Lectin-Based Affinity Enrichment and Characterization of N-Glycoproteins from Human Tear Film by Mass Spectrometry
Abstract
1. Introduction
2. Results
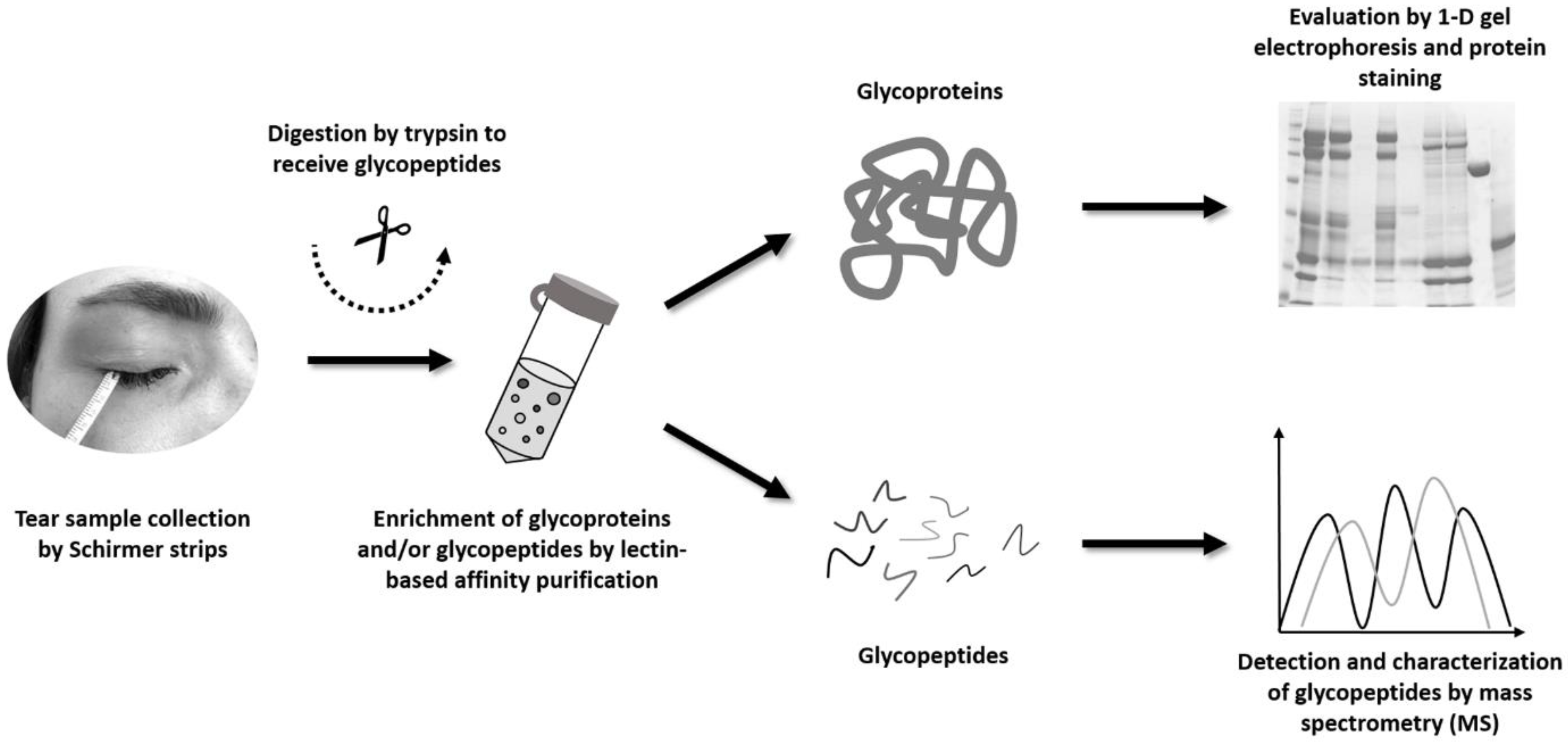
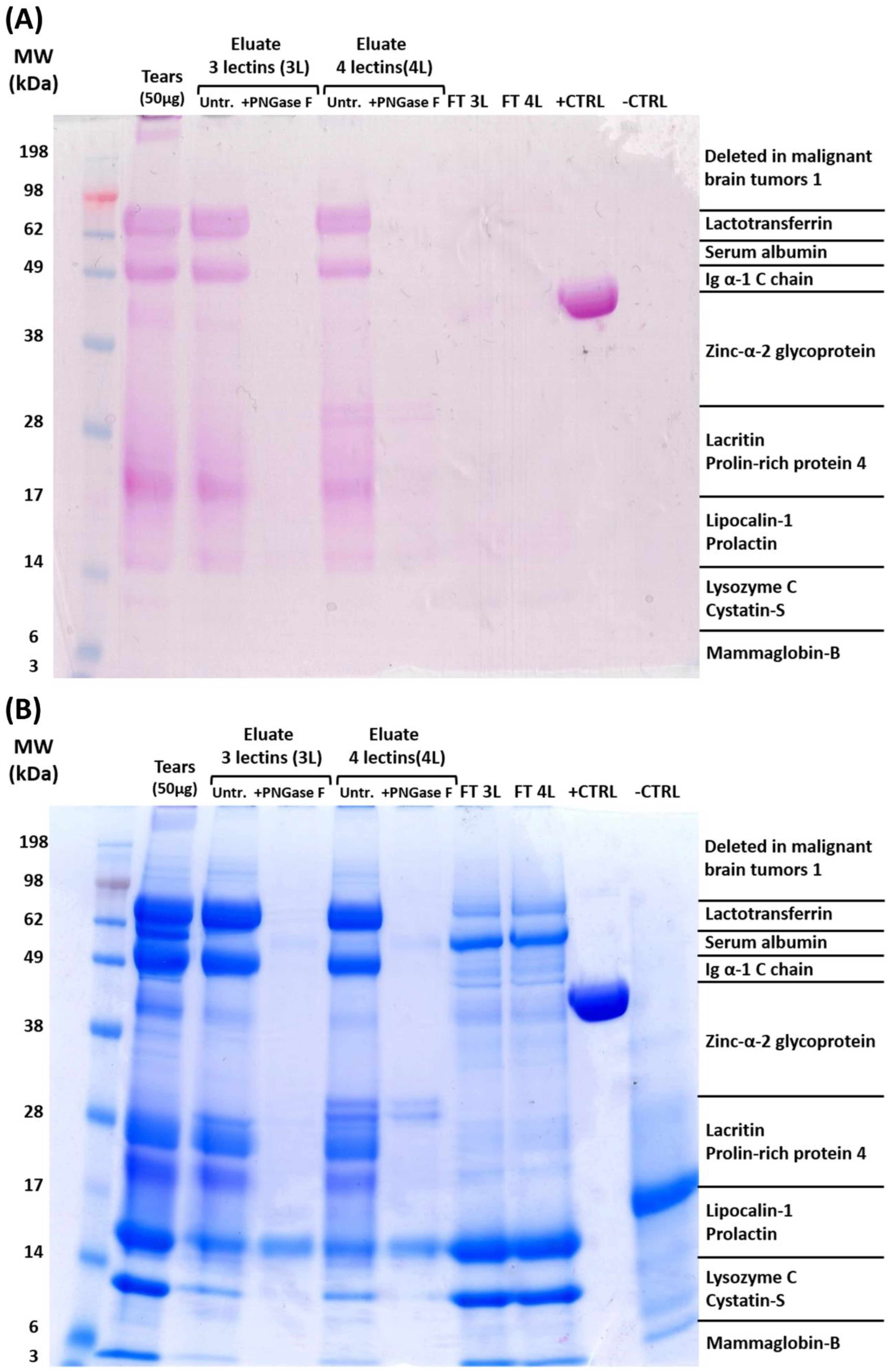
2.1. Enrichment of Tear Glycoproteins and Affinity Method Development
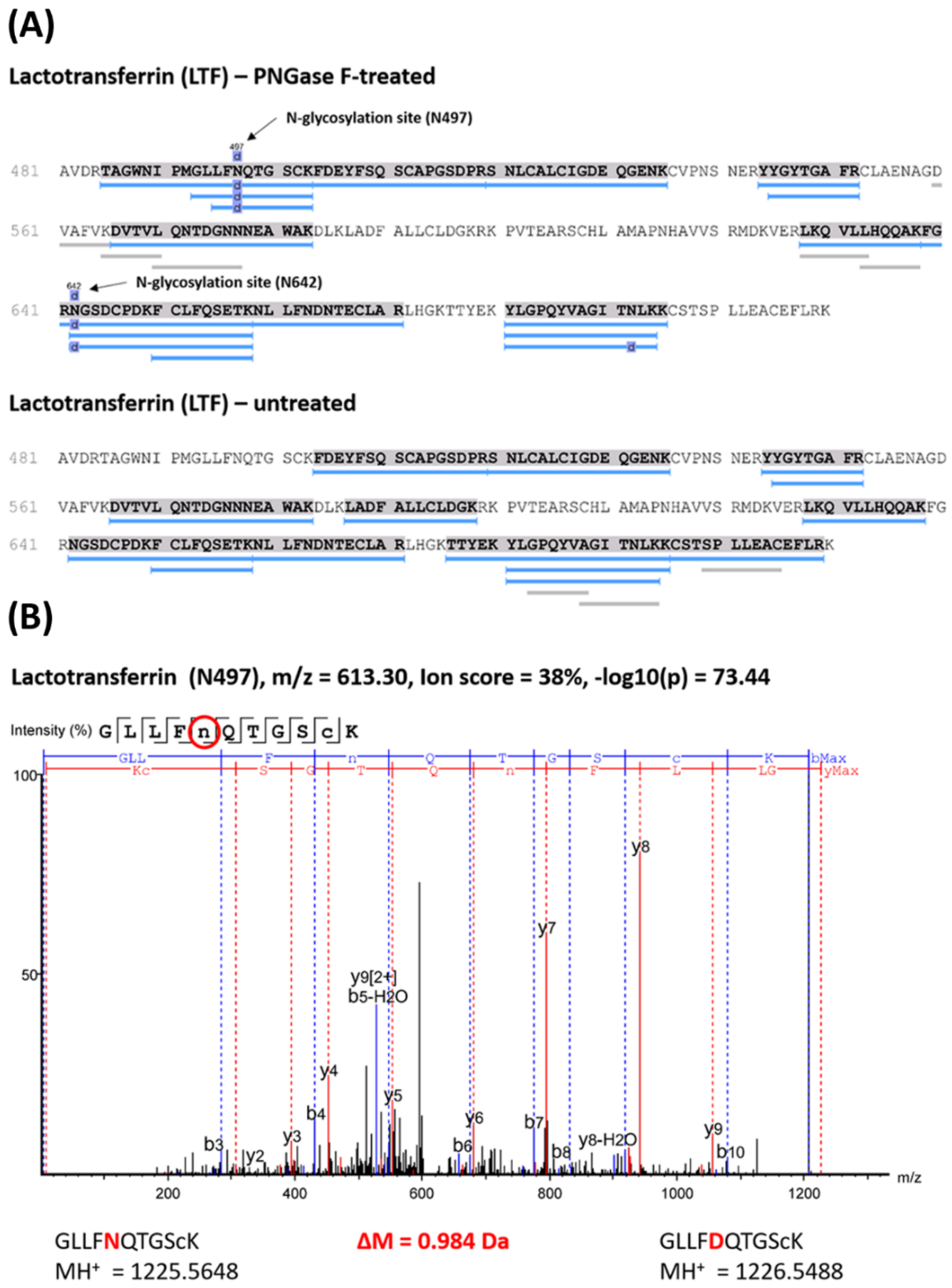
2.2. Enrichment of Tear Glycopeptides and MS Analysis
3. Discussion
4. Materials and Methods
4.1. Study Samples
4.2. Tear Sample Preparation and Pooling
4.3. Lectin-Based Affinity Enrichment
4.4. Sample Preparation for the Enrichment of Tear Glycoproteins
4.4.1. PNGase F Digest of Glycoproteins
4.4.2. 1D Gel Electrophoresis
4.4.3. Protein Stainings (Glycoproteins and Coomassie)
4.5. Sample Preparation for the Enrichment of Tear Glycopeptides
4.5.1. In-Solution Trypsin Digestion
4.5.2. Peptide Purification and Concentration Determination
4.5.3. PNGase F Digest of Glycopeptides
4.5.4. MS Analysis
4.5.5. Data Analysis and Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Masoudi, S. Biochemistry of human tear film: A review. Exp. Eye Res. 2022, 220, 109101. [Google Scholar] [CrossRef]
- Bron, A.J.; Tiffany, J.M.; Gouveia, S.M.; Yokoi, N.; Voon, L.W. Functional aspects of the tear film lipid layer. Exp. Eye Res. 2004, 78, 347–360. [Google Scholar] [CrossRef]
- Mcmonnies, C.W. Tear instability importance, mechanisms, validity and reliability of assessment. J. Optom. 2018, 11, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Posa, A.; Bräuer, L.; Schicht, M.; Garreis, F.; Beileke, S.; Paulsen, F. Schirmer strip vs. capillary tube method: Non-invasive methods of obtaining proteins from tear fluid. Ann. Anat.-Anat. Anz. 2013, 195, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Perumal, N.; Funke, S.; Pfeiffer, N.; Grus, F.H. Proteomics analysis of human tears from aqueous-deficient and evaporative dry eye patients. Sci. Rep. 2016, 6, 29629. [Google Scholar] [CrossRef]
- Zhou, L.; Beuerman, R.W.; Chan, C.M.; Zhao, S.Z.; Li, X.R.; Yang, H.; Tong, L.; Liu, S.; Stern, M.E.; Tan, D. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J. Proteome Res. 2009, 8, 4889–4905. [Google Scholar] [CrossRef]
- Fomo, K.N.; Schmelter, C.; Pfeiffer, N.; Grus, F.H. Tear Film-specific Biomarkers in Glaucoma Patients. Klin. Monbl. Augenheilkd. 2022, 239, 165–168. [Google Scholar] [PubMed]
