Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro
Abstract
1. Introduction
2. Results
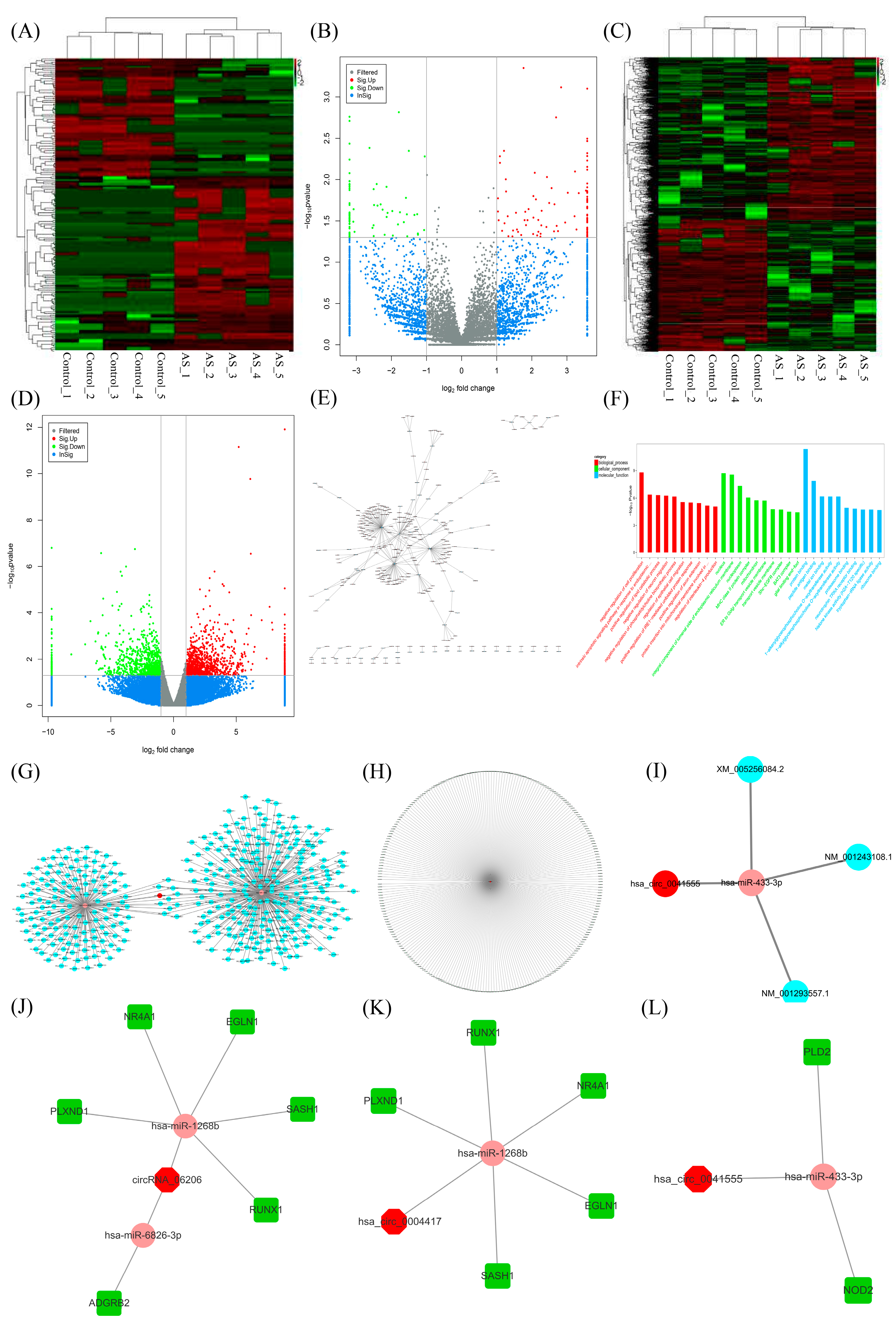
2.1. CircRNA Screening and circRNA-miRNA-mRNA Network Establishment
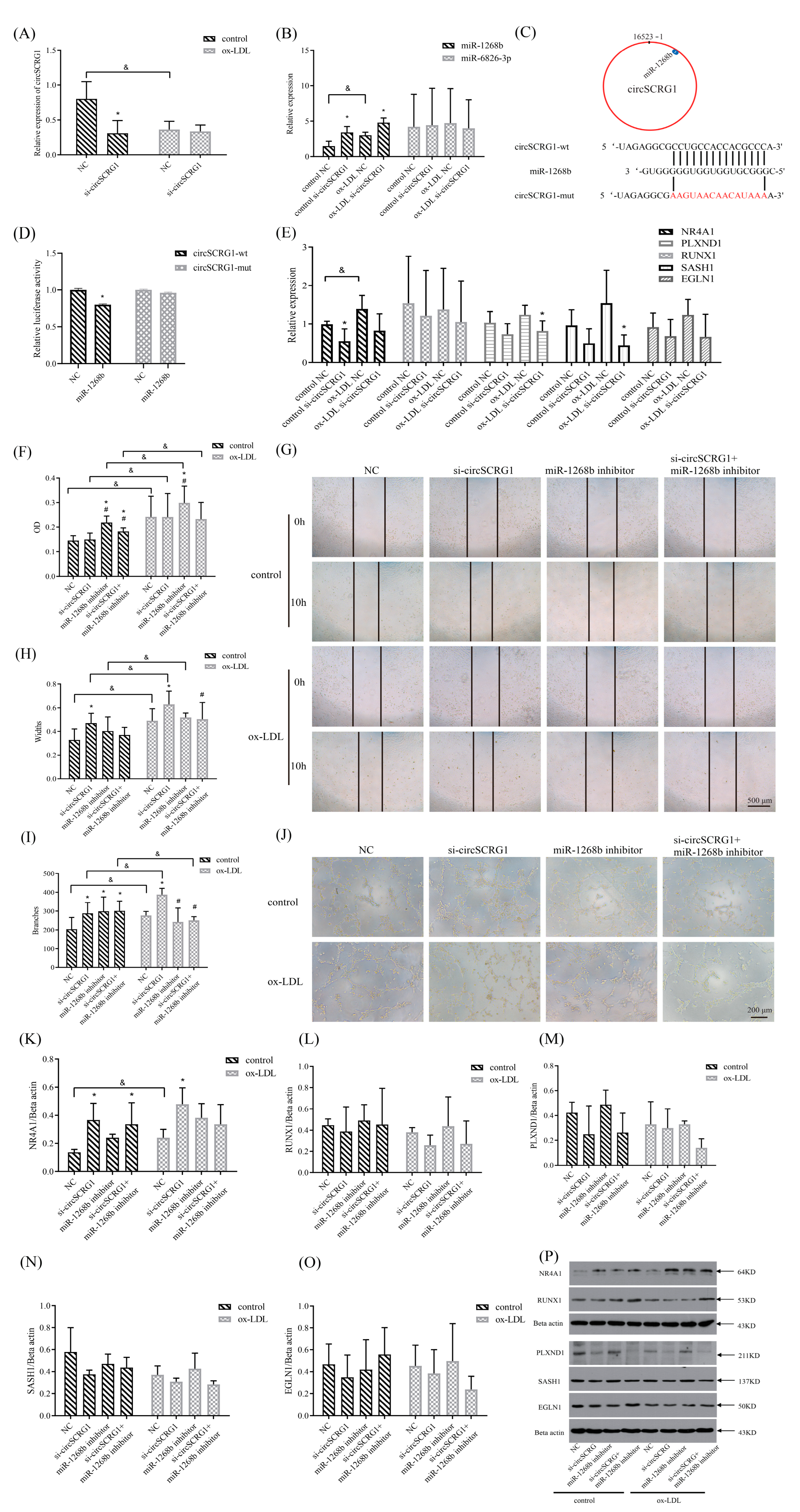
2.2. Verification of a Target circRNA and Its Role in ox-LDL-Induced Angiogenesis
2.3. The circSCRG1/miR-1268b/Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Signalling-Regulated Angiogenesis in Atherosclerosis
2.4. TMP-PF Suppressed ox-LDL-Induced Angiogenesis by Regulating the circSCRG1/miR-1268b/NR4A1 Axis
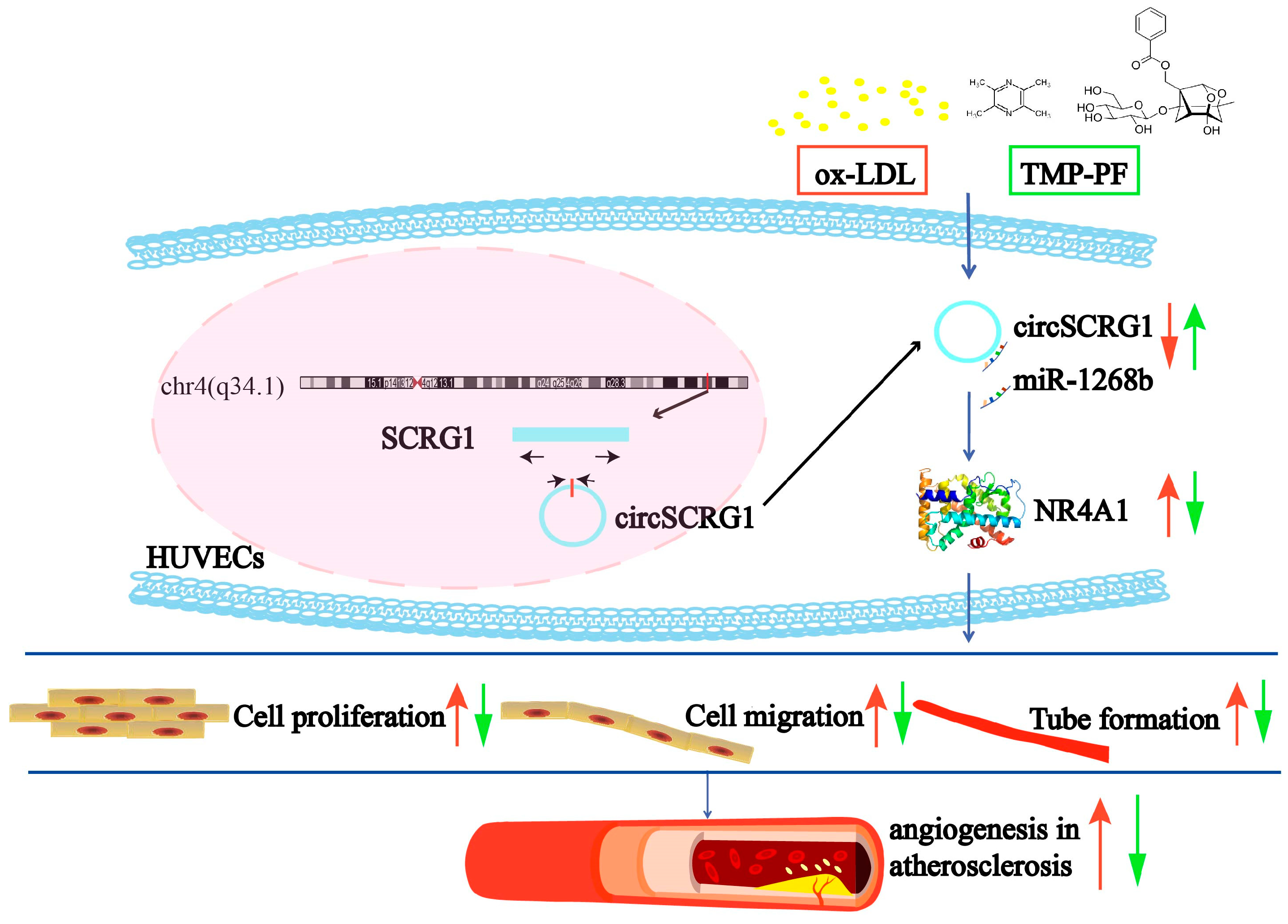
3. Discussion
4. Materials and Methods
4.1. RNA-Seq Data Analysis
4.2. CircRNA-miRNA-mRNA Network Construction Based on Bioinformatic Analysis
4.3. Cell Culture and Treatment
4.4. Target circRNA Screening
4.5. Cell Transfection
4.6. Cell Proliferation Assay
4.7. Cell Migration Assay
4.8. Matrigel Tube Formation Assay
4.9. Luciferase Reporter Assay
4.10. RT-qPCR
4.11. Western Blotting
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cademartiri, F.; Balestrieri, A.; Cau, R.; Punzo, B.; Cavaliere, C.; Maffei, E.; Saba, L. Insight from imaging on plaque vulnerability: Similarities and differences between coronary and carotid arteries-implications for systemic therapies. Cardiovasc. Diagn. Ther. 2020, 10, 1150–1162. [Google Scholar] [CrossRef] [PubMed]
- Baganha, F.; de Jong, R.; Peters, E.A.; Voorham, W.; Jukema, J.W.; Delibegovic, M.; de Vries, M.R.; Quax, P. Atorvastatin pleiotropically decreases intraplaque angiogenesis and intraplaque haemorrhage by inhibiting angpt2 release and ve-cadherin internalization. Angiogenesis 2021, 24, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, C.; Gu, X.; Liu, X.; Wang, X.; Wang, X.; Xie, Z.; Tian, J.; Yu, B. Intraplaque neovascularization attenuated statin benefit on atherosclerotic plaque in cad patients: A follow-up study with combined imaging modalities. Atherosclerosis 2019, 287, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, S.; Xu, T.; He, X.; Sun, Y.; Chen, X.; Cao, S.; Si, X.; Liao, W.; Liao, Y.; et al. Therapeutic ultrasound combined with microbubbles improves atherosclerotic plaque stability by selectively destroying the intraplaque neovasculature. Theranostics 2020, 10, 2522–2537. [Google Scholar] [CrossRef]
