Abstract
Phenols are very soluble in water; as a result, they can pollute a massive volume of fresh water, wastewater, groundwater, oceans, and soil, negatively affecting plant germination and animal and human health. For the detoxification and bioremediation of phenol in wastewater, phenol biodegradation using novel bacteria isolated from sewage sludge was investigated. Twenty samples from sewage sludge (SS) were collected, and bacteria in SS contents were cultured in the mineral salt agar (MSA) containing phenol (500 mg/L). Twenty colonies (S1 up to S20) were recovered from all the tested SS samples. The characteristics of three bacterial properties, 16S rDNA sequencing, similarities, GenBank accession number, and phylogenetic analysis showed that strains S3, S10, and S18 were Pseudomonas aeruginosa, Klebsiella pneumoniae, and Klebsiella variicola, respectively. P. aeruginosa, K. pneumoniae, and K. variicola were able to degrade 1000 mg/L phenol in the mineral salt medium. The bacterial strains from sewage sludge were efficient in removing 71.70 and 74.67% of phenol at 1000 mg/L within three days and could tolerate high phenol concentrations (2000 mg/L). The findings showed that P. aeruginosa, K. pneumoniae, and K. variicola could potentially treat phenolic water. All soybean and faba bean seeds were germinated after being treated with 250, 500, 750, and 1000 mg/L phenol in a mineral salt medium inoculated with these strains. The highest maximum phenol removal and detoxification rates were P. aeruginosa and K. variicola. These strains may help decompose and detoxify phenol from industrial wastewater with high phenol levels and bioremediating phenol-contaminated soils.
1. Introduction
Various living organisms, including microbes, plants, animals, and people, are highly vulnerable to phenol toxicity. Due to the expanding industrial sector, aromatic compounds are environmental contaminants in many places, such as freshwater, marine, and land [1]. Acute and high phenol concentrations can induce central nervous system abnormalities, cardiac depression, ocular irritation, edema, corneal bleaching, blindness, cardiovascular disorders, and gastrointestinal damage [2,3]. The primary pollutant in wastewater is the manufacture and utilization of phenol and phenolic compounds in industrial locations, such as oil refineries, coking plants, medicines, and plastics [4,5]. The maximum limit of phenol in the environment and tap water is 100 µg/L and 1–2 µg/L, respectively, based on WHO, because of its high toxicity [6].
Biodegradation is the ideal technique to dispose of phenol since it is inexpensive, environmentally beneficial, and simple to manage [7,8]. Nevertheless, various treatment approaches are available, including adsorption [9], solvent extraction [10], wet oxidation, and hydrogen peroxide. Fenton’s reagent, chemical oxidation, and combustion have removed phenol from contaminated samples [11,12]. However, these procedures are complicated, expensive, and not environmentally friendly [13,14]. Bacteria oxidize phenol into CO2 and H2O during metabolic processes [15] and use it as a sole source of carbon energy for development [16,17]; hence, these bacteria can degrade phenol [18].
Phenol is an artificial and naturally occurring aromatic compound and an essential intermediate in the biodegradation of natural and industrial aromatic compounds. Whereas many microorganisms capable of aerobic phenol degradation have been isolated, only a few phenol-degrading anaerobic organisms have been described to date. In this study, three novel nitrate-reducing microorganisms capable of using phenol as a sole source of carbon were isolated and characterized. Phenol-degrading denitrifying pure cultures were obtained by enrichment culture from sewage sludge. The three strains were shown to be different from each other based on physiologic and metabolic properties. Sequence analysis of 16S ribosomal DNA indicated that the phenol-degrading isolates were closely related to members of Pseudomonas and Klebsiella. In addition, the quantitative real-time PCR (RT-PCR) demonstrated the expression of all the genes required in the meta-cleavage pathway of phenol in three identified strains. The results of this study add three new members to the genus Pseudomonas and Klebsiella, which previously comprised only nitrogen-fixing species. So, discovering phylogenetically closely related species for phenol degradation is crucial; these strains can be exploited in real-world systems and aid in treating industrial pollutants [19,20,21]. The whole genome of A. lwoffii NL1 was sequenced, yielding 3499 genes on one circular chromosome and three plasmids. Enzyme activity analysis showed that A. lwoffii NL1 degraded phenol via the ortho-cleavage rather than the meta-cleavage pathway. The essential genes for phenol hydroxylase and catechol 1,2-dioxygenase were found to be separated by mobile genetic elements on a megaplasmid (pNL1); their function was verified by heterologous production in Escherichia coli and quantitative real-time PCR. [22]. Using these isolated microbes is more cost-effective and practical, reducing the environmental load by creating an efficient and cost-effective technology [23].
Microbial degradation has been considered an effective and eco-friendly technique for phenols removal. Various microorganisms isolated from contaminated sites can degrade phenols under aerobic conditions. The recent advances in the microbial treatment of phenol have improved our understanding of bioremediation [24,25]. Acinetobacter calcoaceticus can effectively treat phenolic wastewater. A. calcoaceticus effectively reduced 91.6% of phenol concentration (0.8 g/L) after two days and can tolerate phenol (1.7 g/L). These results revealed that A. calcoaceticus could effectively treat phenolic wastewater [26]. A. lwoffii NL1 could degrade 0.5 g/L phenol within 12 h and tolerate a maximum of 1.1 g/L phenol, and A. lwoffii NL1 can potentially be used for efficient phenol degradation in heavy metal wastewater treatment [22]. The bacteria of the genus Pseudomonas have been used as typical phenol-degrading microorganisms. P. putida can degrade phenol (1 g/L) in 162 h (6.17 mg/L per hour) [27], whereas P. cepacia isolated from industrial wastewaters can degrade 2.5 g/L phenol in 144 h (17.36 mg/L per hour) [28]. A new Rhodococcus aetherivorans strain has 35.7 mg/L per hour degradation rates at 0.5 g/L phenol [29]. The mutant M1 of Rhodococcus ruber SD3 can degrade 98% of 2 g/L phenol in 72 h by cell immobilization (27.2 mg/L per hour) [30]. The highest phenol degradation by Serratia sp. (accession number KT693287) was achieved at pH 7.5, the temperature of 30 °C and ammonium sulfate as a nitrogen source at the concentration of 0.4 g/L as well as a sodium chloride concentration of 0.15 g/L [31]. Klebsiella oxytoca degraded phenol (100 ppm) within 72 h. In comparison, at a phenol concentration above 400 ppm, the cells could not degrade the substrate efficiently due to the increasing phenol concentration in the medium. The optimum solution pH and temperature were 6.8 and 37 °C, respectively [32]. Aerobic phenol decomposition by bacterial strains requires many steps to increase the degradation rate and figure out how different physicochemical parameters and degradation processes affected the decomposition rate [33]. In addition, Sachan et al. [34] isolated two bacterial isolates from the effluent of a pulp and paper mill coded as SP-4 and SP-8 which showed luxuriant growth on phenol-amended minimal salt medium (MSM) in the presence of 1% glucose (w/v). In contrast, no growth has been observed in the absence of glucose. Both the strains showed fast and luxuriant growth at phenol concentrations of 0–1000 mg/L. These isolates can tolerate the phenol up to a phenol concentration of 1600 and 1800 mg/L, respectively. However, no growth has been observed in both bacterial isolates at a 2000 mg/L phenol concentration. Furthermore, Pseudomonas putida strains with intense degrading activity at considerable doses of phenol (8.5 mM) and chlorine-substituted derivatives such as O, M, P-chlorophenol (1.56 mM, 1.56 mM, and 2.34 mM, respectively) and 2,4-dichlorophenol (30 mg/L) have been described [27,35,36].
The temperature and pH are essential factors for bacterial growth and phenol degradation. In the typical mesophilic growth range of 20 °C to 30 °C, bacterial growth rates nearly double for every (10 °C) increase in temperature, and it does not alter between 35 and 40 °C. Still, the denaturation process for protein at higher temperatures decelerates growth rates for mesophilic microorganisms [37].