- Gijs, M.; Ramakers, I.H.; Visser, P.J.; Verhey, F.R.J.; van de Waarenburg, M.P.H.; Schalkwijk, C.G.; Nuijts, R.M.; Webers, C.A.B. Association of tear fluid amyloid and tau levels with disease severity and neurodegeneration. Sci. Rep. 2021, 11, 22675. [Google Scholar] [CrossRef] [PubMed]
- Boerger, M.; Funke, S.; Leha, A.; Roser, A.-E.; Wuestemann, A.-K.; Maass, F.; Bähr, M.; Grus, F.; Lingor, P. Proteomic analysis of tear fluid reveals disease-specific patterns in patients with Parkinson’s disease—A pilot study. Park. Relat. Disord. 2019, 63, 3–9. [Google Scholar] [CrossRef]
- Hümmert, M.W.; Wurster, U.; Bönig, L.; Schwenkenbecher, P.; Sühs, K.-W.; Alvermann, S.; Gingele, S.; Skripuletz, T.; Stangel, M. Investigation of oligoclonal IgG bands in tear fluid of multiple sclerosis patients. Front. Immunol. 2019, 10, 1110. [Google Scholar] [CrossRef]
- Lebrecht, A.; Boehm, D.; Schmidt, M.; Koelbl, H.; Schwirz, R.L.; Grus, F.H. Diagnosis of breast cancer by tear proteomic pattern. Cancer Genom. Proteom. 2009, 6, 177–182. [Google Scholar]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43R–56R. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Rodriguez Benavente, M.C.; Argüeso, P. Glycosylation pathways at the ocular surface. Biochem. Soc. Trans. 2018, 46, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Khuong, T.; Everest-Dass, A.V.; Kautto, L.; Zhao, Z.; Willcox, M.D.P.; Packer, N.H. Glycomic characterization of basal tears and changes with diabetes and diabetic retinopathy. Glycobiology 2015, 25, 269–283. [Google Scholar] [CrossRef]
- Messina, A.; Palmigiano, A.; Tosto, C.; Romeo, D.A.; Sturiale, L.; Garozzo, D.; Leonardi, A. Tear N-glycomics in vernal and atopic keratoconjunctivitis. Allergy 2021, 76, 2500–2509. [Google Scholar] [CrossRef] [PubMed]
- An, H.J.; Froehlich, J.W.; Lebrilla, C.B. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 2009, 13, 421–426. [Google Scholar] [CrossRef]
- Mann, A.C.; Self, C.H.; Turner, G.A. A general method for the complete deglycosylation of a wide variety of serum glycoproteins using peptide-N-glycosidase-F. Glycosylation Dis. 1994, 1, 253–261. [Google Scholar] [CrossRef]
- Kronewitter, S.R.; de Leoz, M.L.A.; Peacock, K.S.; McBride, K.R.; An, H.J.; Miyamoto, S.; Leiserowitz, G.S.; Lebrilla, C.B. Human serum processing and analysis methods for rapid and reproducible N-glycan mass profiling. J. Proteome Res. 2010, 9, 4952–4959. [Google Scholar] [CrossRef]
- Wilkinson, H.; Saldova, R. Current methods for the characterization of O-glycans. J. Proteome Res. 2020, 19, 3890–3905. [Google Scholar] [CrossRef]