- Di, M.; Zhang, Y.; Zeng, R.; Liu, X.; Chen, W.; Zhang, M.; Zhang, C.; Li, M.; Zhang, M. The pro-angiogenesis effect of mir33a-5p/ets-1/dkk1 signaling in ox-ldl induced huvecs. Int. J. Biol. Sci. 2021, 17, 4122–4139. [Google Scholar] [CrossRef]
- de Vries, M.R.; Parma, L.; Peters, H.; Schepers, A.; Hamming, J.F.; Jukema, J.W.; Goumans, M.; Guo, L.; Finn, A.V.; Virmani, R.; et al. Blockade of vascular endothelial growth factor receptor 2 inhibits intraplaque haemorrhage by normalization of plaque neovessels. J. Intern. Med. 2019, 285, 59–74. [Google Scholar] [CrossRef]
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular rnas open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514. [Google Scholar] [CrossRef]
- Saaoud, F.; Drummer, I.V.C.; Shao, Y.; Sun, Y.; Lu, Y.; Xu, K.; Ni, D.; Jiang, X.; Wang, H.; Yang, X. Circular rnas are a novel type of non-coding rnas in ros regulation, cardiovascular metabolic inflammations and cancers. Pharmacol. Ther. 2020, 220, 107715. [Google Scholar] [CrossRef]
- Yang, Q.; Li, F.; He, A.T.; Yang, B.B. Circular rnas: Expression, localization, and therapeutic potentials. Mol. Ther. 2021, 29, 1683–1702. [Google Scholar] [CrossRef]
- Rai, A.K.; Lee, B.; Hebbard, C.; Uchida, S.; Garikipati, V. Decoding the complexity of circular rnas in cardiovascular disease. Pharmacol. Res. 2021, 171, 105766. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular rnas. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular rnas. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular rnas. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Chen, L.L. Circular rnas: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Chen, L.; Wang, S.; Wang, Z.; Liu, Y.; Xu, Y.; Yang, S.; Xue, G. Construction and analysis of competing endogenous rna network and patterns of immune infiltration in abdominal aortic aneurysm. Front. Cardiovasc. Med. 2022, 9, 955838. [Google Scholar] [CrossRef]
