; Kammarchedu, V.; Butler, D.; Soltan Khamsi, P.; Ebrahimi, A. Electrochemical sensors based on MOS x -functionalized laser-induced graphene for real-time monitoring of phenazines produced by Pseudomonas aeruginosa. Adv. Health Mat. 2022, 11, 2200773. [Google Scholar] [CrossRef]
- Kern, S.E.; Newman, D.K. Measurement of phenazines in bacterial cultures. Methods Mol. Biol. 2014, 1149, 303–310. [Google Scholar] [PubMed]
- Simionato, A.S.; Navarro, M.O.; de Jesus, M.L.; Barazetti, A.R.; da Silva, C.S.; Simões, G.C.; Balbi-Peña, M.I.; de Mello, J.C.; Panagio, L.A.; de Almeida, R.S.; et al. The effect of phenazine-1-carboxylic acid on mycelial growth of botrytis cinerea produced by Pseudomonas aeruginosa LV strain. Front. Microbiol. 2017, 8, 1102. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S.; Pierson, E.A. Phenazine antibiotic production in Pseudomonas aureofaciens: Role in rhizosphere ecology and pathogen suppression. FEMS Microbiol. Lett. 1996, 136, 101–108. [Google Scholar] [CrossRef]
- Castaldi, S.; Masi, M.; Sautua, F.; Cimmino, A.; Isticato, R.; Carmona, M.; Tuzi, A.; Evidente, A. Pseudomonas fluorescens showing antifungal activity against Macrophomina phaseolina, a severe pathogenic fungus of soybean, produces phenazine as the main active metabolite. Biomolecules 2021, 11, 1728. [Google Scholar] [CrossRef]
- Raio, A.; Puopolo, G. Pseudomonas chlororaphis metabolites as biocontrol promoters of plant health and improved crop yield. World J. Microbiol. Biotechnol. 2021, 37, 99. [Google Scholar] [CrossRef] [PubMed]
- Halicki, P.C.B.; da Silva, E.N.; Jardim, G.A.D.M.; Almeida, R.G.D.; Vicenti, J.R.D.M.; Gonçalves, B.L.; da Silva, P.E.A.; Ramos, D.F. Benzo[a]phenazine derivatives: Promising scaffolds to combat resistant Mycobacterium tuberculosis. Chem. Biol. Drug Des. 2021, 98, 352–362. [Google Scholar] [CrossRef]
- Huigens, R.W.; Abouelhassan, Y.; Yang, H. Phenazine antibiotic-inspired discovery of bacterial biofilm-eradicating agents. Chem. Bio. Chem. 2019, 20, 2885–2902. [Google Scholar] [CrossRef]
- Saosoong, K.; Wongphathanakul, W.; Poasiri, C.; Ruangviriyachai, C. Isolation and analysis of antibacterial substance produced from P. aeruginosa TISTR 781. KKU Sci. J. 2009, 37, 163–172. [Google Scholar]
- Giddens, S.R.; Bean, D.C. Investigations into the in vitro antimicrobial activity and mode of action of the phenazine antibiotic D-alanylgriseoluteic acid. Int. J. Antimicrob. Agents 2007, 29, 93–97. [Google Scholar] [CrossRef]
- Park, G.K. Elucidation of antifungal metabolites produced by Pseudomonas aurantiaca IB5-10 with broad-spectrum antifungal activity. J. Microbiol. Biotechnol. 2014, 22, 326–330. [Google Scholar] [CrossRef]
- George, E.; Kumar, S.N.; Jacob, J.; Bommasani, B.; Lankalapalli, R.S.; Morang, P.; Kumar, B.S. Characterization of the bioactive metabolites from a plant growth-promoting rhizobacteria and their exploitation as antimicrobial and plant growth-promoting agents. App. Biochem. Biotechnol. 2015, 176, 529–546. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Norrman, A.; Ponomarenko, S.A.; Friberg, A.T. Coherence lattices in surface plasmon Polariton Fields. Opt. Lett. 2018, 43, 3429. [Google Scholar] [CrossRef]
- Liu, T.T.; Ye, F.C.; Pang, C.P.; Yong, T.Q.; Tang, W.D.; Xiao, J.; Shang, C.H.; Lu, Z.J. Isolation and identification of bioactive substance 1-hydroxyphenazine from Pseudomonas aeruginosa and its antimicrobial activity. Lett. App. Microbiol. 2020, 71, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Karmegham, N.; Vellasamy, S.; Natesan, B.; Sharma, M.P.; Al Farraj, D.A.; Elshikh, M.S. Characterization of antifungal metabolite phenazine from rice rhizosphere fluorescent pseudomonads (FPS) and their effect on sheath blight of rice. Saudi J. Biol. Sci. 2020, 27, 3313–3326. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Jacobs, N.J.; Rajamani, S.; Krishnamurthy, M.; Cubillos-Ruiz, J.R.; Hogan, D.A. Antifungal mechanisms by which a novel pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol. Microbiol. 2010, 78, 1379–1392. [Google Scholar] [CrossRef]