- Zauner, G.; Kozak, R.P.; Gardner, R.A.; Fernandes, D.L.; Deelder, A.M.; Wuhrer, M. Protein O-glycosylation analysis. Biol. Chem. 2012, 393, 687–708. [Google Scholar] [CrossRef]
- Perumal, N.; Funke, S.; Wolters, D.; Pfeiffer, N.; Grus, F.H. Characterization of human reflex tear proteome reveals high expression of lacrimal proline-rich protein 4 (PRR4). Proteomics 2015, 15, 3370–3381. [Google Scholar] [CrossRef] [PubMed]
- Perumal, N.; Funke, S.; Pfeiffer, N.; Grus, F.H. Characterization of lacrimal proline-rich protein 4 (PRR4) in human tear proteome. Proteomics 2014, 14, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Picariello, G.; Ferranti, P.; Mamone, G.; Roepstorff, P.; Addeo, F. Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics 2008, 8, 3833–3847. [Google Scholar] [CrossRef]
- Zhou, L.; Beuerman, R.W.; Chew, A.P.; Koh, S.K.; Cafaro, T.A.; Urrets-Zavalia, E.A.; Urrets-Zavalia, J.A.; Li, S.F.Y.; Serra, H.M. Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J. Proteome Res. 2009, 8, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009, 8, 651–661. [Google Scholar] [CrossRef]
- Haridas, M.; Anderson, B.F.; Baker, E.N. Structure of human diferric lactoferrin refined at 2.2 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1995, 51, 629–646. [Google Scholar] [CrossRef]
- Ramachandran, P.; Boontheung, P.; Xie, Y.; Sondej, M.; Wong, D.T.; Loo, J.A. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006, 5, 1493–1503. [Google Scholar] [CrossRef]
- Liu, T.; Qian, W.-J.; Gritsenko, M.A.; Camp, D.G.; Monroe, M.E.; Moore, R.J.; Smith, R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005, 4, 2070–2080. [Google Scholar] [CrossRef]
- Jia, W.; Lu, Z.; Fu, Y.; Wang, H.-P.; Wang, L.-H.; Chi, H.; Yuan, Z.-F.; Zheng, Z.-B.; Song, L.-N.; Han, H.-H. A Strategy for Precise and Large Scale Identification of Core Fucosylated Glycoproteins* S. Mol. Cell. Proteom. 2009, 8, 913–923. [Google Scholar] [CrossRef]
- Nilsson, J.; Rüetschi, U.; Halim, A.; Hesse, C.; Carlsohn, E.; Brinkmalm, G.; Larson, G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 2009, 6, 809–811. [Google Scholar] [CrossRef]
- Halim, A.; Nilsson, J.; Rüetschi, U.; Hesse, C.; Larson, G. Human urinary glycoproteomics; attachment site specific analysis of N-and O-linked glycosylations by CID and ECD. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef]