- Sun, X.; Deng, K.; Zang, Y.; Zhang, Z.; Zhao, B.; Fan, J.; Huang, L. Exploring the regulatory roles of circular rnas in the pathogenesis of atherosclerosis. Vasc. Pharmacol. 2021, 141, 106898. [Google Scholar] [CrossRef]
- Cai, Y.; Pan, J.; Li, Z. Mathematical modeling of intraplaque neovascularization and hemorrhage in a carotid atherosclerotic plaque. Biomed. Eng. Online 2021, 20, 42. [Google Scholar] [CrossRef]
- Maurea, N.; Coppola, C.; Piscopo, G.; Galletta, F.; Riccio, G.; Esposito, E.; De Lorenzo, C.; De Laurentiis, M.; Spallarossa, P.; Mercuro, G. Pathophysiology of cardiotoxicity from target therapy and angiogenesis inhibitors. J. Cardiovasc. Med. (Hagerstown) 2016, 17 (Suppl. S1), S19–S26. [Google Scholar] [CrossRef]
- Jiang, Z.; Wang, L.; Liu, X.; Chen, C.; Wang, B.; Wang, W.; Hu, C.; Yu, K.; Qi, Z.; Liu, Q.; et al. Discovery of a highly selective vegfr2 kinase inhibitor chmfl-vegfr2-002 as a novel anti-angiogenesis agent. Acta Pharm. Sin. B 2020, 10, 488–497. [Google Scholar] [CrossRef]
- Yuan, R.; Wang, Y.; Cong, W.H.; Chen, K.J. Treatment of cardiovascular disease with xiongshao capsule. Zhongguo Zhong Yao Za Zhi 2017, 42, 640–643. [Google Scholar]
- Yuan, R.; Shi, W.; Xin, Q.; Cong, W.; Chen, K. Progress on rhizoma chuanxiong-radix paeoniae rubra herb pair. Glob. Tradit. Chin. Med. 2019, 12, 808–811. [Google Scholar]
- Qiao, L.; Chen, W. Atheroprotective effects and molecular targets of bioactive compounds from traditional chinese medicine. Pharmacol. Res. 2018, 135, 212–229. [Google Scholar] [CrossRef]
- Lin, J.; Wang, Q.; Zhou, S.; Xu, S.; Yao, K. Tetramethylpyrazine: A review on its mechanisms and functions. Biomed. Pharmacother. 2022, 150, 113005. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Gong, X.H.; Zhang, H.; Peng, C. A review on the pharmacokinetics of paeoniflorin and its anti-inflammatory and immunomodulatory effects. Biomed. Pharmacother. 2020, 130, 110505. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Shi, W.; Xin, Q.; Yang, B.; Hoi, M.P.; Lee, S.M.; Cong, W.; Chen, K. Tetramethylpyrazine and paeoniflorin inhibit oxidized ldl-induced angiogenesis in human umbilical vein endothelial cells via vegf and notch pathways. Evid.-Based Complement. Altern. Med. 2018, 2018, 3082507. [Google Scholar] [CrossRef]
