Glycoproteomic Analysis of Urinary Extracellular Vesicles for Biomarkers of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results and Discussion
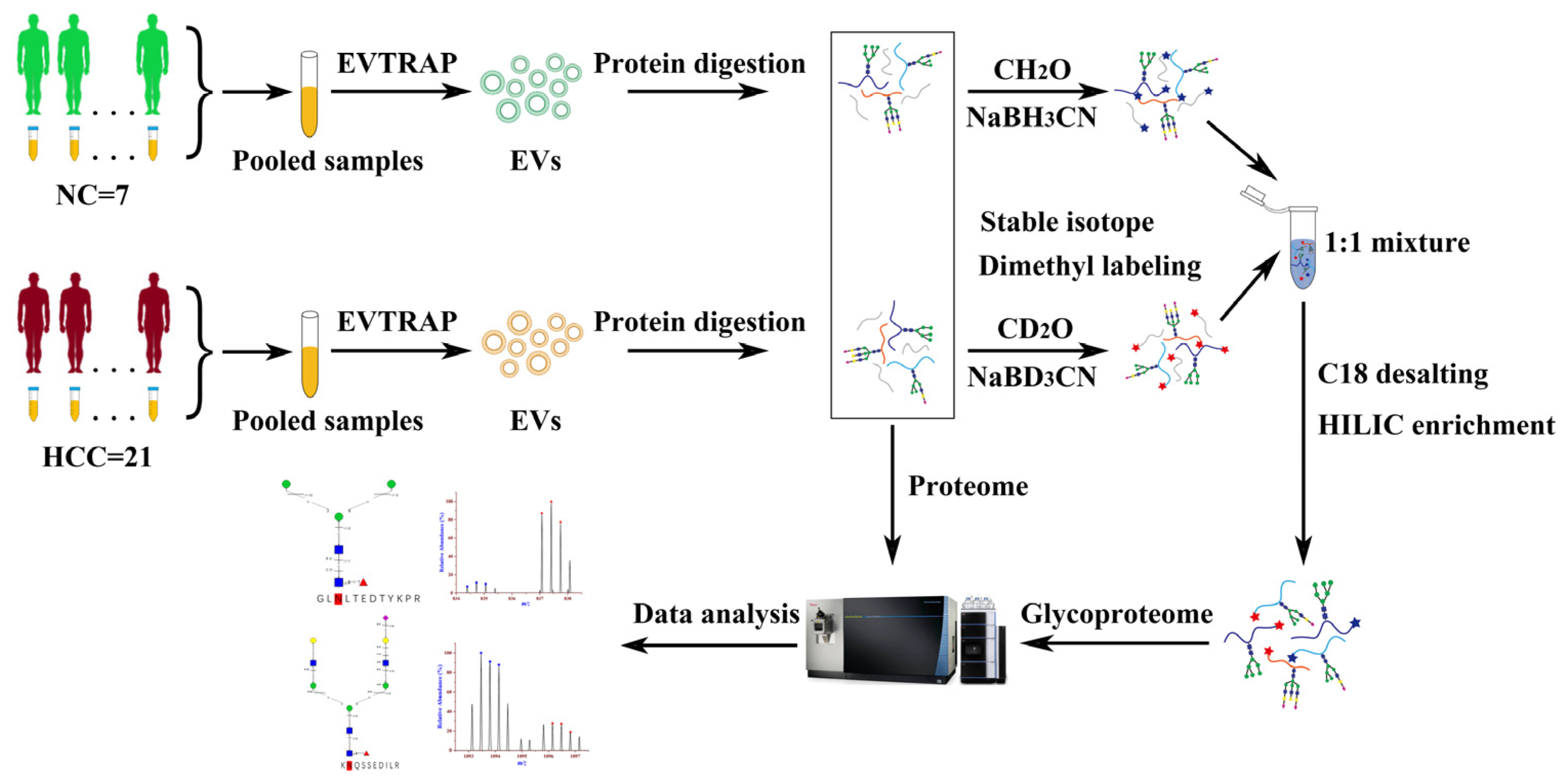
2.1. Workflow of EVs Proteomics and Glycoproteomics from Urine Samples
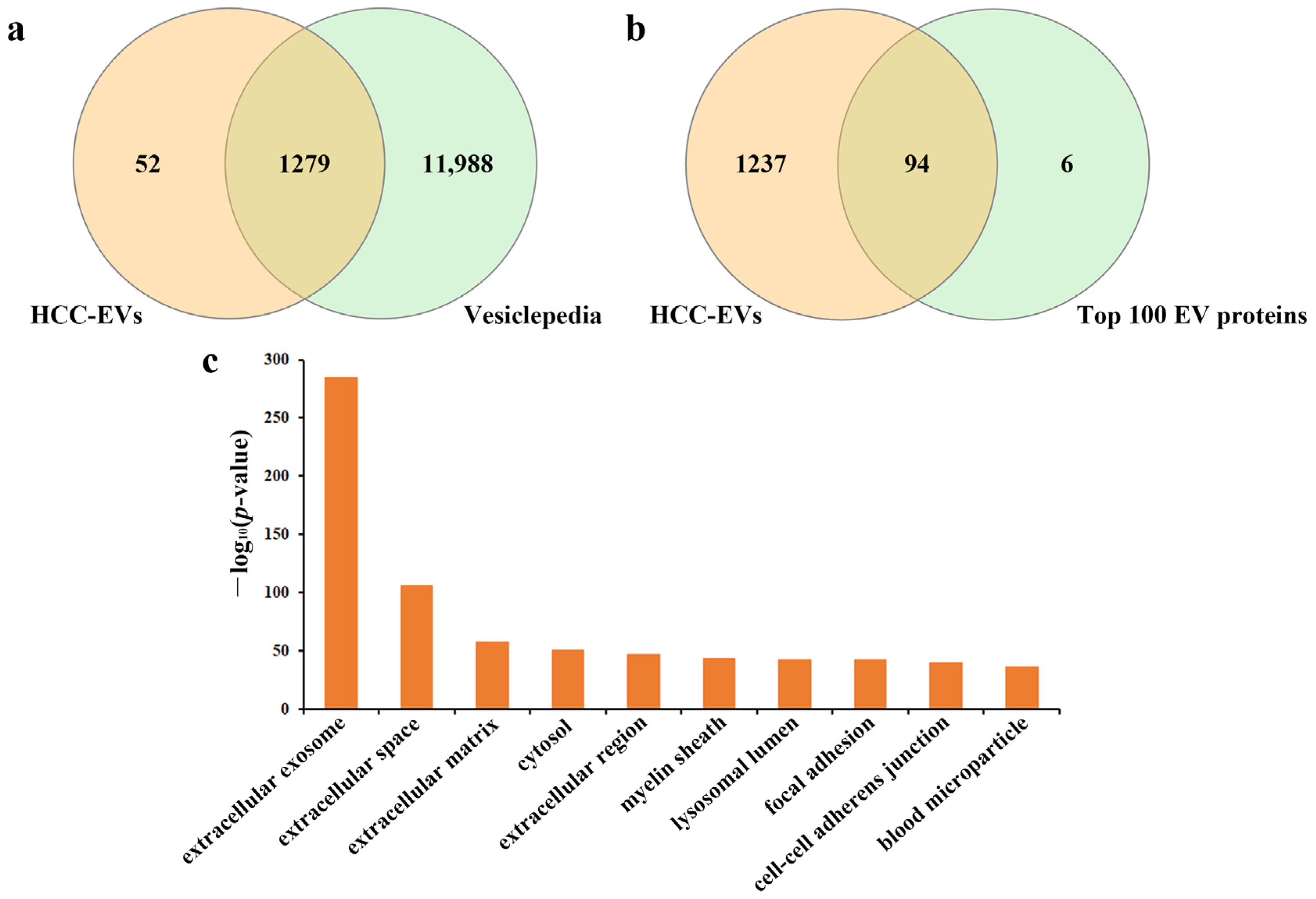
2.2. General Description of Extracellular Vesicles Isolated by EVTRAP
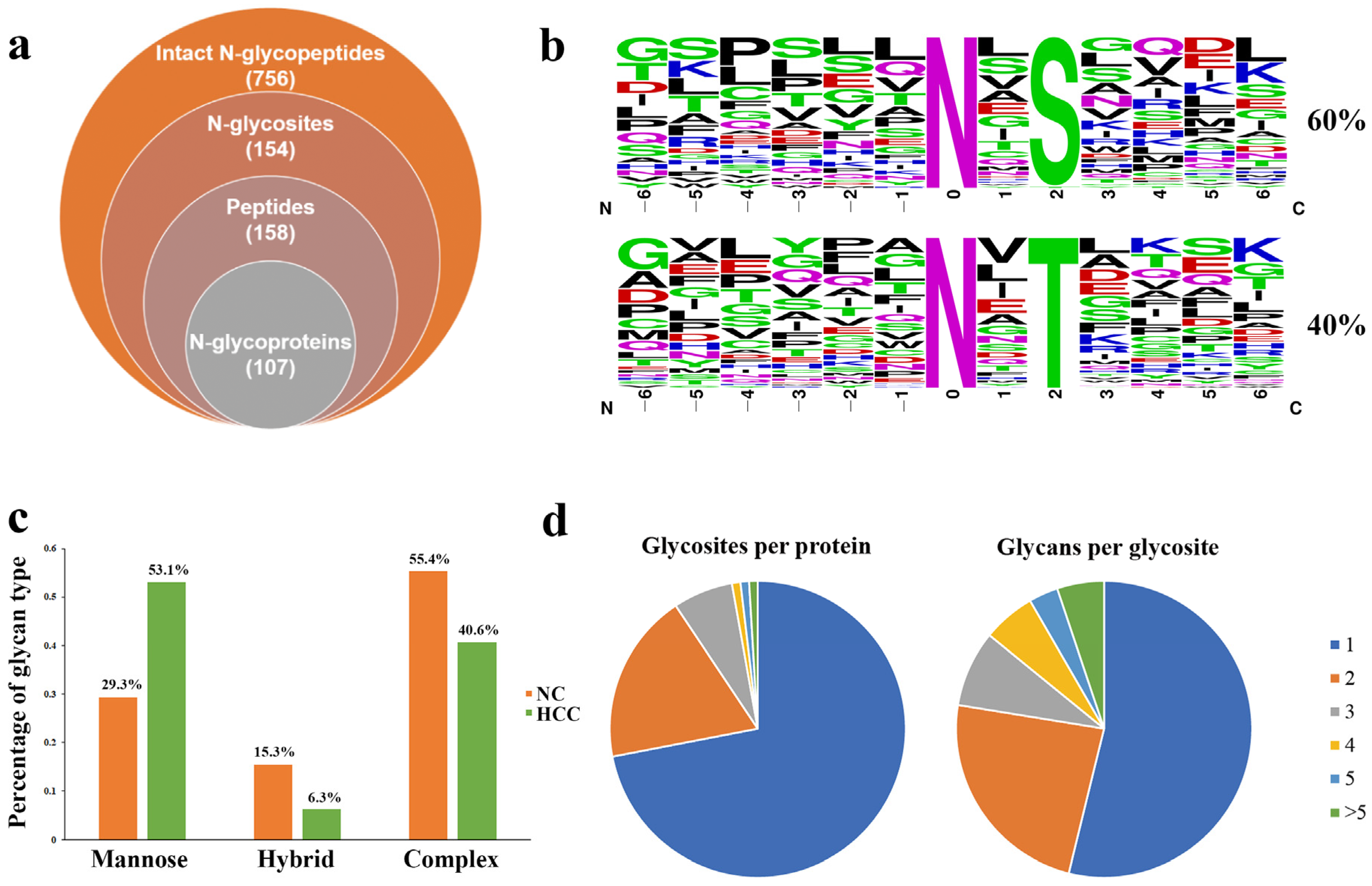
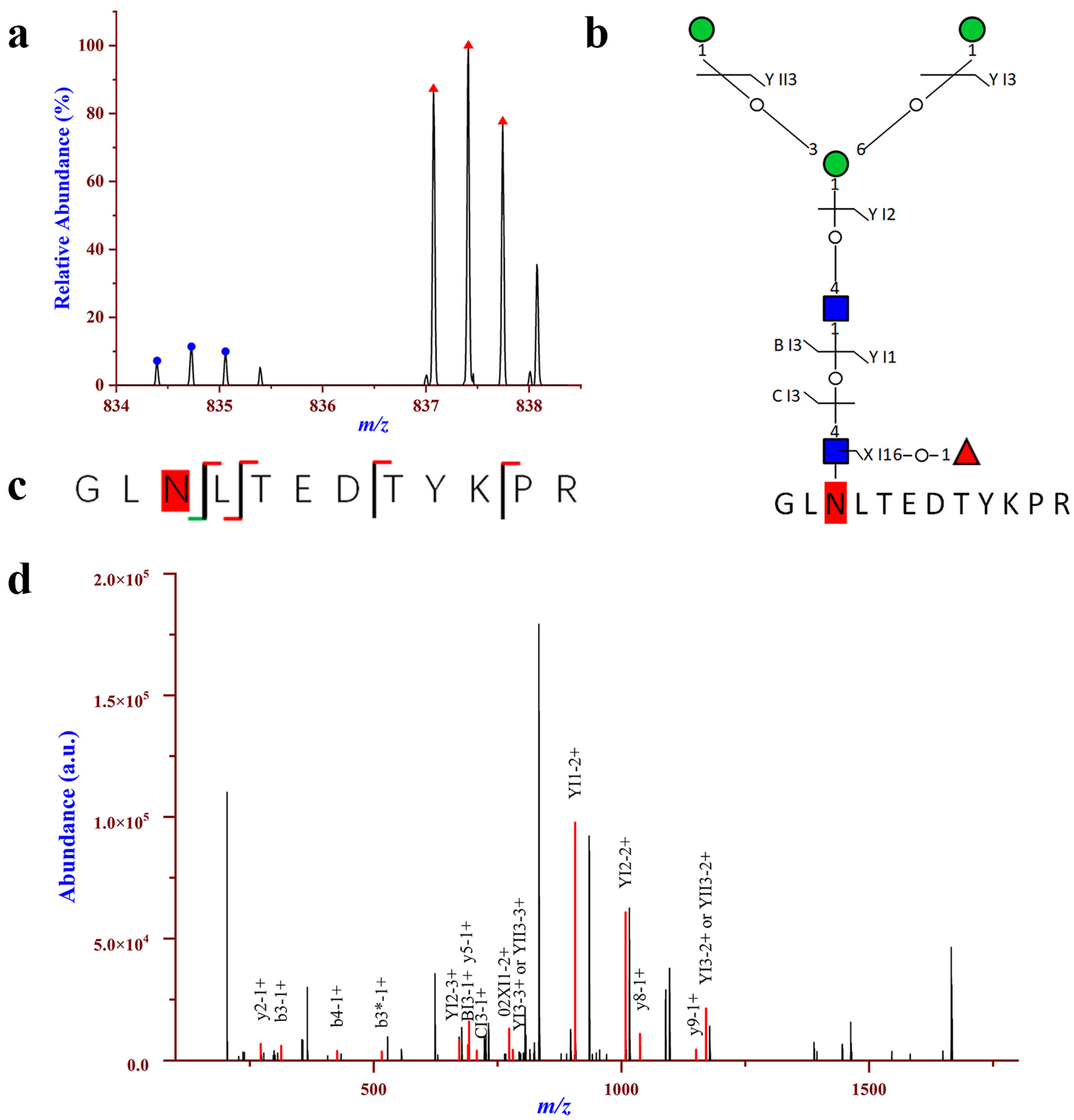
2.3. Comprehensive Analysis of Intact N-glycopeptides in the Urinary EVs
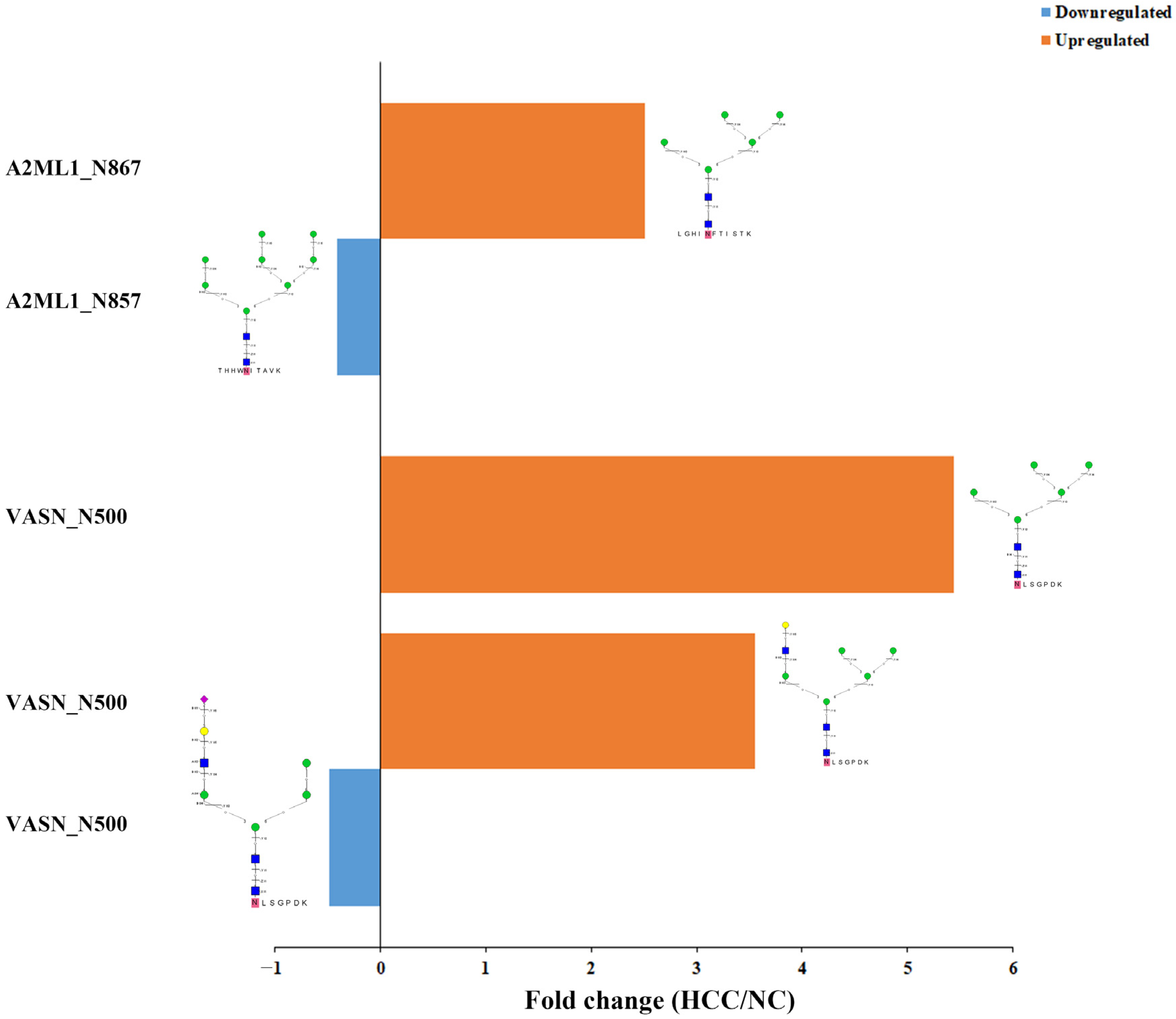
2.4. Relatively Quantification Analysis of the Site-Specific Glycans in the Urinary EVs
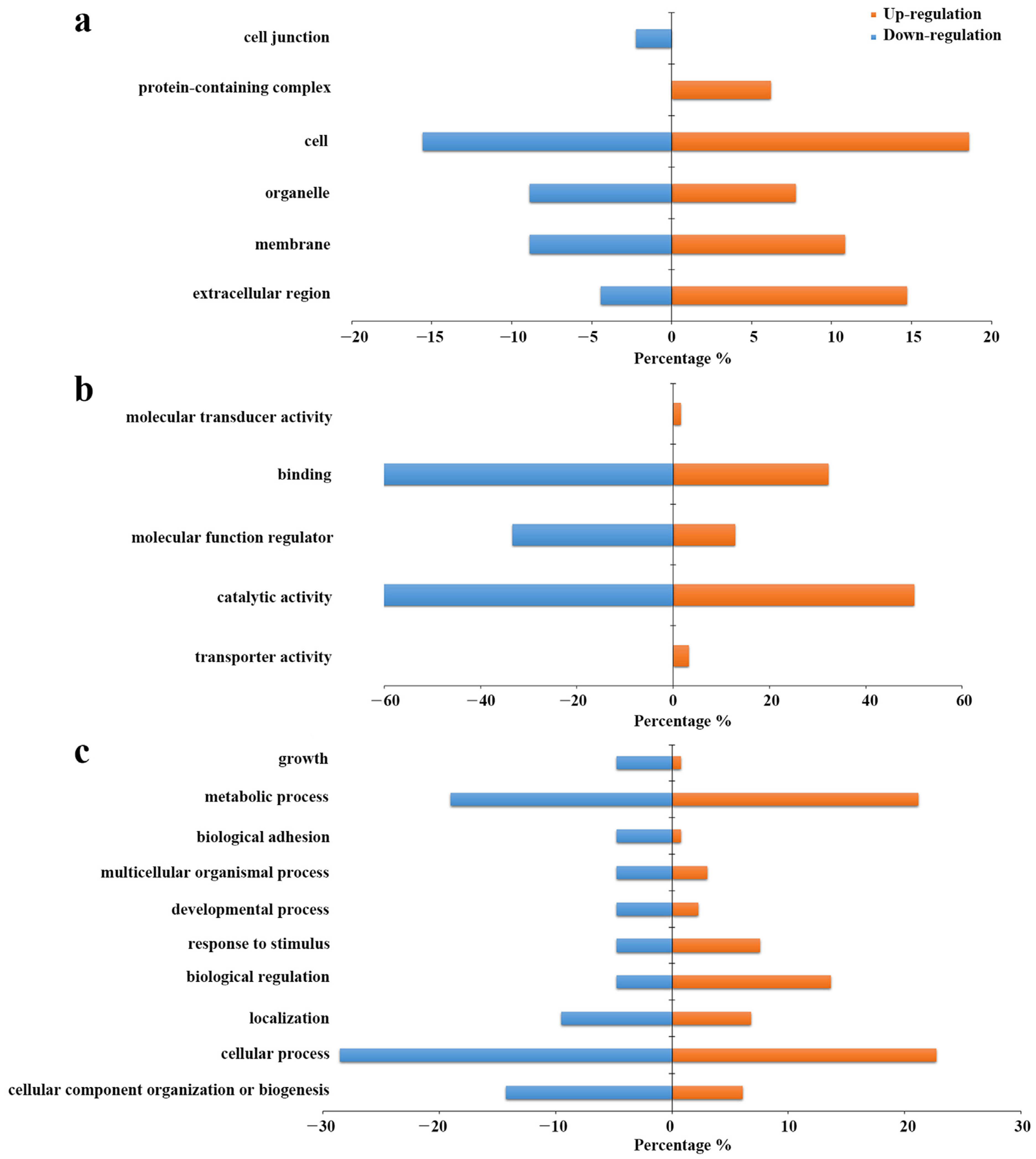
2.5. Gene Ontology Analysis of the Differentianl Expressed Intact N-glycopeptides
3. Materials and Methods
3.1. Patients and Sample Collection
3.2. Isolation of Extracellular Vesicles from Urine by EVTRAP
3.3. Enzyme Digestion and Protein Extraction of Urinary EVs
3.4. Stable Isotopic Dimethyl Labeling and Enrichment of N-glycopeptides from Urinary EVs
3.5. LC−MS/MS Analysis
3.6. Protein Identification
3.7. Intact N-glycopeptideIdentification and Quantification
3.8. Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 10573–10583. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Lok, A.S. Newer markers for hepatocellular carcinoma. Gastroenterology 2004, 127 (Suppl. 1), S113–S119. [Google Scholar] [CrossRef] [PubMed]
- Maluccio, M.; Covey, A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J. Clin. 2012, 62, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Babayan, A.; Pantel, K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Kogure, A.; Yamamoto, T.; Urabe, F.; Usuba, W.; Prieto-Vila, M.; Ochiya, T. Exploiting the message from cancer: The diagnostic value of extracellular vesicles for clinical applications. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Yuana, Y.; Sturk, A.; Nieuwland, R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013, 27, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2021, 7, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.F.; Wei, S.N.; Geng, N.; Qin, W.W.; He, X.; Wang, X.H.; Qi, Y.P.; Song, S.; Wang, P. Evaluation of circulating small extracellular vesicle-derived miRNAs as diagnostic biomarkers for differentiating between different pathological types of early lung cancer. Sci. Rep. 2022, 12, 17201. [Google Scholar] [CrossRef]
- Herreros-Villanueva, M.; Bujanda, L. Glypican-1 in exosomes as biomarker for early detection of pancreatic cancer. Ann. Transl. Med. 2016, 4, 64. [Google Scholar]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Lin, J.; Lin, W.; Bai, Y.; Liao, Y.; Lin, Q.; Chen, L.; Wu, Y. Identification of exosomal hsa-miR-483-5p as a potential biomarker for hepatocellular carcinoma via microRNA expression profiling of tumor-derived exosomes. Exp. Cell Res. 2022, 417, 113232. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.Y.; Bailey, U.M.; Jamaluddin, M.F.; Mahmud, S.H.; Raman, S.C.; Schulz, B.L. Sequence-based protein stabilization in the absence of glycosylation. Nat. Commun. 2014, 5, 3099. [Google Scholar] [CrossRef] [PubMed]
- Croci, D.O.; Cerliani, J.P.; Dalotto-Moreno, T.; Mendez-Huergo, S.P.; Mascanfroni, I.D.; Dergan-Dylon, S.; Toscano, M.A.; Caramelo, J.J.; Garcia-Vallejo, J.J.; Ouyang, J.; et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 2014, 156, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Kolarich, D.; Lepenies, B.; Seeberger, P.H. Glycomics, glycoproteomics and the immune system. Curr. Opin. Chem. Biol. 2012, 16, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Badr, H.A.; Alsadek, D.M.; Darwish, A.A.; Elsayed, A.I.; Bekmanov, B.O.; Khussainova, E.M.; Zhang, X.; Cho, W.C.; Djansugurova, L.B.; Li, C.Z. Lectin approaches for glycoproteomics in FDA-approved cancer biomarkers. Expert Rev. Proteom. 