In vitro findings on phenol degradation have been carried out at an optimum temperature of 30 °C [38,39]; however, Tengku-Mazuki, [40] found that Rhodococcus sp. strain AQ5-14 has excellent potential for the bioremediation of phenol. The optimum conditions for phenol degradation (83.90%) were pH 7.0 and 0.4 g l-1 NaCl at 25 °C. In addition, S S, et al. [41] reported that no phenol degradation was noticed when the temperature was raised from 30 to 34 °C, indicating that phenol decomposition is a temperature-dependent process. Furthermore, Kotresha and Vidyasagar [42] found P. aeruginosa MTCC 4996 degraded phenol at temperatures ranging from 15 to 45 °C, with an optimal of 37 °C, and pH ranging from 6.0 to 10.0, with optimal of 7.0 to 7.5. Furthermore, Samimi and Moghadam [39] showed that the optimal condition for 97% phenol removal was pH 8, 35 °C, C: N ratio of 100: 30 (g/L), and salinity of 35 g/L.
Regarding pH factor, Rhodococcus sp. strain AQ5-14 preferred growth at the near-neutral condition. The optimum pH value for accurate phenol degradation was pH 7.0, which corresponded with their growth. The internal growth factors affecting an all-living cell are approximate. Most organisms cannot withstand pH below 4.0 or above 9.0 [40]. The acids or bases can easily penetrate bacterial cells since they prefer to live in a non-dissociated state under these conditions, and electrostatic forces cannot prevent them from entering cells [43].
All detected phenol-degrading isolates in the previous studies can tolerate phenol concentrations less than 2g/L. Therefore, in this study, we isolated, characterized, and evaluated a novel phenol-degrading bacteria (Pseudomonas aeruginosa, Klebsiella pneumoniae, and Klebsiella variicola) from sewage sludge (SS). The capability of the bacterial strains to degrade and detoxify phenols at a high level > (2 g/L) and their effect on bean seed germination have been evaluated.
2. Results and Discussion
2.1. Isolation of Phenol-Degrading Bacteria
Table 1 shows the OD of bacterial growth in MSM supplemented with different phenol concentrations (0.0, 500, 750, 1000, 1500, 2000 mg/L) at 600 nm. The sewage sludge sample (10 g) was added aseptically to 90 mL of MSM media supplemented with different levels of phenol and incubated in a shaking incubator (120 rpm; 35 °C) [38]. The culture broths were examined at an OD600 nm after incubation for three days. The noticeable increase in optical density (OD) in the control compared to all the samples. The higher value of OD was obtained from the SS3, SS10, and SS18 compared to the control at all the phenol concentrations. Thus, from these samples, 10 mL of the culture broth was transferred to a new shaking flask containing 90 mL of fresh MSM containing 500 mg/L phenol and incubated for three days. This culture broth was streaked onto MSM agar plates containing 0.5 g/L phenol (Figure 1). After 3–4 days of incubation at 35 °C, colonies on agar plates were harvested and subcultured repeatedly to obtain pure cultures [39]. The bacterial isolates of S3, S10, and S18 were stored in LB agar slants at (4 °C) for the following experiments.

Table 1.
Bacterial populations (OD600 ± SD) from sewage sludge (SS) can grow on MSM supplemented with different concentrations of phenol (500 to 2000 mg/L).

Figure 1.
Bacterial growth of Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10), and Klebsiella variicola (S18) onto MSM plates amended with 500 mg/L phenol.
2.2. Identification and Characterization of Phenol-Degrading Bacterial Isolates
The samples (10 mL) were put into a mineral salt broth containing 500 mg/L phenol to enrich phenol-degrading bacteria in a 250-mL flask. The phenol-utilizing bacterial isolates were subsequently grown on mineral salt agar (MSA) containing 500 mg/mL of phenol. All colonies that develop on the phenol-containing solid medium can use phenol as their primary carbon and energy source. These isolates displayed more significant growth in phenol-containing media containing up to 2 g/L phenol (Table 2). Regarding the physical and biochemical properties, Gram staining, motility, acid formation of glucose, oxidase, catalase, oxidative fermentation, and growth under different conditions were on these bacteria. The bacterial strains S3, S10, and S18 from sewage sludge samples may tolerate phenol concentrations up to 2000 mg/L. Among the twenty established isolates, these strains were shown to be the most effective in terms of phenol tolerance and incubation period (Table 2). Using these isolated microbes is more cost-effective and practical, reducing environmental load by developing efficient and cost-effective technologies [21]. These bacteria are essential to soil and capable of degrading polycyclic aromatic hydrocarbons. Still, it is frequently identified in water reservoirs contaminated by animals and people, such as sewage and sinks in and around hospitals.

Table 2.
Bacterial isolates from sewage sludge can grow on different concentrations of phenol.
Table 3 revealed that all bacteria were G− rods, aerobic, motile, positive for oxidase- and catalase, and produced acid from glucose. Three isolates were Gram-negative, strictly aerobic, motile, and rod-shaped. Table 3 shows the colonies’ growth and morphological, microscopic, and biochemical features. The detected bacteria, Pseudomonas spp., and Klebsiella spp., were aerobic, positive for oxidase and catalase, and produced acid from glucose (Table 3). The phenol-degrading isolates S3, S10, and S18 could grow at temperatures ranging from 25 to 40 °C and pH levels of 5 to 9. The optimal growing conditions were between 30 and 35 °C and pH 7.0. In this work, three Gram-negative isolates were plated on a solid MSM medium supplemented with different levels of phenol (Figure 1). The species of Pseudomonas and Klebsiella have been characterized by Stover et al. [44]. Pseudomonas and Klebsiella species can typically thrive on MSM medium containing a single carbon and energy source. P. aeruginosa develops well around 37 °C. However, it can also survive in a wide range of temperatures between 4 and 42 °C. Pseudomonas and Klebsiella species may decompose phenol [35,45].

Table 3.
Preliminary identification of bacterial isolate by conventional microbiological methods.
2.3. 16S rDNA Gene Sequence Analysis of the Three Isolates
Figure 2 shows that each isolate was identified based on its morphology and 16S rDNA gene sequence analysis. The development of a single band in the PCR result of the isolates (Figure 2) suggests that PCR effectively amplified the 16S rRNA gene sequence at 1500 bp.
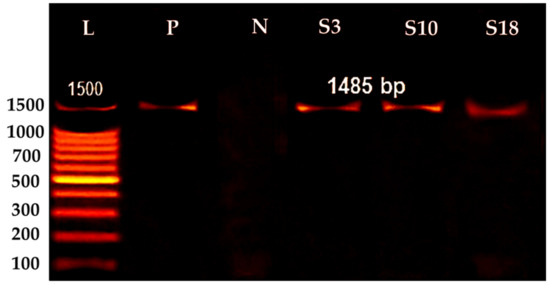
Figure 2.
16S rRNA genes of bacterial isolates, Lane 1, Ladder [(L) 100–1500 bp], Positive control (P, Pesudomonas areginosa), Negative control (N, Aeromonas hydrophila) identified isolates (S3, S10, and S18) at (1500 bp).
In this work, 16S ribosomal DNA sequence analysis revealed that phenol-degrading isolates were closely related to Pseudomonas and Klebsiella species. In addition, quantitative real-time PCR (RT-PCR) revealed the expression of all genes necessary for the phenol meta-cleavage process in three discovered strains. The results of this study add three new members to the genera Pseudomonas and Klebsiella, which previously constituted exclusively nitrogen-fixing bacteria. Therefore, it is essential to discover phylogenetically related species for phenol degradation; these strains may be utilized in real-world systems and help treat industrial contaminants [19,20,21]. A. lwoffii NL1’s whole genome was sequenced, revealing 3499 genes on one circular chromosome and three plasmids. A. lwoffii NL1 degraded phenol by the ortho-cleavage pathway rather than the meta-cleavage pathway, as determined by enzyme activity analysis. Critical genes for phenol-hydroxylase and catechol-1,2-dioxygenase were identified on a giant plasmid (pNL1) and were discovered to be separated by mobile genetic elements; their activity was verified by heterologous production in Escherichia coli and quantitative real-time PCR [22].