- Zeng, Z.; Xia, L.; Fan, S.; Zheng, J.; Qin, J.; Fan, X.; Liu, Y.; Tao, J.; Liu, Y.; Li, K.; et al. Circular rna circmap3k5 acts as a microrna-22-3p sponge to promote resolution of intimal hyperplasia via tet2-mediated smooth muscle cell differentiation. Circulation 2021, 143, 354–371. [Google Scholar] [CrossRef]
- Miao, R.; Qi, C.; Fu, Y.; Wang, Y.; Lang, Y.; Liu, W.; Zhang, Y.; Zhang, Z.; Liu, A.; Chai, H.; et al. Silencing of circarhgap12 inhibits the progression of atherosclerosis via mir-630/ezh2/timp2 signal axis. J. Cell. Physiol. 2022, 237, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, D.; Zhou, T.; Zhang, J.; Liu, C.; Cao, F.; Dong, N. Melatonin ameliorates aortic valve calcification via the regulation of circular rna circric3/mir-204-5p/dpp4 signaling in valvular interstitial cells. J. Pineal Res. 2020, 69, e12666. [Google Scholar] [CrossRef]
- Wen, Y.; Chun, Y.; Lian, Z.Q.; Yong, Z.W.; Lan, Y.M.; Huan, L.; Xi, C.Y.; Juan, L.S.; Qing, Z.W.; Jia, C.; et al. Circrna-0006896-mir1264-dnmt1 axis plays an important role in carotid plaque destabilization by regulating the behavior of endothelial cells in atherosclerosis. Mol. Med. Rep. 2021, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Lavenniah, A.; Luu, T.; Li, Y.P.; Lim, T.B.; Jiang, J.; Ackers-Johnson, M.; Foo, R.S. Engineered circular rna sponges act as mirna inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 2020, 28, 1506–1517. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, Y.; Deng, K.; Liang, Y.; Zhang, G.; Gao, X.; El-Samahy, M.A.; Zhang, Y.; Deng, M.; Wang, F. Circular rna circusp13 sponges mir-29c to promote differentiation and inhibit apoptosis of goat myoblasts by targeting igf1. FASEB J. 2022, 36, e22097. [Google Scholar] [CrossRef] [PubMed]