2014, 11, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lin, Z.; Wu, J.; Yin, H.; Dai, J.; Feng, Z.; Marrero, J.; Lubman, D.M. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J. Proteome Res. 2014, 13, 2986–2997. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, Y.; Kim, J.H.; Kim, H.S. Evaluation of revisited fucosylated alpha-fetoprotein (AFP-L3) with an autoanalyzer muTAS in a clinical laboratory. Clin. Chim. Acta 2012, 413, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Costa, J. Glycoconjugates from extracellular vesicles: Structures, functions and emerging potential as cancer biomarkers. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.M.; Ramos, C.C.; Freitas, D.; Reis, C.A. Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells 2021, 10, 109. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Sakaue, T.; Koga, H.; Iwamoto, H.; Nakamura, T.; Ikezono, Y.; Abe, M.; Wada, F.; Masuda, A.; Tanaka, T.; Fukahori, M.; et al. Glycosylation of ascites-derived exosomal CD133: A potential prognostic biomarker in patients with advanced pancreatic cancer. Med. Mol. Morphol. 2019, 52, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2015, 7, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.; Gomes-Alves, P.; Carvalho, S.B.; Peixoto, C.; Alves, P.M.; Altevogt, P.; Costa, J. Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures. Biomolecules 2015, 5, 1741–1761. [Google Scholar] [CrossRef] [PubMed]
- Surman, M.; Hoja-Lukowicz, D.; Szwed, S.; Kedracka-Krok, S.; Jankowska, U.; Kurtyka, M.; Drozdz, A.; Litynska, A.; Stepien, E.; Przybylo, M. An Insight into the Proteome of Uveal Melanoma-Derived Ectosomes Reveals the Presence of Potentially Useful Biomarkers. Int. J. Mol. Sci. 2019, 20, 3789. [Google Scholar] [CrossRef]