The 16S rRNA gene sequences for Pseudomonas aeruginosa ON880415.1, Klebsiella pneumoniae ON0414.1, and Klebsiella variicola ON880418.1 have been submitted to GenBank. All sequencing results were compared to the 16S rRNA gene sequence in the NCBI GenBank database. Ten (50%) of the identified bacteria were Pseudomonas spp., and ten (50%) were Klebsiella spp. Based on consensus sequences for the 16S rRNA gene, a phylogenetic tree was created by comparing sequences from 29 GenBank strains to strain S3 and typical Gram-negative Pseudomonas spp.
The blast was used to compare our data to reference sequences from the NCBI Gene Bank Data Base. The phylogenetic analysis revealed that the 29 phenol-degrading bacteria had identical 16S rDNA gene sequences and belonged to the same cluster. This cluster’s members showed a significantly greater specificity and could be separated into two distinct subclusters. The Pseudomonas species exhibited tighter relationships, with a bootstrap score between 98 and 99%, confirming their placement within P. aeruginosa ON88415.1 (S3). In addition, sequences from 16 GenBank strains were compared to those of strains S10 and S18 and typical Gram-negative Klebsiella species. Klebsiella spp. has a bootstrap value between 97 and 89% and was well-established within two clusters. Cluster 1 is comprised of K. pneumonia, while Cluster 2 is comprised of K. variicola. Global alignment was utilized for pairwise sequence alignment and the generation of similarity values in the EzBio-Cloud database [46].
This molecular taxonomic investigation determined the strains to be Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10), and Klebsiella variicola (S18). The evolutionary history was inferred using the neighbor-joining and maximum likelihood techniques with 1000 repetitions of MEGA version 7’s bootstrap values [40]. The nucleotide sequence from this study has been added to the NCBI nucleotide sequence databases with the Accession numbers: ON880415.1, ON880414.1, and ON880418.1, respectively, and these strains are 97–99% similar to the strains in the GenBank, P. aeruginosa JN995663.1, K. pneumoniae MT516162.1, and K. variicola ON597432.1, respectively (Figure 3).
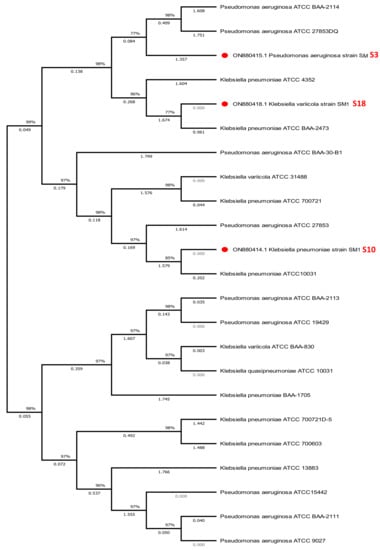
Figure 3.
Phylogenetic tree of Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10) and Klebsiella variicola (S18).
2.4. Evaluation of the Bacterial Isolates for Phenol Degradation
This research aims to isolate particular bacteria from a diverse population of naturally existing microorganisms. Twenty isolates were selected on LB media after 24 h incubation. After 24 h of growth on LB agar plates containing 100 µL of a 103–106-fold dilution of the EM after three weeks of enrichment and one week of bacterial isolation. These isolates were evaluated for growth in MSM supplemented with varying amounts of phenol (Table 1). The phenol-degrading abilities of P. aeruginosa (S3), K. pneumoniae (S10), and K. variicola (S18) at different starting phenol concentrations (0.0, 500, 750, 1000, 1500, and 2000 mg/L) were studied by measuring the phenol concentration and cell growth at OD600 (Figure 4).
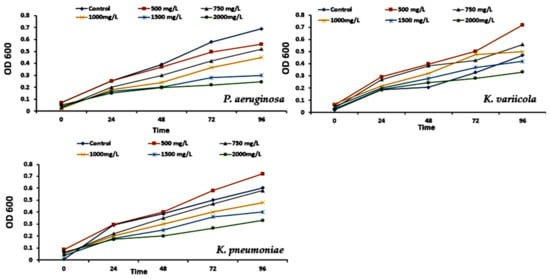
Figure 4.
Effect of different phenol concentrations (0, 500, 750, 1000, 1500, and 2000 mg/L) on the growth of Pseudomonas aeruginosa, Klebsiella pneumoniae, and Klebsiella variicola.
Maximum phenol degradation was reported at an initial 1000 mg/L value (Figure 5). The three strains survived at phenol concentrations up to 1500 mg/L with a degradation rate of 39.82%. Concerning the pH and temperature, the bacterial strain can populate at a pH range of 6–8 to degrade 75% of phenol (Figure 5). Figure 5 demonstrates the bacterial growth of strains S3, S10, and S18 and the 75% phenol breakdown at 35 °C. Conversely, the phenol degradation decreased substantially at temperatures between 25 and 40 °C. Therefore, the ideal growing temperature for strains S3, S10, and S18 was 35 °C. Shourian et al. [47] and Kotresha and Vidyasagar [42] discovered that Pseudomonas aeruginosa could degrade phenol at temperatures ranging from 15 °C to 45 °C, with an optimal of 37 °C and pH values ranging from 6.0 to 8.0, with an optimal of 7.0 to 7.5 (phosphate buffer). The optimal temperature and pH levels were 35 °C and 7.0, respectively. The mechanism of phenol degradation by isolated strains can be correlated with these pathways. The P-nitro phenol degradation pathway involves the formation of benentroid, which contains aromatic ring cleavage that results in the biodegradation of P-nitro phenol. The p-nitro phenol was degraded to benzoquinone (BQ) and hydroquinone (HQ) by the Arthobacter strain [48]. Gram-negative bacteria, such as Burkholderia spp. and Moraxella spp., convert hydroquinone into maleyl acetate [49]. While G+ bacteria, such as Bacillus spp. and Arthrobacter spp., converted 4-nitro-phenol into 4-nitro catechol and hydroxyquinol [50]. Consequently, several studies conducted on 4-nitro phenol degradation have been reported. In association with the hydroquinone pathway, the 4-nitro phenol degradation genes were cloned from the Pseudomonas sp. strain and Pseudomonas putida. In the case of the hydroxyquinol pathway, no 4-nitro phenol catabolic gene has been reported. The individual nucleotide sequence of 4-nitro phenol degradation genes (nphA1A2) was reported using Rhodococcus sp. strain PN1 [51].
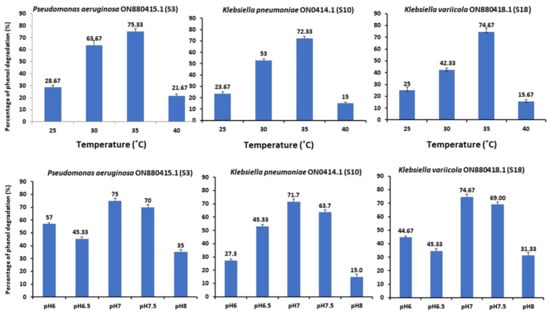
Figure 5.
Effect of pH and temperature on phenol biodegradation by Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10), and Klebsiella variicola (S18) (initial phenol concentration was 1000 mg/L).
Peptone and yeast extract are the most effective organic nitrogen sources for phenolic growth and breakdown. In addition, a maximum of 98.93% phenol degradation was observed at an initial concentration of 1000 mg/L at 35 °C and pH 7.0 for 96 h [52], in agreement with previous studies that stated P. aeruginosa MTCC 4997 strain isolated from effluents of petrochemical companies completely degraded phenol at 15–45 °C range, with the optimal temperature being 37 °C [42]. Mishra and Kumar [53] found that P. aeruginosa KBM13 needs 40 °C to degrade the phenol in the medium. The susceptibility of these bacteria to higher phenol concentrations may require phenol acclimation before its destruction [54].