- Militello, G.; Weirick, T.; John, D.; Döring, C.; Dimmeler, S.; Uchida, S. Screening and validation of lncrnas and circrnas as mirna sponges. Brief. Bioinform. 2017, 18, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Ebbesen, K.K.; Sokol, M.; Jakobsen, T.; Korsgaard, U.; Eriksen, A.C.; Hansen, T.B.; Kjems, J.; Hager, H. Spatial expression analyses of the putative oncogene cirs-7 in cancer reshape the microrna sponge theory. Nat. Commun. 2020, 11, 4551. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and functional rna circularization in localized prostate cancer. Cell 2019, 176, 831–843. [Google Scholar] [CrossRef]
- Su, Q.; Lv, X. Revealing new landscape of cardiovascular disease through circular rna-mirna-mrna axis. Genomics 2020, 112, 1680–1685. [Google Scholar] [CrossRef]
- Zheng, G.H.; Liu, J.P.; Chu, J.F.; Mei, L.; Chen, H.Y. Xiongshao for restenosis after percutaneous coronary intervention in patients with coronary heart disease. Cochrane Database Syst. Rev. 2013, 5, CD009581. [Google Scholar] [CrossRef]
- Chen, C.; Li, Y.; Hou, S.; Bourbon, P.M.; Qin, L.; Zhao, K.; Ye, T.; Zhao, D.; Zeng, H. Orphan nuclear receptor tr3/nur77 biologics inhibit tumor growth by targeting angiogenesis and tumor cells. Microvasc. Res. 2020, 128, 103934. [Google Scholar] [CrossRef]
- Rodríguez-Calvo, R.; Tajes, M.; Vázquez-Carrera, M. The nr4a subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert Opin. Ther. Targets 2017, 21, 291–304. [Google Scholar] [CrossRef]
- Crean, D.; Murphy, E.P. Targeting nr4a nuclear receptors to control stromal cell inflammation, metabolism, angiogenesis, and tumorigenesis. Front. Cell Dev. Biol. 2021, 9, 589770. [Google Scholar] [CrossRef]
- Ye, T.; Peng, J.; Liu, X.; Hou, S.; Niu, G.; Li, Y.; Zeng, H.; Zhao, D. Orphan nuclear receptor tr3/nur77 differentially regulates the expression of integrins in angiogenesis. Microvasc. Res. 2019, 122, 22–33. [Google Scholar] [CrossRef]
- Kominek, J.; Doering, D.T.; Opulente, D.A.; Shen, X.X.; Zhou, X.; Devirgilio, J.; Hulfachor, A.B.; Groenewald, M.; Mcgee, M.A.; Karlen, S.D.; et al. Eukaryotic acquisition of a bacterial operon. Cell 2019, 176, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Yu, M. Construction of Circrnas Expression Profiles Based on the Characteristics of Blood Stasis in Patients with Myocardial Infarction; Beijing Univ Chin Med: Beijing, China, 2017; Volume 1, pp. 41–67. [Google Scholar]