- Costa, J.; Gatermann, M.; Nimtz, M.; Kandzia, S.; Glatzel, M.; Conradt, H.S. N-Glycosylation of Extracellular Vesicles from HEK-293 and Glioma Cell Lines. Anal. Chem. 2018, 90, 7871–7879. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Wang, Z.; Li, F.; Zhang, Y.; Lu, H. Reverse capture for selectively and sensitively revealing the N-glycome of serum exosomes. Chem. Commun. 2019, 55, 14339–14342. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Wang, L.; Wang, S.; Li, W.; Liu, Y.; Zhang, J. Knockdown of ST6Gal-I expression in human hepatocellular carcinoma cells inhibits their exosome-mediated proliferation- and migration-promoting effects. IUBMB Life 2021, 73, 1378–1391. [Google Scholar] [CrossRef]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Tian, Z. GPSeeker Enables Quantitative Structural N-Glycoproteomics for Site- and Structure-Specific Characterization of Differentially Expressed N-Glycosylation in Hepatocellular Carcinoma. J. Proteome Res. 2019, 18, 2885–2895. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Iacobelli, S.; Arno, E.; D’Orazio, A.; Coletti, G. Detection of antigens recognized by a novel monoclonal antibody in tissue and serum from patients with breast cancer. Cancer Res. 1986, 46, 3005–3010. [Google Scholar] [PubMed]
- Linsley, P.S.; Horn, D.; Marquardt, H.; Brown, J.P.; Hellstrom, I.; Hellstrom, K.E.; Ochs, V.; Tolentino, E. Identification of a novel serum protein secreted by lung carcinoma cells. Biochemistry 1986, 25, 2978–2986. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.S.; Weng, D.S.; Wang, Q.J.; Pan, K.; Zhang, Y.J.; Li, Y.Q.; Li, J.J.; Zhao, J.J.; He, J.; Lv, L.; et al. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J. Transl. Med. 2014, 12, 273. [Google Scholar] [CrossRef] [PubMed]