Concerning time and acclimatization, it has also been discovered that bacterial growth develops in the phenol medium, showing that the bacteria eventually adapt to the compound. Sachan et al. [34] isolated competent bacterial cells from phenol-formaldehyde resin factory effluent. Numerous studies suggest that the pH range (7–10) was optimal for the growth and phenol degradation of Pseudomonas sp., based on the bacterial origin [55,56]. Mishra and Kumar [53] demonstrated that the optimal pH for P. aeruginosa to degrade phenol was eight. For instance, the optimal pH range for Klebsiella oxytoca to degrade phenol was 6.8 [57]. The pH’s influence on phenol decomposition may be attributable to its effects on transport, stimulation of enzyme activity, and nutrient solubility [58]. The pH substantially affects the metabolic processes necessary for phenol breakdown. According to reports, the pH influences the surface charge of the activated sludge biomass cells [59].
As shown in Figure 6, the effect of various nitrogen sources on deterioration has been examined. The influence of three organic nitrogen sources, peptone, yeast extract, and urea, and three inorganic nitrogen sources, sodium nitrate, potassium nitrate, and ammonium chloride, on the development and breakdown of phenol by Strains S3, S10, and S18 were considered. The N sources were added to the MS medium containing (1 g/L) phenol and incubated for four days at 35 °C and pH 7 in triplicate at a temperature of 35 °C. The choice of nitrogen supply influenced the development of S3, S10, and S18 strains and phenol degradation. More than 75% of the nitrogen sources, peptone and yeast extract, demonstrated strong growth and phenol degradation rates. The addition of yeast extract increased the percentage of phenol degradation to 75%, whereas urea resulted in the lowest growth and phenol degradation (35%). Consequently, yeast extract was the best of the evaluated organic nitrogen sources for maximal growth and phenol degradation.
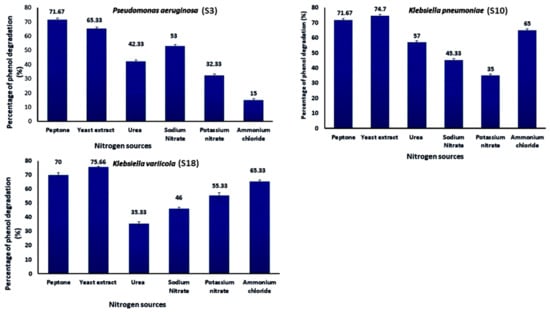
Figure 6.
Effect of N sources on phenol biodegradation by Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10), and Klebsiella variicola (S18) (initial phenol concentration was 1000 mg/L).
The present investigation proved that adding yeast extract to the medium resulted in the maximum phenol elimination efficiency and promoted the development of strain S3. The nature of yeast extract, easily obtainable as amino acids in a medium containing mineral salts, is needed for the phenol breakdown process by bacterial cells [60,61]. Mishra and Kumar [53] have shown that yeast extract is the optimum nitrogen source for growth and phenol decomposition. Ammonium sulfate was established to be the best nitrogen source at a concentration of 0.4 g/L and a sodium chloride concentration of 0.15 g/L. The different nitrogen sources, such as peptone and yeast extract, were used to get the optimum growth conditions for phenol degradation [62,63], which were selected to do a single factor test for further study of the optimum conditions for bacterial growth, which was an ability to degrade phenol. K. pneumoniae (S10) and K. variicola (S18) showed maximum phenol degradation in the presence of yeast extract at pH 7. However, in the case of P. aeruginosa (S3), peptone was the best nitrogen source of phenol degradation. Replacement of yeast extract with tryptone and urea affected phenol degradation efficiency [64]. The phenol biodegradation rate may significantly affect by the presence of a high load of nitrogen pollutants. Ammonia is known to be toxic to aquatic life and creates an excellent oxygen demand in receiving waters. The highest phenol degradation by Serratia sp. (accession number KT693287) was achieved at pH 7.5, the temperature of 30 °C and ammonium sulfate as a nitrogen source at the concentration of 0.4 g/L as well as a sodium chloride concentration of 0.15 g/L [31].
The microbial phenol degradation, with an emphasis on aerobic degradation utilizing bacterial strains, various strategies for improving/enhancing the phenol degradation rate, impacts of various physicochemical parameters on the degradation process, and degradation processes [33]. The three bacterial strains, S3, S10, and S18, were cultured on MSM medium with phenol (1000 mg/L) at a temperature of 35 °C and a pH of 7 (phosphate buffer). This combination was kept in Erlenmeyer flasks of 250 mL capacity. The cultures were shaken on a shaker (120 rpm) at the specified temperature. The phenol degradation was assessed at 96 h using HPLC and is presented in Supplementary Figure S1).
2.5. Effect of K. variicola on Soybean Seedlings
We conducted a trial on three isolates as phenol degraders, we found that S3 and S18 were more effective in phenol degradation, so we tested these isolates (S3 and S18) with a seed germination test in phenol.
Seed germination results (Figure 7 and Table 4) showed higher inhibitory activity of the different phenol concentrations. According to germination percentage, the group observed: control without bacteria and treated with different phenol concentrations (250 to 1000 mg/L) showed total inhibition in seed germination. In contrast, group Pseudomonas aeruginosa and group Klebsiella pneumonia in the presence of phenol concentrations (up to 1000 mg/L) showed 100% germination. The seeds in the control without bacteria and treated with all phenol concentrations appear dark in color. The dark seeds increase as phenol concentration increases, as shown in Table 4 and Figure 7. Thus, soaked soybean seeds in 250 up to 1000 mg/L phenol stress significantly decreased (p < 0.05) the percentage of germination and the seedling growth rate in the soybean seedlings and inhibited the germination by 100%. However, using the bacterial suspension of P. aeruginosa and K. variicola significantly increased (p < 0.05) the percentage of germination and the seedling growth rate in the soybean seedlings in the presence of phenol stress.
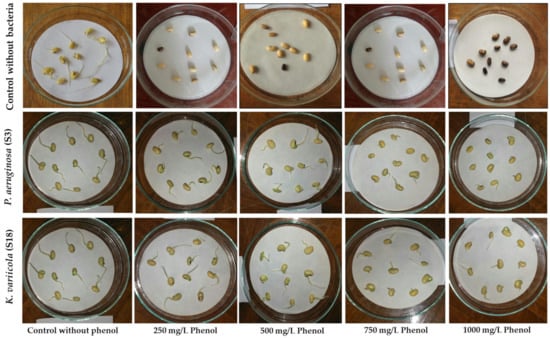
Figure 7.
Effect of Pseudomonas aeruginosa (S3) and Klebsiella variicola (S18) on soybean seed germination after bioremediation and detoxification of water contaminated with different concentrations of phenol (0.0, 250, 500, 750, and 1000 mg/L).

Table 4.
Effect of Pseudomonas aeruginosa, Klebsiella variicola, and Klebsiella pneumonia on germination and seedling growth (Mean ± SD) of different beans at phenol concentration regimes.
In a saline-alkaline environment, K. variicola promoted the development of maize seedlings effectively [65]. In our experiment, primary roots reached a maximum length (2.83 ± 0.65 cm) in the control and 2.89 ± 0.32 in the K. variicola. The primary roots in the P. aeruginosa group range from 2.83 ± 0.45 cm at 250 mg/L to 2.41 ± 0.24 cm at 1000 mg/L. As discovered in spinach, phenol can modify mitochondrial and chloroplast membranes, impeding the energy transfer required for ion transport. Singh et al. [62] demonstrated that 100 µM of caffeic acid applied to the soil completely inhibited the germination of mung bean seedlings. Plant height and the fresh weight of shoots dropped as phenolic chemical concentrations in the soil increased. At doses of 50 and 100 mg/L, phenol, coumarin, ferulic acid, and naringenin inhibited soybean seed germination and the development of seed-borne fungus significantly. P. aeruginosa (S3) and K. variicola (S18) have the capacity to remove phenol. Based on a separate investigation, K. variicola may generate indole acetic acid. According to the studies, indole acetic acid is one of the primary hormones regulating the growth and development of plant cells, including cell proliferation and division, organ development, and directional movement [66,67].