- Hassan, M.I.; Bilgrami, S.; Kumar, V.; Singh, N.; Yadav, S.; Kaur, P.; Singh, T.P. Crystal structure of the novel complex formed between zinc α2-glycoprotein (ZAG) and prolactin-inducible protein (PIP) from human seminal plasma. J. Mol. Biol. 2008, 384, 663–672. [Google Scholar] [CrossRef]
- Wiegandt, A.; Behnken, H.N.; Meyer, B. Unusual N-type glycosylation of salivary prolactin-inducible protein (PIP): Multiple LewisY epitopes generate highly-fucosylated glycan structures. Glycoconj. J. 2018, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Kaji, H.; Saito, H.; Yamauchi, Y.; Shinkawa, T.; Taoka, M.; Hirabayashi, J.; Kasai, K.; Takahashi, N.; Isobe, T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003, 21, 667–672. [Google Scholar] [CrossRef]
- Yang, Z.; Hancock, W.S. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J. Chromatogr. A 2004, 1053, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Totten, S.M.; Adusumilli, R.; Kullolli, M.; Tanimoto, C.; Brooks, J.D.; Mallick, P.; Pitteri, S.J. Multi-lectin affinity chromatography and quantitative proteomic analysis reveal differential glycoform levels between prostate cancer and benign prostatic hyperplasia sera. Sci. Rep. 2018, 8, 6509. [Google Scholar] [CrossRef]
- Kullolli, M.; Hancock, W.S.; Hincapie, M. Preparation of a high-performance multi-lectin affinity chromatography (HP-M-LAC) adsorbent for the analysis of human plasma glycoproteins. J. Sep. Sci. 2008, 31, 2733–2739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, S.; Hancock, W.S. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap—Fourier transform mass spectrometry. Glycobiology 2006, 16, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Cho, W.; Regnier, F.E. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J. Proteome Res. 2009, 8, 643–650. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Owen, J.B. Lectin-affinity chromatography brain glycoproteomics and Alzheimer disease: Insights into protein alterations consistent with the pathology and progression of this dementing disorder. Proteom. Clin. Appl. 2011, 5, 50–56. [Google Scholar] [CrossRef]
- Lee, L.Y.; Hincapie, M.; Packer, N.; Baker, M.S.; Hancock, W.S.; Fanayan, S. An optimized approach for enrichment of glycoproteins from cell culture lysates using native multi-lectin affinity chromatography. J. Sep. Sci. 2012, 35, 2445–2452. [Google Scholar] [CrossRef] [PubMed]