- Correale, M.; Giannuzzi, V.; Iacovazzi, P.A.; Valenza, M.A.; Lanzillotta, S.; Abbate, I.; Quaranta, M.; Caruso, M.L.; Elba, S.; Manghisi, O.G. Serum 90K/MAC-2BP glycoprotein levels in hepatocellular carcinoma and cirrhosis. Anticancer Res. 1999, 19, 3469–3472. [Google Scholar] [PubMed]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Aguero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Zhao, L.; Shi, J.; Chang, L.; Wang, Y.; Liu, S.; Li, Y.; Zhang, T.; Zuo, T.; Fu, B.; Wang, G.; et al. Serum-Derived Exosomal Proteins as Potential Candidate Biomarkers for Hepatocellular Carcinoma. ACS Omega 2021, 6, 827–835. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Chen, X.; Yang, Z. Polymeric immunoglobulin receptor (PIGR) exerts oncogenic functions via activating ribosome pathway in hepatocellular carcinoma. Int. J. Med. Sci. 2021, 18, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Tang, Q.; Wu, Y.; Xu, Y.; Feng, T.; Zhou, R.; Chen, Y.; Gao, X.; Zhu, Q.; Yue, X.; et al. The role of polymeric immunoglobulin receptor in inflammation-induced tumor metastasis of human hepatocellular carcinoma. J. Natl. Cancer Inst. 2011, 103, 1696–1712. [Google Scholar] [CrossRef]
- Tey, S.K.; Wong, S.W.K.; Tung Chan, J.Y.; Mao, X.; Ng, T.H.; Yeung, C.L.S.; Leung, Z.; Fung, H.L.; Tang, A.H.N.; Wong, D.K.H.; et al. Patient pIgR-enriched extracellular vesicles drive cancer stemness, tumorigenesis and metastasis in hepatocellular carcinoma. J. Hepatol. 2022, 76, 883–895. [Google Scholar] [CrossRef]