K. variicola played a role in indole acetic acid, acetoin, ammonia, phosphorus, potassium production, and nitrogen fixation. A high level of colonization was observed in the rhizosphere soil of maize seedlings. Following the application of K. variicola in neutral and saline-alkali soils, the nutrient composition of the rhizosphere soil of maize seedlings increased in varying degrees, more notably in saline-alkali soil. The content of organic matter, alkali-hydrolyzable nitrogen, available phosphorus, available potassium, alkaline phosphatase, sucrase, urease, and catalase increased by 64.22%, 117.39%, 175.64%, 28.63%, 146.08%, 76.77%, 86.60%, and 45.29%, respectively, in saline-alkali soil [65]. Thus, K. variicola could be a promised strain with more biological functions, such as removing phenol from our study.
3. Material and Methods
3.1. Methods
The minimal salt medium (MSM) and Luria–Bertani (LB) medium were used in this study. MSM consisted of 10 g of NaCl, 0.42 g of MgSO4.7H2O, 0.29 g KCl, 0.83 g KH2PO4, 1.25 g Na2HPO4, 0.42 g NaNO3, and 20g agar in one liter of distilled water, while the LB medium consisted of NaCl (5g), tryptone (10 g), and yeast extract (5 g) g/L [34]. The media was autoclaved for 15 min at 121 °C and 15 pounds per square inch (psi) and then cooled. The MSM medium was supplemented with phenol or other nitrogen sources. The pH was adjusted (pH 6.8–7.0), and various phenol concentrations (0.5–2 g/L) were added. A 0.45 µm filter syringe was used to add 50 µg/L phenol directly from a stock of phenol to medium working solutions. On a rotary shaker (120 rpm), cells were incubated in flasks at 35 °C at 120 rpm. Periodically, samples were collected to determine bacterial growth and phenol concentration.
3.2. Sample Collection and Sources of Bacterial Isolates
The phenol-degrading bacterial isolates were obtained from sewage sludge (SS) at SWTP, Egypt. Before usage, the sludge was kept in sealed containers at 4 °C. The sample collection occurred between May and October of 2021. First, 10 g of SS were weighed into 90 mL of sterile peptone saline (8.5 g/L NaCl) to generate 10−1 dilution. The mixture was agitated for one minute to facilitate the separation of microbial cells from sludge particles. One mL (1 mL) from the 10−1 dilution was transferred to a sterile test tube containing 9 mL of sterile physiological saline to prepare a 10−2 dilution. One mL was transferred into a 9 mL sterile physiological saline to achieve the 10−3 (always use a pipette to transfer). The content of the 10−3 dilution test tube was shaken, and dilution continued until the 10−8 dilution was obtained. After that, an aliquot of 0.1 mL of 10−5, 10−6, 10−7, and 10−8 dilutions were plated in triplicates on mineral salt agar (MSA) supplemented with 500 mg/mL of phenol. The plates were then incubated at 30 °C for 24 h for bacterial growth. The pure colonies have been taken and purified onto LB agar plates.
3.3. Isolation of Phenol-Degrading Bacteria
Each bacterial culture (10 mL) from SS1 up to SS20 samples was added aseptically to 90 mL of MSM media supplemented with different levels of phenol (0.5–2 g/L) and incubated in a shaking incubator (120 rpm; 35 °C) [68]. The optical density of the bacterial cultures was examined at a wavelength of 600 nm throughout the incubation period. After 2–3 days of a noticeable increase in optical density (OD), 10 mL of the culture broth was transferred to a new shaking flask containing 90 mL of fresh medium. The final culture broth was serially diluted and distributed on LB agar and MSM agar plates containing 0.5 g/L phenol. After 3–4 days of incubation at 35 °C, colonies on agar plates were harvested and subcultured repeatedly to obtain pure cultures. A microscopic examination confirmed the cleanliness of the artifact. By optical microscopy, the morphological features of the isolated colonies were examined. Gram staining, motility, starch hydrolysis, and gelatinase activity are common physio-chemical characteristics of bacterial strains that were carefully determined using Bergey’s manual [69]. As indicated earlier, the indole, methyl red, and hydrogen sulfide tests were also conducted [70]. All bacterial isolates were selected for phenol removal assays at various phenol concentrations (0.0, 0.5, 1, 1.5, and 2 g/L) by Klibanov et al. [71]. S3, S10, and S18 isolates were stored in LB agar slants at 4 °C and in LB supplemented with 20% sterile glycerol at 20 °C.
3.4. Characterization of the Most Effective Bacterial Isolates by 16S rDNA and Phylogenetic Analysis
The genomic DNA of bacterial cells was recovered by phenol-chloroform after treating them with achromopeptidase, lysostaphin, and mutanolysin [72]. DNA was separated by electrophoresis in a 0.7 or 1.5% agarose gel with TBE buffer, stained with ethidium bromide, and seen under ultraviolet light. DNA fragment sizes were determined by comparing them to a 50 bp ladder (Fermentas). PFf 5′ AGGGATGTATTTATTAGATAAAAAATCAA 3′ and PFr 5′ CGCAGTAGTTTCTTCAGTAAATC 3′ are the primer sequences used for PCR amplification. 16S rRNA gene was amplified by PCR and then partially sequenced by Mannerová et al. [73]. The gene sequence was rebuilt using RNAmmer version 1.2 from whole-genome shotgun (WGS) data [74] and compared to other Pseudomonas and Klebsiella species. In the EzBioCloud database, a global alignment technique allows pairwise sequence alignment and the creation of similarity values [46]. The evolutionary history was estimated using the neighbor-joining and maximum likelihood techniques with 1000 bootstrap values from MEGA version 7 [75].
3.5. Preparing for Bacterial Inoculation
Each strain of P. aeruginosa (S3), K. pneumoniae (S10), and K. variicola (S18) was inoculated into LB broth, and the culture flasks were incubated at 35 °C and 120 rpm for 24 h in a shaker incubator. The bacterial culture was centrifuged for 15 min at 6000× g; the residue was obtained, washed, and resuspended in sterilized distillate water. This procedure was carried out three times. The sample was diluted to 1 × 102, 1 × 104, and 1 × 106 CFU/mL, following the experiment’s specifications.
3.6. Evaluation of the Three Bacterial Isolates on Phenol Degradation
Referring to the one variable at a time (OVAT) technique, all factors were held constant in these experiments, with only the targeted variable being changed to optimize process variables. Briefly, the optical density of prepared cultures of each isolate, S3, S10, and S18, were adjusted at (1.0) at a wavelength of 600 nm. The inocula (2%) were inoculated into the phenol (1 g/L)-MSM medium as the only carbon source. The bacterial growth was monitored at 0, 1, 2, 3, and 4 days. Additionally, phenol degradation was recorded after four days of incubation in a shaking incubator (120 rpm) at different conditions of temperatures (25, 30, 35, and 40 °C), pH values (6, 7, and 8), and different nitrogen sources in growth medium MSM that contain peptone, yeast extract, urea extract, NaNO3, KNO3, and NH4Cl at a concentration of 2 g/L. Buffer solutions with different pH ranges, i.e., acetate buffer for pH 6.0, phosphate buffer for pH 6.5, 7.0, and 7.5, and Tris-HCl buffer for pH 9 were used to achieve different pH values [31].
The phenol concentrations were measured at 520 nm using 4-amino antipyrine following American Public Health Association-recommended procedures [6]. In summary, 0.3 mL of 2% aqueous 4-amino antipyrine solution and 1 mL of 2 N NH4OH were added to a 5 mL sample. After adequately mixing the contents, 1 mL of 2% K3FeCN6 is added. At pH 10, the OH group reacts with 4-amino antipyrine and K4Fe(CN)6 to produce a purple-red color. Quantitative analysis of phenol degradation was conducted by measuring the phenol content in the supernatant of each sample and calculating degradation using the following equation
% Degradation = (A − B)/A × 100
The phenol content was determined by comparing the absorbance at 520 nm to the phenol standard curve. Where A is the initial concentration of phenol (Zero h. sample), and B is the phenol concentration in the sample taken at various time intervals.
