A Review on Recent Approaches on Molecular Docking Studies of Novel Compounds Targeting Acetylcholinesterase in Alzheimer Disease
Abstract
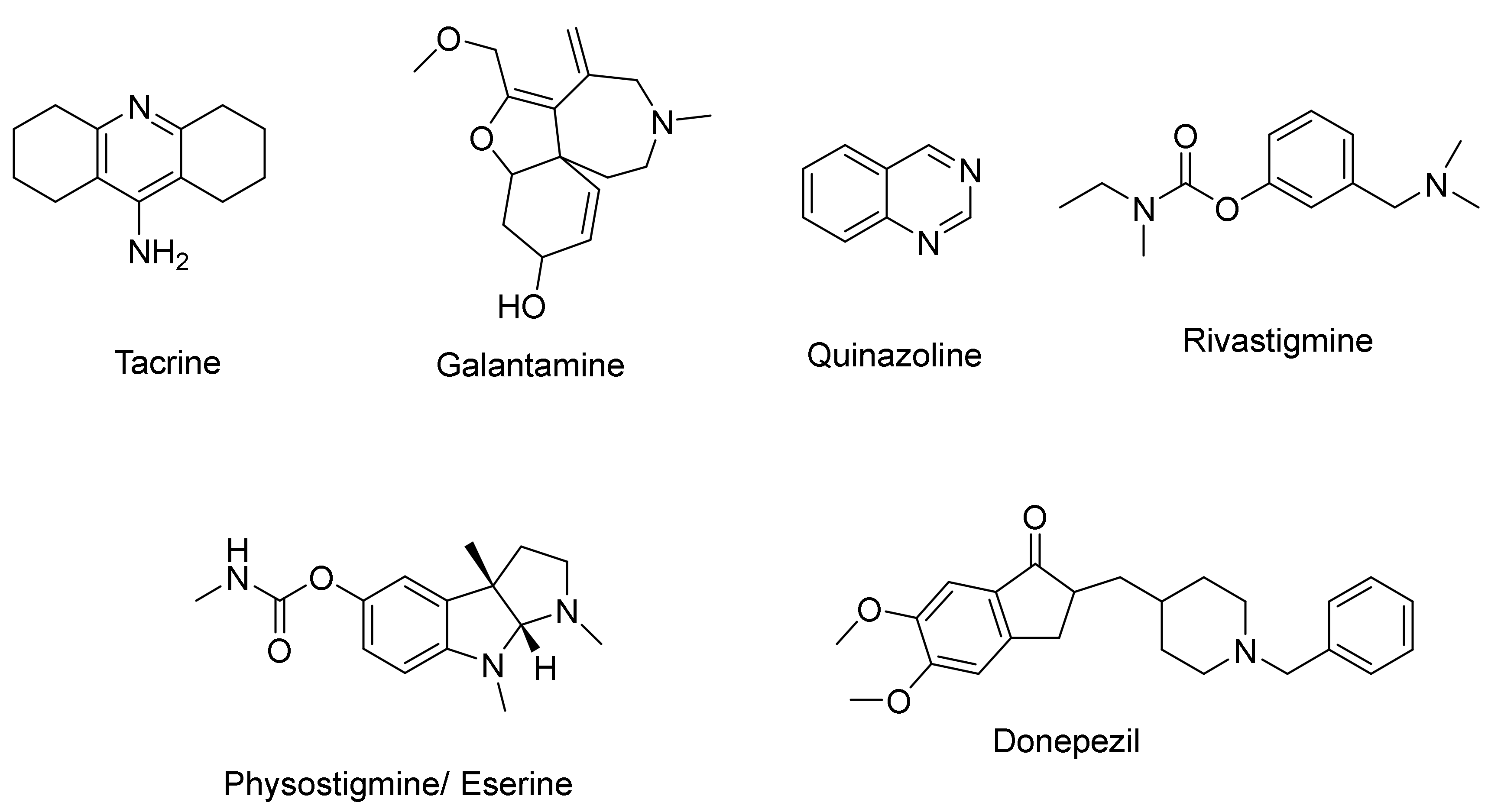