- Gasymov, O.K.; Abduragimov, A.R.; Yusifov, T.N.; Glasgow, B.J. Interaction of tear lipocalin with lysozyme and lactoferrin. Biochem. Biophys. Res. Commun. 1999, 265, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Sondej, M.; Denny, P.A.; Xie, Y.; Ramachandran, P.; Si, Y.; Takashima, J.; Shi, W.; Wong, D.T.; Loo, J.A.; Denny, P.C. Glycoprofiling of the human salivary proteome. Clin. Proteom. 2009, 5, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Nättinen, J.; Aapola, U.; Jylhä, A.; Vaajanen, A.; Uusitalo, H. Comparison of capillary and Schirmer strip tear fluid sampling methods using SWATH-MS proteomics approach. Trans. Vis. Sci. Technol. 2020, 9, 16. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Hu, Y.; Ao, M.; Shah, P.; Chen, J.; Yang, W.; Jia, X.; Tian, Y.; Thomas, S.; Zhang, H. N-GlycositeAtlas: A database resource for mass spectrometry-based human N-linked glycoprotein and glycosylation site mapping. Clin. Proteom. 2019, 16, 35. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, G.; Melo-Braga, M.N.; Engholm-Keller, K.; Parker, B.L.; Larsen, M.R. Chemical deamidation: A common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 2012, 11, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Fullard, R.J.; Tucker, D. Tear protein composition and the effects of stimulus. In Lacrimal Gland, Tear Film, and Dry Eye Syndromes; Springer: Boston, MA, USA, 1994; pp. 309–314. [Google Scholar]
- González-Chávez, S.A.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin: Structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, 301.e1–301.e8. [Google Scholar] [CrossRef]
- Karav, S.; German, J.B.; Rouquié, C.; Le Parc, A.; Barile, D. Studying lactoferrin N-glycosylation. Int. J. Mol. Sci. 2017, 18, 870. [Google Scholar] [CrossRef] [PubMed]
- Kautto, L.; Nguyen-Khuong, T.; Everest-Dass, A.; Leong, A.; Zhao, Z.; Willcox, M.D.P.; Packer, N.H.; Peterson, R. Glycan involvement in the adhesion of Pseudomonas aeruginosa to tears. Exp. Eye Res. 2016, 145, 278–288. [Google Scholar] [CrossRef]
- Srinivasan, S.; Thangavelu, M.; Zhang, L.; Green, K.B.; Nichols, K.K. iTRAQ quantitative proteomics in the analysis of tears in dry eye patients. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5052–5059. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Ishida, R.; Kojima, T.; Goto, E.; Matsumoto, Y.; Watanabe, K.; Ishida, N.; Nakata, K.; Takeuchi, T.; Tsubota, K. Abnormal protein profiles in tears with dry eye syndrome. Am. J. Ophthalmol. 2003, 136, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Vagge, A.; Senni, C.; Bernabei, F.; Pellegrini, M.; Scorcia, V.; Traverso, C.E.; Giannaccare, G. Therapeutic effects of lactoferrin in ocular diseases: From dry eye disease to infections. Int. J. Mol. Sci. 2020, 21, 6668. [Google Scholar] [CrossRef]