- Li, X.; Yang, Y.; Liang, L.; Fan, M.; Li, X.; Feng, N.; Pan, Y.; Tan, Q.; Xu, Q.; Xie, Y.; et al. Effect of xbp1 deficiency in cartilage on the regulatory network of lncrna/circrna-mirna-mrna. Int. J. Biol. Sci. 2022, 18, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Beck, R.; Bertolino, S.; Abbot, S.E.; Aaronson, P.I.; Smirnov, S.V. Modulation of arachidonic acid release and membrane fluidity by albumin in vascular smooth muscle and endothelial cells. Circ. Res. 1998, 83, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Yu, J.; Shan, G.; Su, L.; Yu, N.; Yang, S. N6-methyladenosine methyltransferase mettl3 promotes angiogenesis and atherosclerosis by upregulating the jak2/stat3 pathway via m6a reader igf2bp1. Front. Cell Dev. Biol. 2021, 9, 731810. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.G.; Zhou, C.F.; Zhang, Y.M.; Yan, R.M.; Wei, W.F.; Chen, X.J.; Yi, H.Y.; Liang, L.J.; Fan, L.S.; Liang, L.; et al. Cancer-derived exosomal mir-221-3p promotes angiogenesis by targeting thbs2 in cervical squamous cell carcinoma. Angiogenesis 2019, 22, 397–410. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, R.; Xin, Q.; Ma, X.; Yu, M.; Miao, Y.; Chen, K.; Cong, W. Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro. Molecules 2023, 28, 1271. https://doi.org/10.3390/molecules28031271
Yuan R, Xin Q, Ma X, Yu M, Miao Y, Chen K, Cong W. Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro. Molecules. 2023; 28(3):1271. https://doi.org/10.3390/molecules28031271
Chicago/Turabian StyleYuan, Rong, Qiqi Xin, Xiaochang Ma, Meng Yu, Yu Miao, Keji Chen, and Weihong Cong. 2023. "Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro" Molecules 28, no. 3: 1271. https://doi.org/10.3390/molecules28031271
APA StyleYuan, R., Xin, Q., Ma, X., Yu, M., Miao, Y., Chen, K., & Cong, W. (2023). Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro. Molecules, 28(3), 1271. https://doi.org/10.3390/molecules28031271