- Sainz, I.M.; Pixley, R.A.; Colman, R.W. Fifty years of research on the plasma kallikrein-kinin system: From protein structure and function to cell biology and in-vivo pathophysiology. Thromb. Haemost. 2007, 98, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Abdel Wahab, A.H.A.; El-Halawany, M.S.; Emam, A.A.; Elfiky, A.; Abd Elmageed, Z.Y. Identification of circulating protein biomarkers in patients with hepatocellular carcinoma concomitantly infected with chronic hepatitis C virus. Biomarkers 2017, 22, 621–628. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Lin, S.; Chen, C.; Wang, C.; Ma, Q.; Jiang, B. Identification of kininogen-1 as a serum biomarker for the early detection of advanced colorectal adenoma and colorectal cancer. PLoS ONE 2013, 8, e70519. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sanda, M.; Comunale, M.A.; Herrera, H.; Swindell, C.; Kono, Y.; Singal, A.G.; Marrero, J.; Block, T.; Goldman, R.; et al. Changes in the Glycosylation of Kininogen and the Development of a Kininogen-Based Algorithm for the Early Detection of HCC. Cancer Epidemiol. Biomark. Prev. 2017, 26, 795–803. [Google Scholar] [CrossRef]
- Trigiante, G.; Lu, X. ASPP [corrected] and cancer. Nat. Rev. Cancer 2006, 6, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Sottocornola, R.; Royer, C.; Vives, V.; Tordella, L.; Zhong, S.; Wang, Y.; Ratnayaka, I.; Shipman, M.; Cheung, A.; Gaston-Massuet, C.; et al. ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev. Cell 2010, 19, 126–137. [Google Scholar] [CrossRef]
- Sullivan, A.; Lu, X. ASPP: A new family of oncogenes and tumour suppressor genes. Br. J. Cancer 2007, 96, 196–200. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, G.; Bu, F.; Lu, B.; Liang, A.; Cao, L.; Tong, X.; Lu, X.; Wu, M.; Guo, Y. Epigenetic silence of ankyrin-repeat-containing, SH3-domain-containing, and proline-rich-region- containing protein 1 (ASPP1) and ASPP2 genes promotes tumor growth in hepatitis B virus-positive hepatocellular carcinoma. Hepatology 2010, 51, 142–153. [Google Scholar] [CrossRef]