3.7. Phenol Degradation Assay by HPLC Analysis
HPLC (Agilent 12,000 series) detected the phenol degradation in MSM by three new bacterial strains. The separation column was C18 column (Eclipse Plus, 4.6 mm × 100 mm i.d., 3.5 m), and the mobile phase was composed of 0.01% formic acid in water (A) and 0.01% formic acid in acetonitrile (B) with a flow rate of 1 mL/min. The mobile phase was sequentially programmed as follows: 0 min (85% A); 0–5 min (50% A); 5–6 min (10% A); 6–7 min (10% A); 7–7.5 min (85% A); and 7.5–10 min (85% A). The standard solutions and samples were injected at a volume of 40 µL, the column temperature was kept at room temperature, and the phenol degradation rate was monitored at 280 nm [76]. The elutes were identified by comparing their retention time to the reference compound.
3.8. Effect of Pseudomonas Aeruginosa (S3) and Klebsiella Variicola (S18) on Soybean Seed Germination
Bean, soy, and faba bean seeds (270 days old) of the same size and without damage were picked, cleaned, soaked, and sterilized with a 10% sodium hypochlorite solution for 10 min; then, the seeds were immersed for six hours in distilled water, then washed several times and dried. The seeds were then placed in an incubator set at 25 °C. A piece of filter paper was used as a negative control, and sterile water was gently poured over it until it was slightly moist. The seeds covered with bacterial suspension were used as the positive control group. The bottom of the germination plate (20 cm) was lined with filter paper. Ten soaking seeds were distributed uniformly on each plate (20 cm). A piece of filter paper was put on top of the seeds and evenly coated with 3 mL of each bacterial suspension grown in MSM containing 1g/L phenol (1.4 × 108 CFU/mL). The germination box was covered with a lid and incubated at 25 °C for 24 h to speed up germination in the dark.
3.9. Statistical Evaluation
Experiments to determine the biodegradation capability of the studied strain were conducted in triplicate, and the findings presented in the figures and tables were the means. The figures shown display the mean and standard deviation values for three replicates. The means of triplicate data were analyzed by ANOVA test, and the LSD test was used as a post-doc test to compare the means.
4. Conclusions
The phenol-degradable bacterial strains of interest were isolated and identified from sewage sludge. The 16S rDNA sequencing and the phylogenetic analysis revealed that the isolates were found to be Pseudomonas aeruginosa (S3) ON88415.1, Klebsiella pneumoniae (S10) ON880414.1, and Klebsiella variicola (S18) ON880418.1. P. aeruginosa (S3), K. pneumoniae (S10), and K. variicola (S18) can thrive in a liquid medium with varying amounts of phenol as the only carbon and energy source (0, 250, 500, 750, 1000, 1250, 1500, and 2000 mg/L). The strains could digest 75.66% of phenol (1000 mg/L while these strains were resistant and tolerant in the presence of phenol up to 2000 mg/L. The ideal growing conditions for each bacterium to degrade phenol were 35 °C, pH 7.5, and the best nitrogen source was peptone and yeast extract. Because native microbial species in polluted settings were more adaptable than non-indigenous microorganisms, their dominance aided the bioremediation of phenol-contaminated ecosystems. K.variicola consummated phenol through removal from media to promote soybean seedlings under phenol stress. K. variicola (S18) strains are promising and might be employed for the bioremediation of phenol-contaminated regions. K. variicola could be a novel strain with more biological function which is friendly to the ecosystem. K. variicola could be applied for the integrated biological treatment of wastewater for the removal of phenol from water.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28031203/s1, Figure S1: HPLC analysis for phenol-degrading bacterial strains of Pseudomonas aeruginosa (S3), Klebsiella pneumoniae (S10), and Klebsiella variicola (S18).
Author Contributions
Conceptualization, S.A.M., S.S.S., H.M.A. and H.I.A.E.-F.; methodology, S.A.M., S.Y.A.Q., S.S.S., H.M.A., M.R., F.S.A., M.F.A., D.A.A.-Q., E.A.M. and H.I.A.E.-F.; software, S.A.M.; validation, S.Y.A.Q., S.S.S., H.M.A., M.R., M.F.A., D.A.A.-Q., F.S.A., E.A.M. and H.I.A.E.-F.; formal analysis, S.S.S. and H.M.A.; investigation, H.I.A.E.-F.; resources, M.F.A.; data curation, D.A.A.-Q. and E.A.M.; writing—original draft preparation, S.A.M. and H.I.A.E.-F.; writing—review and editing, S.A.M., H.M.A., F.S.A. and H.I.A.E.-F.; visualization, S.A.M.; funding acquisition, M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Umm Al-Qura University with grant code (23UQU4290559DSR008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank the Deanship of scientific research at Umm Al-Qura University for supporting this work by grant code (23UQU4290559DSR008).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Sample Availability
Samples of the compounds and identified strains are available from the authors.
References
- Deng, T.; Wang, H.; Yang, K. Phenol biodegradation by isolated Citrobacter strain under hypersaline conditions. Water Sci. Technol. 2018, 77, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Govindarajalu, K.; Govindarajalu, K. Industrial effluent and health status: A case study of Noyyal river basin. In Proceedings of the Third International Conference on Environment and Health, Chennai, India, 15–17 December 2003; pp. 150–157. [Google Scholar]
- Naresh, B.; Honey, P.; Vaishali, S. Biodegradation of phenol by a bacterial strain isolated from a phenol contaminated site in India. Res. J. Environ. Sci. 2012, 1, 46–49. [Google Scholar]
- McCall, I.C.; Betanzos, A.; Weber, D.A.; Nava, P.; Miller, G.W.; Parkos, C.A. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol. Appl. Pharmacol. 2009, 241, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Brown, M.T.; Han, T. Phenol toxicity to the aquatic macrophyte Lemna paucicostata. Aquat. Toxicol. 2012, 106, 182–188. [Google Scholar] [CrossRef]
- Rice, E.W.; Baird, R.B.; Eaton, A.D.; Clesceri, L.S. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2012; Volume 10. [Google Scholar]
- Basha, K.M.; Rajendran, A.; Thangavelu, V. Recent advances in the biodegradation of phenol: A review. Asian J. Exp. Biol. Sci. 2010, 1, 219–234. [Google Scholar]
- Tay, S.T.-L.; Moy, B.Y.-P.; Maszenan, A.M.; Tay, J.-H. Comparing activated sludge and aerobic granules as microbial inocula for phenol biodegradation. Appl. Microbiol. Biotechnol. 2005, 67, 708–713. [Google Scholar] [CrossRef]
- Carmona, M.; De Lucas, A.; Valverde, J.L.; Velasco, B.; Rodriguez, J.F. Combined adsorption and ion exchange equilibrium of phenol on Amberlite IRA-420. Chem. Eng. J. 2006, 117, 155–160. [Google Scholar] [CrossRef]
- Lazarova, Z.; Boyadzhieva, S. Treatment of phenol-containing aqueous solutions by membrane-based solvent extraction in coupled ultrafiltration modules. Chem. Eng. J. 2004, 100, 129–138. [Google Scholar] [CrossRef]