- Kawashima, M.; Kawakita, T.; Inaba, T.; Okada, N.; Ito, M.; Shimmura, S.; Watanabe, M.; Shinmura, K.; Tsubota, K. Dietary lactoferrin alleviates age-related lacrimal gland dysfunction in mice. PLoS One 2012, 7, e33148. [Google Scholar] [CrossRef] [PubMed]
- McKown, R.L.; Wang, N.; Raab, R.W.; Karnati, R.; Zhang, Y.; Williams, P.B.; Laurie, G.W. Lacritin and other new proteins of the lacrimal functional unit. Exp. Eye Res. 2009, 88, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Karnati, R.; Laurie, D.E.; Laurie, G.W. Lacritin and the tear proteome as natural replacement therapy for dry eye. Exp. Eye Res. 2013, 117, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Vijmasi, T.; Chen, F.Y.T.; Balasubbu, S.; Gallup, M.; McKown, R.L.; Laurie, G.W.; McNamara, N.A. Topical administration of lacritin is a novel therapy for aqueous-deficient dry eye disease. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5401–5409. [Google Scholar] [CrossRef]
- Samudre, S.; Lattanzio, F.A.; Lossen, V.; Hosseini, A.; Sheppard, J.D.; McKown, R.L.; Laurie, G.W.; Williams, P.B. Lacritin, a novel human tear glycoprotein, promotes sustained basal tearing and is well tolerated. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6265–6270. [Google Scholar] [CrossRef]
- Asrani, A.C.; Odrich, M.G.; Carpenter, M.A.; Logan, M.B.; Laurie, G.W. Topical lacritin C-terminal peptide ’LacripepTM’ significantly reduces both inferior corneal staining and burning/stinging in primary Sjögren’s Syndrome dry eye. Investig. Ophthalmol. Vis. Sci. 2021, 62, 1266. [Google Scholar]
- Chen, F.Y.T.; Stephens, D.; Ge, S.; Vijmasi, T.; Laurie, G.W.; Knox, S.; McNamara, N.A. Lacritin’s active C-terminal peptide, ‘Lacripep’, as an efficient and innovative therapeutic for the treatment of aqueous-deficient dry eye. Investig. Ophthalmol. Vis. Sci. 2015, 56, 300. [Google Scholar]
- Georgiev, G.A.; Gh, M.S.; Romano, J.; Teixeira, K.L.D.; Struble, C.; Ryan, D.S.; Sia, R.K.; Kitt, J.P.; Harris, J.M.; Hsu, K.-L. Lacritin proteoforms prevent tear film collapse and maintain epithelial homeostasis. J. Biol. Chem. 2021, 296. [Google Scholar] [CrossRef]
- McKown, R.L.; Frazier, E.V.C.; Zadrozny, K.K.; Deleault, A.M.; Raab, R.W.; Ryan, D.S.; Sia, R.K.; Lee, J.K.; Laurie, G.W. A cleavage-potentiated fragment of tear lacritin is bactericidal. J. Biol. Chem. 2014, 289, 22172–22182. [Google Scholar] [CrossRef]