- Chen, R.; Wang, H.; Liang, B.; Liu, G.; Tang, M.; Jia, R.; Fan, X.; Jing, W.; Zhou, X.; Wang, H.; et al. Downregulation of ASPP2 improves hepatocellular carcinoma cells survival via promoting BECN1-dependent autophagy initiation. Cell Death Dis. 2016, 7, e2512. [Google Scholar] [CrossRef]
- Wang, Y.; Bu, F.; Royer, C.; Serres, S.; Larkin, J.R.; Soto, M.S.; Sibson, N.R.; Salter, V.; Fritzsche, F.; Turnquist, C.; et al. ASPP2 controls epithelial plasticity and inhibits metastasis through beta-catenin-dependent regulation of ZEB1. Nat. Cell Biol. 2014, 16, 1092–1104. [Google Scholar] [CrossRef]
- Li, S.; Li, H.; Yang, X.; Wang, W.; Huang, A.; Li, J.; Qin, X.; Li, F.; Lu, G.; Ding, H.; et al. Vasorin is a potential serum biomarker and drug target of hepatocarcinoma screened by subtractive-EMSA-SELEX to clinic patient serum. Oncotarget 2015, 6, 10045–10059. [Google Scholar] [CrossRef]
- Huang, A.; Dong, J.; Li, S.; Wang, C.; Ding, H.; Li, H.; Su, X.; Ge, X.; Sun, L.; Bai, C.; et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 2015, 11, 961–969. [Google Scholar] [CrossRef]
- Andaluz Aguilar, H.; Iliuk, A.B.; Chen, I.H.; Tao, W.A. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat. Protoc. 2020, 15, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, X.; Guo, Z.; Zhang, X.; Liang, X. Hydrophilic interaction chromatography based enrichment of glycopeptides by using click maltose: A matrix with high selectivity and glycosylation heterogeneity coverage. Chemistry 2009, 15, 12618–12626. [Google Scholar] [CrossRef]
- Xiao, K.; Wang, Y.; Shen, Y.; Han, Y.; Tian, Z. Large-scale identification and visualization of N-glycans with primary structures using GlySeeker. Rapid Commun. Mass Spectrom. 2018, 32, 142–148. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]