- Lin, S.; Chuang, T. Combined treatment of phenolic wastewater by wet air oxidation and activated sludge. Toxicol. Environ. Chem. 1994, 44, 243–258. [Google Scholar] [CrossRef]
- Wu, Z. Study on degrading phenol by immobilized Ralstonia metallidurans CH34. Microbiology 1992, 32, 31–36. [Google Scholar]
- Yan, J.; Jianping, W.; Jing, B.; Daoquan, W.; Zongding, H. Phenol biodegradation by the yeast Candida tropicalis in the presence of m-cresol. Biochem. Eng. J. 2006, 29, 227–234. [Google Scholar] [CrossRef]
- Zhai, Z.; Wang, H.; Yan, S.; Yao, J. Biodegradation of phenol at high concentration by a novel bacterium: Gulosibacter sp. YZ4. J. Chem. Technol. Biotechnol. 2012, 87, 105–111. [Google Scholar] [CrossRef]
- Loh, K.-C.; Chua, S.-S. Ortho pathway of benzoate degradation in Pseudomonas putida: Induction of meta pathway at high substrate concentrations. Enzyme Microb. Technol. 2002, 30, 620–626. [Google Scholar] [CrossRef]
- Geng, A.; Soh, A.E.W.; Lim, C.J.; Loke, L.C.T. Isolation and characterization of a phenol-degrading bacterium from an industrial activated sludge. Appl. Microbiol. Biotechnol. 2006, 71, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Tuah, P.M.; Rashid, N.A.A.; Salleh, M.M. Degradation Pathway of Phenol Through-Cleavage by RETL-Cr1. Borneo Sci. 2009, 24, 432–567. [Google Scholar]
- Nair, C.I.; Jayachandran, K.; Shashidhar, S. Biodegradation of phenol. Afr. J. Biotechnol. 2008, 7, 25. [Google Scholar]
- Abu-Dief, A.M.; El-Metwaly, N.M.; Alzahrani, S.O.; Alkhatib, F.; Abumelha, H.M.; El-Dabea, T.; Ali El-Remaily, M.A.E.A.A. Structural, conformational and therapeutic studies on new thiazole complexes: Drug-likeness and MOE-simulation assessments. Res. Chem. Intermed. 2021, 47, 1979–2002. [Google Scholar] [CrossRef]
- Abumelha, H.M.; Alkhatib, F.; Alzahrani, S.; Abualnaja, M.; Alsaigh, S.; Alfaifi, M.Y.; Althagafi, I.; El-Metwaly, N. Synthesis and characterization for pharmaceutical models from Co (II), Ni (II) and Cu (II)-thiophene complexes; apoptosis, various theoretical studies and pharmacophore modeling. J. Mol. Liq. 2021, 328, 115483. [Google Scholar] [CrossRef]
- Adam, M.S.S.; Abu-Dief, A.M.; Makhlouf, M.; Shaaban, S.; Alzahrani, S.O.; Alkhatib, F.; Masaret, G.S.; Mohamed, M.A.; Alsehli, M.; El-Metwaly, N.M. Tailoring, structural inspection of novel oxy and non-oxy metal-imine chelates for DNA interaction, pharmaceutical and molecular docking studies. Polyhedron 2021, 201, 115167. [Google Scholar] [CrossRef]
- Xu, N.; Qiu, C.; Yang, Q.; Zhang, Y.; Wang, M.; Ye, C.; Guo, M. Analysis of Phenol Biodegradation in Antibiotic and Heavy Metal Resistant Acinetobacter lwoffii NL1. Front. Microbiol. 2021, 10, 2670. [Google Scholar] [CrossRef]
- Kujur, R.R.A.; Das, S.K. Pseudomonas phenolilytica sp. nov., a novel phenol-degrading bacterium. Arch. Microbiol. 2022, 204, 320. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ghosh, A. Fractional-order identification and synthesis of equivalent circuit for electrochemical system based on pulse voltammetry. In Fractional-Order Design; Elsevier: Amsterdam, The Netherlands, 2022; pp. 373–402. [Google Scholar]
- Malhotra, M.; Gupta, D.; Sahani, J.; Singh, S. Microbial Degradation of Phenol and Phenolic Compounds. In Recent Advances in Microbial Degradation; Springer: Berlin/Heidelberg, Germany, 2021; pp. 297–312. [Google Scholar]
- Liu, Z.; Xie, W.; Li, D.; Peng, Y.; Li, Z.; Liu, S. Biodegradation of phenol by bacteria strain Acinetobacter calcoaceticus PA isolated from phenolic wastewater. Int. J. Environ. Res. Public Health 2016, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, S.; Kumar, S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem. Eng. J. 2005, 22, 151–159. [Google Scholar] [CrossRef]
- Arutchelvan, V.; Kanakasabai, V.; Nagarajan, S.; Muralikrishnan, V. Isolation and identification of novel high strength phenol degrading bacterial strains from phenol-formaldehyde resin manufacturing industrial wastewater. J. Hazard. Mater. 2005, 127, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Nogina, T.; Fomina, M.; Dumanskaya, T.; Zelena, L.; Khomenko, L.; Mikhalovsky, S.; Podgorskyi, V.; Gadd, G.M. A new Rhodococcus aetherivorans strain isolated from lubricant-contaminated soil as a prospective phenol-biodegrading agent. Appl. Microbiol. Biotechnol. 2020, 104, 3611–3625. [Google Scholar] [CrossRef]
- Peng, R.; Yang, G.; Du, Y. Immobilized Mutants M1 of Rhodococcus ruber SD3 and Its Application in Phenol Degradation. CN Patent CN103160491A, 15 April 2013. [Google Scholar]
- Aisami, A.; Yasid, N.; Johari, W.; Ahmad, S.; Shukor, M. Effect of temperature and ph on phenol biodegradation by a newly identified Serratia sp. AQ5-03. Open J. Biosci. Res. 2020, 1, 28–43. [Google Scholar] [CrossRef]
- Shawabkeh, R.; Khleifat, K.M.; Al-Majali, I.; Tarawneh, K. Rate of biodegradation of phenol by Klebsiella oxytoca in minimal medium and nutrient broth conditions. Bioremediat. J. 2007, 11, 13–19. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Gupta, A.; Kaur, A.; Malik, D. Remediation of phenol using microorganisms: Sustainable way to tackle the chemical pollution menace. Curr. Org. Chem. 2018, 22, 370–385. [Google Scholar] [CrossRef]
- Sachan, P.; Madan, S.; Hussain, A. Isolation and screening of phenol-degrading bacteria from pulp and paper mill effluent. Appl. Water Sci. 2019, 9, 100. [Google Scholar] [CrossRef]
- Ahmad, N.; Ahmed, I.; Shahzad, A.; Khalid, N.; Mehboob, F.; Ahad, K.; Muhammad Ali, G. Molecular identification and characterization of Pseudomonas sp. NCCP-407 for phenol degradation isolated from industrial waste. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 341–346. [Google Scholar] [CrossRef]
- Ahmad, S.A.; Shamaan, N.A.; Arif, N.M.; Koon, G.B.; Shukor, M.Y.A.; Syed, M.A. Enhanced phenol degradation by immobilized Acinetobacter sp. strain AQ5NOL 1. World J. Microbiol. Biotechnol. 2012, 28, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Auma, E.O. Anaerobic Co-Digestion of Water Hyacinth (Eichhornia crassipes) with Ruminal Slaughterhouse Waste under Mesophilic Conditions. Ph.D. Thesis, University of Nairobi, Nairobi, Kenya, 2020. [Google Scholar]
- Heilbuth, N.M.; Linardi, V.R.; Monteiro, A.S.; da Rocha, R.A.; Mimim, L.A.; Santos, V.L. Estimation of kinetic parameters of phenol degradation by bacteria isolated from activated sludge using a genetic algorithm. J. Chem. Technol. Biotechnol. 2015, 90, 2066–2075. [Google Scholar] [CrossRef]
- Samimi, M.; Shahriari Moghadam, M. Phenol biodegradation by bacterial strain O-CH1 isolated from seashore. Glob. J. Environ. Sci. Manag. 2020, 6, 109–118. [Google Scholar]
- Tengku-Mazuki, T.A.; Subramaniam, K.; Zakaria, N.N.; Convey, P.; Khalil, K.A.; Lee, G.L.Y.; Zulkharnain, A.; Shaharuddin, N.A.; Ahmad, S.A. Optimization of phenol degradation by Antarctic bacterium Rhodococcus sp. Antarct. Sci. 2020, 32, 486–495. [Google Scholar] [CrossRef]
- Sreeja Mole, S.S.; Vijayan, D.; Anand, M.; Ajona, M.; Jarin, T. Biodegradation of P-nitro phenol using a novel bacterium Achromobacter denitrifacians isolated from industrial effluent water. Water Sci. Technol. 2021, 84, 3334–3345. [Google Scholar]