- Xu, S.; Ma, L.; Evans, E.; Okamoto, C.T.; Hamm-Alvarez, S.F. Polymeric immunoglobulin receptor traffics through two distinct apically targeted pathways in primary lacrimal gland acinar cells. J. Cell. Sci. 2013, 126, 2704–2717. [Google Scholar] [CrossRef]
- Willcox, M.D.P.; Lan, J. Secretory immunoglobulin A in tears: Functions and changes during contact lens wear. Clin. Exp. Optom. 1999, 82, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Qu, Y.; Zhang, Z.; Wang, Z.; Prytkova, I.; Wu, S. Intact glycopeptide characterization using mass spectrometry. Expert Rev. Proteom. 2016, 13, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H.; Grus, F. Comparison of Two Solid-Phase Extraction (SPE) Methods for the Identification and Quantification of Porcine Retinal Protein Markers by LC-MS/MS. Int. J. Mol. Sci. 2018, 19, 3847. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, C.; Fomo, K.N.; Perumal, N.; Pfeiffer, N.; Grus, F.H. Regulation of the HTRA2 Protease Activity by an Inhibitory Antibody-Derived Peptide Ligand and the Influence on HTRA2-Specific Protein Interaction Networks in Retinal Tissues. Biomedicines 2021, 9, 1013. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]

| Protein ID | Protein Name | Gene Name | N-glycosylation Sites | Present in R1 | Present in R2 | Present in R3 | CTRL Group | Reported in Literature | Peptide Score |
|---|---|---|---|---|---|---|---|---|---|
| P02788 | Lactotransferrin | LTF | N497 | √ | √ | √ | - | [24,25,26,27] | 103 |
| N642 | √ | √ | √ | - | 87 | ||||
| P01833 | Polymeric immunoglobulin receptor | PIGR | N83 | √ | √ | √ | - | [24,25,28,29] | 106 |
| N90 | √ | √ | √ | - | 105 | ||||
| N135 | √ | √ | √ | - | 73 | ||||
| N186 | √ | √ | √ | - | 96 | ||||
| N421 | √ | √ | - | - | 112 | ||||
| N469 | √ | √ | √ | - | 108 | ||||
| N499 | √ | √ | - | - | 108 | ||||
| P01877 | Immunoglobulin heavy constant alpha 2 | IGHA2 | N47 | √ | √ | √ | - | [24,26,30] | 54 |
| N92 | - | √ | - | - | 102 | ||||
| N131 | √ | √ | √ | - | 108 | ||||
| N205 | √ | √ | √ | - | 81 | ||||
| P01876 | Immunoglobulin heavy constant alpha 1 | IGHA1 | N144 | √ | √ | √ | - | [25,29,31,32] | 108 |
| N340 | √ | √ | √ | - | 101 | ||||
| P25311 | Zinc-α2-glycoprotein | AZGP1 | N109 | √ | √ | √ | - | [24,25,26] | 120 |
| N112 | √ | √ | - | - | 77 | ||||
| N128 | √ | √ | √ | - | 70 | ||||
| Q9GZZ8 | Extracellular glycoprotein lacritin | LACRT | N119 | √ | √ | √ | √ | [25,28] | 88 |