| Glycosite | Peptide Sequence | Composition | Glycan-Linkage | GF Score | Structural Diagnostic Ions |
|---|---|---|---|---|---|
| 81 | SYAGFLTVNK | N2H4F0S0 | 01Y41Y41M(31M21M)61M | 9 | BII2,BI2,YI1,YI2,BI3,YII3,YI3,YII4,MH,YI1 |
| N2H5F0S0 | 01Y41Y41M(31M)61M(31M)61M | 11 | 24AI4,ZI1,YI1,YI2,YI3,BI4, YI4,YII4,YIII3,MH,02XI1,ZI1,YI1 | ||
| N2H6F0S0 | 01Y41Y41M(31M21M)61M(31M)61M | 3 | BI2,YI3,YI3 | ||
| 307 | LLDGDLTSDPSYFQNVTGCSNYYNFLR | N2H5F0S0 | 01Y41Y41M(31M)61M(31M)61M | 3 | BI2,BI4,YI1 |
| 346 | QAIHVGNQTFNDGTIVEK | N2H3F0S0 | 01Y41Y41M(31M)61M | 5 | BI3,YI1,YI2,ZI1,YI1 |
| N2H4F0S0 | 01Y41Y41M(31M21M)61M | 12 | BII2,BI2,YI1,YI2,BI3,YII3,YI3,YII4,MH,02XI1,ZI1,YI1,YI2 | ||
| N2H4F1S0 | 01Y(61F)41Y41M(31M)61M61M | 4 | BI4,YI3,YI4,YII3,YI1 | ||
| N2H5F0S0 | 01Y41Y41M(31M)61M(31M)61M | 12 | ZI1,YI1,YI2,YI3,BI4,YI4,YII4,YIII3,02XI1,MH,ZI1,YI1,YI2,YI3 | ||
| N2H6F0S0 | 01Y41Y41M(31M)61M(31M21M)61M | 2 | BI2,YI3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Jia, S.; Wang, S.; Hu, L. Glycoproteomic Analysis of Urinary Extracellular Vesicles for Biomarkers of Hepatocellular Carcinoma. Molecules 2023, 28, 1293. https://doi.org/10.3390/molecules28031293
Li D, Jia S, Wang S, Hu L. Glycoproteomic Analysis of Urinary Extracellular Vesicles for Biomarkers of Hepatocellular Carcinoma. Molecules. 2023; 28(3):1293. https://doi.org/10.3390/molecules28031293
Chicago/Turabian StyleLi, Dejun, Shengnan Jia, Shuyue Wang, and Lianghai Hu. 2023. "Glycoproteomic Analysis of Urinary Extracellular Vesicles for Biomarkers of Hepatocellular Carcinoma" Molecules 28, no. 3: 1293. https://doi.org/10.3390/molecules28031293
APA StyleLi, D., Jia, S., Wang, S., & Hu, L. (2023). Glycoproteomic Analysis of Urinary Extracellular Vesicles for Biomarkers of Hepatocellular Carcinoma. Molecules, 28(3), 1293. https://doi.org/10.3390/molecules28031293