- Kotresha, D.; Vidyasagar, G. Degradation of phenol by novel strain Pseudomonas aeruginosa MTCC 4997 isolated from petrochemical industrial effluent. Int. J. Microbiol. Res. Technol. 2014, 2, 7–15. [Google Scholar]
- Rajani, V. Microbial degradation of phenol: A review. Int. J. Res. Rev. 2015, 2, 46–54. [Google Scholar]
- Stover, C.K.; Pham, X.Q.; Erwin, A.; Mizoguchi, S.; Warrener, P.; Hickey, M.; Brinkman, F.; Hufnagle, W.; Kowalik, D.; Lagrou, M. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Mahiudddin, M.; Fakhruddin, A. Degradation of phenol via meta cleavage pathway by Pseudomonas fluorescens PU1. ISRN Microbiol. 2012, 2012, 741820. [Google Scholar] [CrossRef]
- Shourian, M.; Noghabi, K.A.; Zahiri, H.S.; Bagheri, T.; Karbalaei, R.; Mollaei, M.; Rad, I.; Ahadi, S.; Raheb, J.; Abbasi, H. Efficient phenol degradation by a newly characterized Pseudomonas sp. SA01 isolated from pharmaceutical wastewaters. Desalination 2009, 246, 577–594. [Google Scholar] [CrossRef]
- Kadiyala, V.; Spain, J.C. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 1998, 64, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Vasantharaj, K.; Jerold, M.; Deepanraj, B.; Velan, M.; Sivasubramanian, V. Assessment of a sulfidogenic system utilizing microalgal biomass of Chlorella pyrenoidosa as an electron donor: Taguchi based grey relational analysis. Int. J. Hydrogen Energy 2017, 42, 26545–26554. [Google Scholar] [CrossRef]
- Jain, R.K.; Dreisbach, J.H.; Spain, J.C. Biodegradation of p-nitrophenol via 1, 2, 4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 1994, 60, 3030–3032. [Google Scholar] [CrossRef]
- Zylstra, G.J.; Bang, S.-W.; Newman, L.M.; Perry, L.L. Microbial degradation of mononitrophenols and mononitrobenzoates. In Biodegradation of Nitroaromatic Compounds and Explosives; Lewis Publishers: Boca Raton, FL, USA, 2000; pp. 145–160. [Google Scholar]
- Sivasubramanian, S.; Namasivayam, S.K.R. Phenol degradation studies using microbial consortium isolated from environmental sources. J. Environ. Chem. Eng. 2015, 3, 243–252. [Google Scholar] [CrossRef]
- Mishra, V.K.; Kumar, N. Microbial degradation of phenol: A review. J. Water Pollut. Purif. Res. 2017, 4, 17–22. [Google Scholar]
- Shahryari, S.; Zahiri, H.S.; Haghbeen, K.; Adrian, L.; Noghabi, K.A. High phenol degradation capacity of a newly characterized Acinetobacter sp. SA01: Bacterial cell viability and membrane impairment in respect to the phenol toxicity. Ecotoxicol. Environ. Saf. 2018, 164, 455–466. [Google Scholar] [CrossRef]
- Sarnaik, S.; Kanekar, P. Bioremediation of colour of methyl violet and phenol from a dye-industry waste effluent using Pseudomonas spp. isolated from factory soil. J. Appl. Bacteriol. 1995, 79, 459–469. [Google Scholar] [CrossRef]
- Shahriari, M.M.; Safaei, N.; Ebrahimipour, G. Optimization of phenol biodegradation by efficient bacteria isolated from petrochemical effluents. Glob. J. Environ. Sci. Manag. 2016, 2, 249–256. [Google Scholar]
- Khleifat, K.M. Biodegradation of phenol by Ewingella americana: Effect of carbon starvation and some growth conditions. Process Biochem. 2006, 41, 2010–2016. [Google Scholar] [CrossRef]
- Lin, L.-X.; Liu, H.; Zhou, N.-Y. MhbR, a LysR-type regulator involved in 3-hydroxybenzoate catabolism via gentisate in Klebsiella pneumoniae M5a1. Microbiol. Res. 2010, 165, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Aksu, Z.; Gönen, F. Biosorption of phenol by immobilized activated sludge in a continuous packed bed: Prediction of breakthrough curves. Process Biochem. 2004, 39, 599–613. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Alkhatib, F.M.; Alzahrani, S.O.; Shafi, M.E.; Abdel-Hamid, S.E.; Taha, T.F.; Ahmed, N.H. Im-pact of mycogenic zinc nanoparticles on performance, behavior, immune response, and microbial load in Oreo-chromis niloticus. Saudi J. Biol. Sci. 2021, 28, 4592–4604. [Google Scholar] [CrossRef]
- El-Beeh, M.E.; El-Badawi, A.A.; Amin, A.H.; Qari, S.H.; Ramadan, M.F.; Filfilan, W.M.; El-Sayyad, H.I. Anti-aging trait of whey protein against brain damage of senile rats. J. Umm Al-Qura Uni. Appl. Sci. 2022, 8, 8–20. [Google Scholar] [CrossRef]
- Rodgers-Vieira, E.A.; Zhang, Z.; Adrion, A.C.; Gold, A.; Aitken, M.D. Identification of anthraquinone-degrading bacteria in soil contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2015, 81, 3775–3781. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Wang, W.; Zhu, M.; Liao, Y.; Yao, Y. Screening and characterization of aroma yeast with thermo-tolerant and salt-tolerance. Food Ferment. Ind. 2016, 42, 92–96. [Google Scholar]
- Veenagayathri, K.; Vasudevan, N. Effect of pH, nitrogen sources and salts on the degradation of phenol by the bacterial consortium under saline conditions. Int. J. Biotechnol. Biochem. 2010, 6, 783–792. [Google Scholar]
- Yang, L.; Yang, K. Biological function of Klebsiella variicola and its effect on the rhizosphere soil of maize seedlings. PeerJ 2020, 8, e9894. [Google Scholar] [CrossRef]
- Toscano, S.; Trivellini, A.; Cocetta, G.; Bulgari, R.; Francini, A.; Romano, D.; Ferrante, A. Effect of preharvest abiotic stresses on the accumulation of bioactive compounds in horticultural produce. Front. Plant Sci. 2019, 10, 1212. [Google Scholar] [CrossRef]
- Timmusk, S.; Abd El-Daim, I.A.; Copolovici, L.; Tanilas, T.; Kännaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenström, E.; Niinemets, Ü. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 2014, 9, e96086. [Google Scholar] [CrossRef]
- Watanabe, K.; Teramoto, M.; Harayama, S. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 1999, 65, 2813–2819. [Google Scholar] [CrossRef]
- Kloos, W.E.; Tornabene, T.G.; Schleifer, K.H. Isolation and characterization of micrococci from human skin, including two new species: Micrococcus lylae and Micrococcus kristinae. Int. J. Syst. Evol. Microbiol. 1974, 24, 79–101. [Google Scholar] [CrossRef]
- Lanyi, B. Classical and rapid identification methods for medically important bacteria. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 1988; Volume 19, pp. 1–67. [Google Scholar]
- Klibanov, A.M.; Alberti, B.; Morris, E.; Felshin, L. Enzymatic removal of toxic phenols and anilines from waste waters. J. Appl. Biochem. 1980, 2, 5. [Google Scholar]
- Freitas, D.B.; Reis, M.P.; Lima-Bittencourt, C.I.; Costa, P.S.; Assis, P.S.; Chartone-Souza, E.; Nascimento, A. Genotypic and phenotypic diversity of Bacillus spp. isolated from steel plant waste. BMC Res. Notes 2008, 1, 92. [Google Scholar] [CrossRef]
- Obaid, R.J. Synthesis and biological evaluation of some new imidazo[1,2-c]pyrimido [5,4-e]pyrimidin-5-amine de-rivatives. J. Umm Al-Qura Uni. Appl. Sci. 2021, 7, 16–22. [Google Scholar]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saad, A.M.; Mohamed, A.S.; El-Saadony, M.T.; Sitohy, M.Z. Palatable functional cucumber juices supplemented with polyphenols-rich herbal extracts. LWT Food Sci. Technol. 2021, 148, 111668. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).