| P01591 | Immunoglobulin J chain | JCHAIN | N67 | √ | √ | √ | - | [24,25,28,29] | 93 |
| P12273 | Prolactin-inducible protein | PIP | N105 | √ | √ | √ | - | [25,28,33,34] | 92 |
| P02787 | Serotransferrin | TF | N630 | n.d. | √ | √ | - | [25,26,28,29] | 90 |
| P00738 | Haptoglobin | HP | N207 | n.d. | √ | n.d. | - | [25,28,29] | 85 |
| N211 | n.d. | √ | n.d. | - | 85 | ||||
| N241 | n.d. | √ | n.d. | - | 103 | ||||
| P01009 | Alpha-1-Antitrypsin | SERPINA1 | N271 | n.d. | √ | n.d. | - | [25,26,29,30] | 94 |
| Lectin | Glycan Specificity |
|---|---|
| Concanavalin (ConA) | α-linked mannose |
| Wheat germ agglutinin (WGA) | N-acetylglucosamine |
| Jacalin (JAC) | galactosyl (ß-1,3) N-acetylgalactosamine |
| Ulex europaeus agglutinin I (UEA I) | α-linked fucose |
| No. | Fragmentation Method | Dynamic Exclusion Enabled | Repeat Count | Repeat Duration (s) | Exclusion Size List | Exclusion Duration (s) | Automatic Gain Control (AGC) |
|---|---|---|---|---|---|---|---|
| 1 | CID | yes | 1 | 30 | 100 | 180 | 1 × 106 |
| 2 | CID | yes | 1 | 30 | 100 | 180 | 5 × 105 |
| 3 | CID | yes | 1 | 30 | 50 | 180 | 1 × 106 |
| 4 | CID | yes | 1 | 30 | 100 | 180 | 1 × 106 |
| 5 | CID | yes | 1 | 30 | 100 | 300 | 1 × 106 |
| 6 | CID | yes | 1 | 30 | 100 | 90 | 1 × 106 |
| 7 | HCD | no | - | - | - | - | 1 × 106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmelter, C.; Brueck, A.; Perumal, N.; Qu, S.; Pfeiffer, N.; Grus, F.H. Lectin-Based Affinity Enrichment and Characterization of N-Glycoproteins from Human Tear Film by Mass Spectrometry. Molecules 2023, 28, 648. https://doi.org/10.3390/molecules28020648
Schmelter C, Brueck A, Perumal N, Qu S, Pfeiffer N, Grus FH. Lectin-Based Affinity Enrichment and Characterization of N-Glycoproteins from Human Tear Film by Mass Spectrometry. Molecules. 2023; 28(2):648. https://doi.org/10.3390/molecules28020648
Chicago/Turabian StyleSchmelter, Carsten, Alina Brueck, Natarajan Perumal, Sichang Qu, Norbert Pfeiffer, and Franz H. Grus. 2023. "Lectin-Based Affinity Enrichment and Characterization of N-Glycoproteins from Human Tear Film by Mass Spectrometry" Molecules 28, no. 2: 648. https://doi.org/10.3390/molecules28020648
APA StyleSchmelter, C., Brueck, A., Perumal, N., Qu, S., Pfeiffer, N., & Grus, F. H. (2023). Lectin-Based Affinity Enrichment and Characterization of N-Glycoproteins from Human Tear Film by Mass Spectrometry. Molecules, 28(2), 648. https://doi.org/10.3390/molecules28020648