- Liu, X.-W.; Liu, N.-Y.; Deng, Y.-Q.; Wang, S.; Liu, T.; Tang, Y.-C.; Chen, Y.-D.; Lu, J.-L. Anticancer activity, topoisomerase I inhibition, DNA ‘light switch’ behavior and molecular docking of two ruthenium complexes containing phenazine ring. J. Biomol. Struct. Dyn. 2020, 39, 5953–5962. [Google Scholar] [CrossRef]
- Hollstein, U.; Van Gemert, R.J. Interaction of phenazines with Polydeoxyribonucleotides. Biochemistry 1971, 10, 497–504. [Google Scholar] [CrossRef]
- Kennedy, R.K.; Naik, P.R.; Veena, V.; Lakshmi, B.S.; Lakshmi, P.; Krishna, R.; Sakthivel, N. 5-methyl phenazine-1-carboxylic acid: A novel bioactive metabolite by a rhizosphere soil bacterium that exhibits potent antimicrobial and anticancer activities. Chem. Biol. Interact. 2015, 231, 71–82. [Google Scholar] [CrossRef]
- Vicker, N.; Burgess, L.; Chuckowree, I.S.; Dodd, R.; Folkes, A.J.; Hardick, D.J.; Hancox, T.C.; Miller, W.; Milton, J.; Sohal, S.; et al. Novel angular benzophenazines: dual Topoisomerase I and Topoisomerase II inhibitors as potential anticancer agents. J. Med. Chem. 2002, 45, 721–739. [Google Scholar] [CrossRef]
- Kato, S.; Shindo, K.; Yamagishi, Y.; Matsuoka, M.; Kawai, H.; Mochizuki, J. Phenazoviridin, a novel free radical scavenger from Streptomyces sp. taxonomy, fermentation, isolation, structure elucidation and biological properties. J. Antibiot. 1993, 46, 1485–1493. [Google Scholar] [CrossRef]
- Laxmi, M.; Bhat, S.G. Characterization of pyocyanin with radical scavenging and antibiofilm properties isolated from Pseudomonas aeruginosa strain BTRY1. 3 Biotech 2016, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Niu, J.; Liu, H.; Xu, Z.; Li, J.; Wu, Q. Synthesis and bioactivities of phenazine-1-carboxylic acid derivatives based on the modification of PCA Carboxyl Group. Bioorg. Med. Chem. Lett. 2017, 27, 2010–2013. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Li, S.; Wang, J.; Chen, C.; Zhou, M. Integrated biological and chemical control of rice sheath blight by bacillus subtilisNJ-18 and pingyangmycin. Pest Manag. Sci. 2013, 70, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yu, L.; Hsiang, T.; Huang, D.; Xu, Z.; Wu, Q.; Du, X.; Li, J. The influence of steric configuration of phenazine-1-carboxylic acid-amino acid conjugates on fungicidal activity and systemicity. Pest Manag. Sci. 2019, 75, 3323–3330. [Google Scholar] [CrossRef]
- Ali, S.; Hameed, S.; Shahid, M.; Iqbal, M.; Lazarovits, G.; Imran, A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol. Res. 2020, 232, 126389. [Google Scholar] [CrossRef]
- Ou, J.; Li, H.; Ou, X.; Yang, Z.; Chen, M.; Liu, K.; Teng, Y.; Xing, B. Degradation, adsorption and leaching of phenazine-1-carboxamide in agrigulture soils. Ecotoxicol. Environ. Saf. 2020, 205, 111375. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serafim, B.; Bernardino, A.R.; Freitas, F.; Torres, C.A.V. Recent Developments in the Biological Activities, Bioproduction, and Applications of Pseudomonas spp. Phenazines. Molecules 2023, 28, 1368. https://doi.org/10.3390/molecules28031368
Serafim B, Bernardino AR, Freitas F, Torres CAV. Recent Developments in the Biological Activities, Bioproduction, and Applications of Pseudomonas spp. Phenazines. Molecules. 2023; 28(3):1368. https://doi.org/10.3390/molecules28031368
Chicago/Turabian StyleSerafim, Bruno, Ana R. Bernardino, Filomena Freitas, and Cristiana A. V. Torres. 2023. "Recent Developments in the Biological Activities, Bioproduction, and Applications of Pseudomonas spp. Phenazines" Molecules 28, no. 3: 1368. https://doi.org/10.3390/molecules28031368
APA StyleSerafim, B., Bernardino, A. R., Freitas, F., & Torres, C. A. V. (2023). Recent Developments in the Biological Activities, Bioproduction, and Applications of Pseudomonas spp. Phenazines. Molecules, 28(3), 1368. https://doi.org/10.3390/molecules28031368