1. Introduction
2. Inhibition
2.1. Coumarin Derivatives
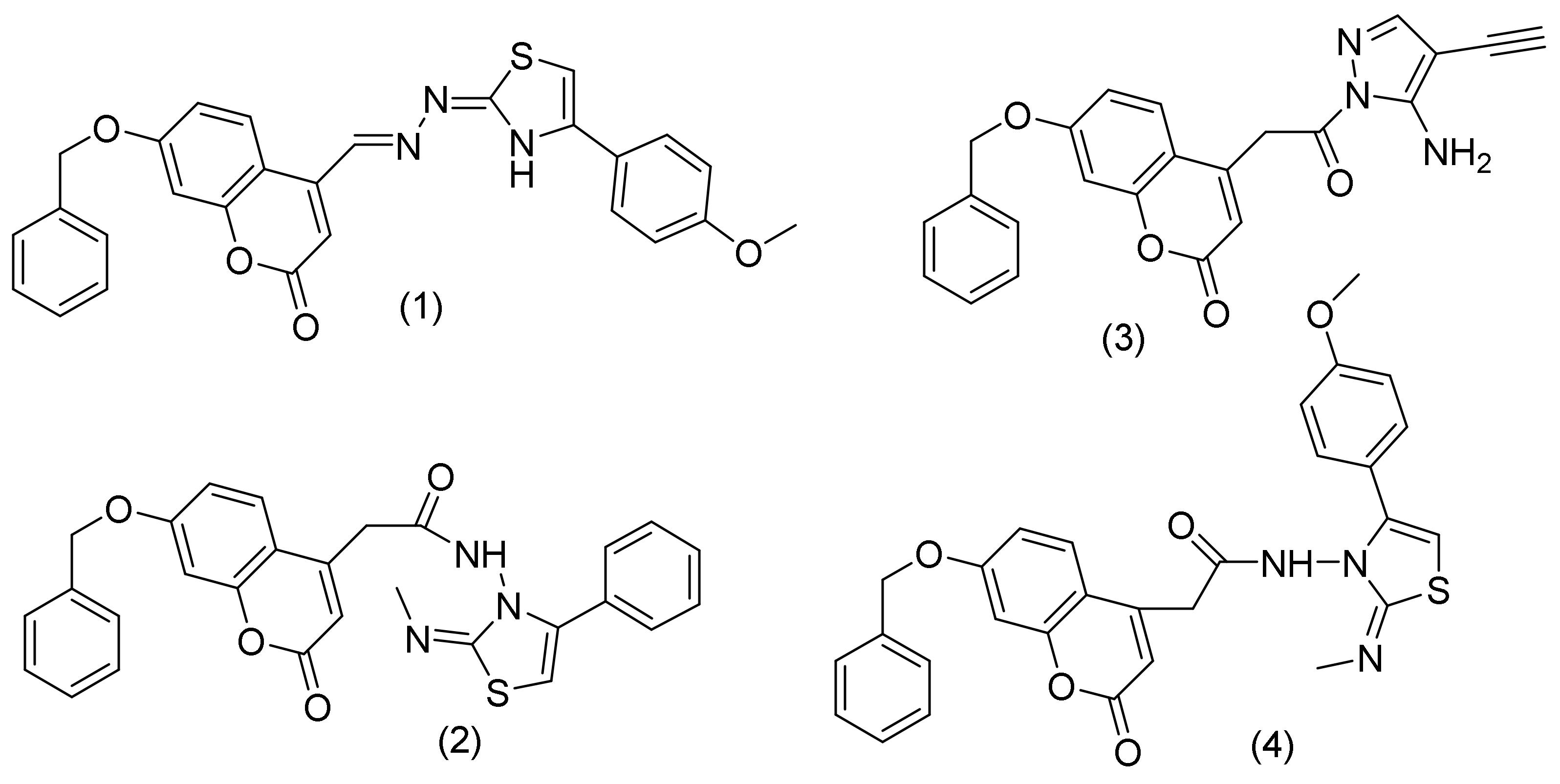
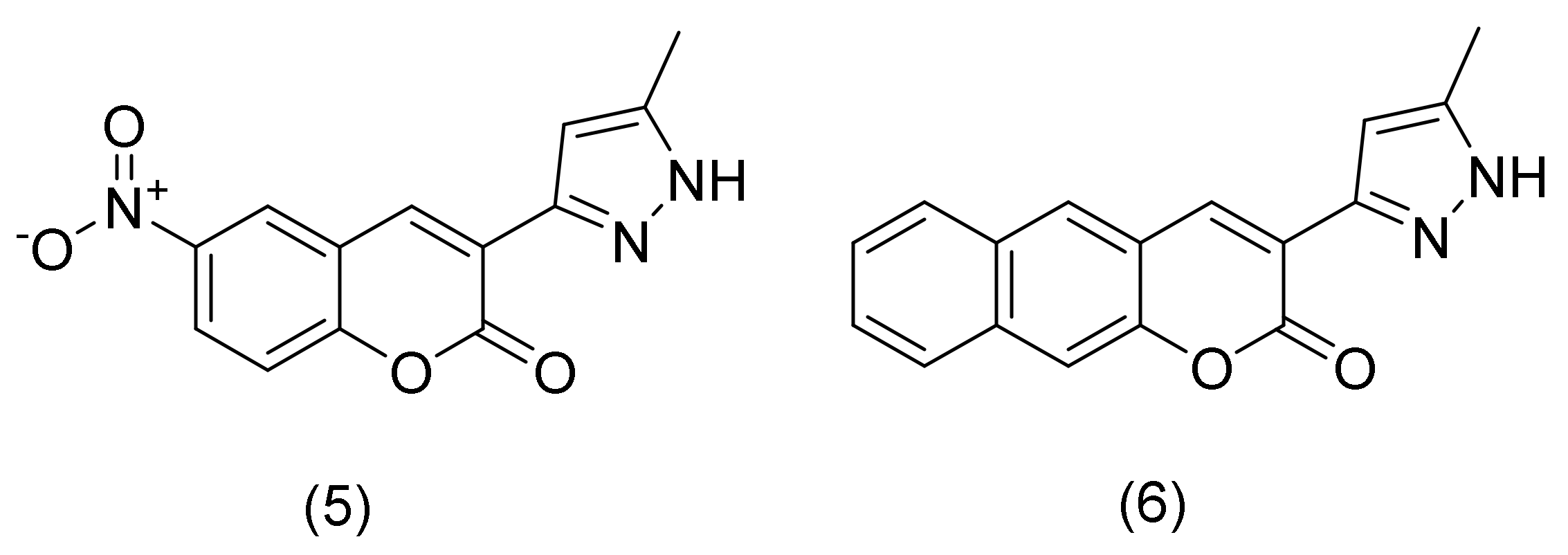
2.1.1. Coumarin-Pyrazoles Hybrids
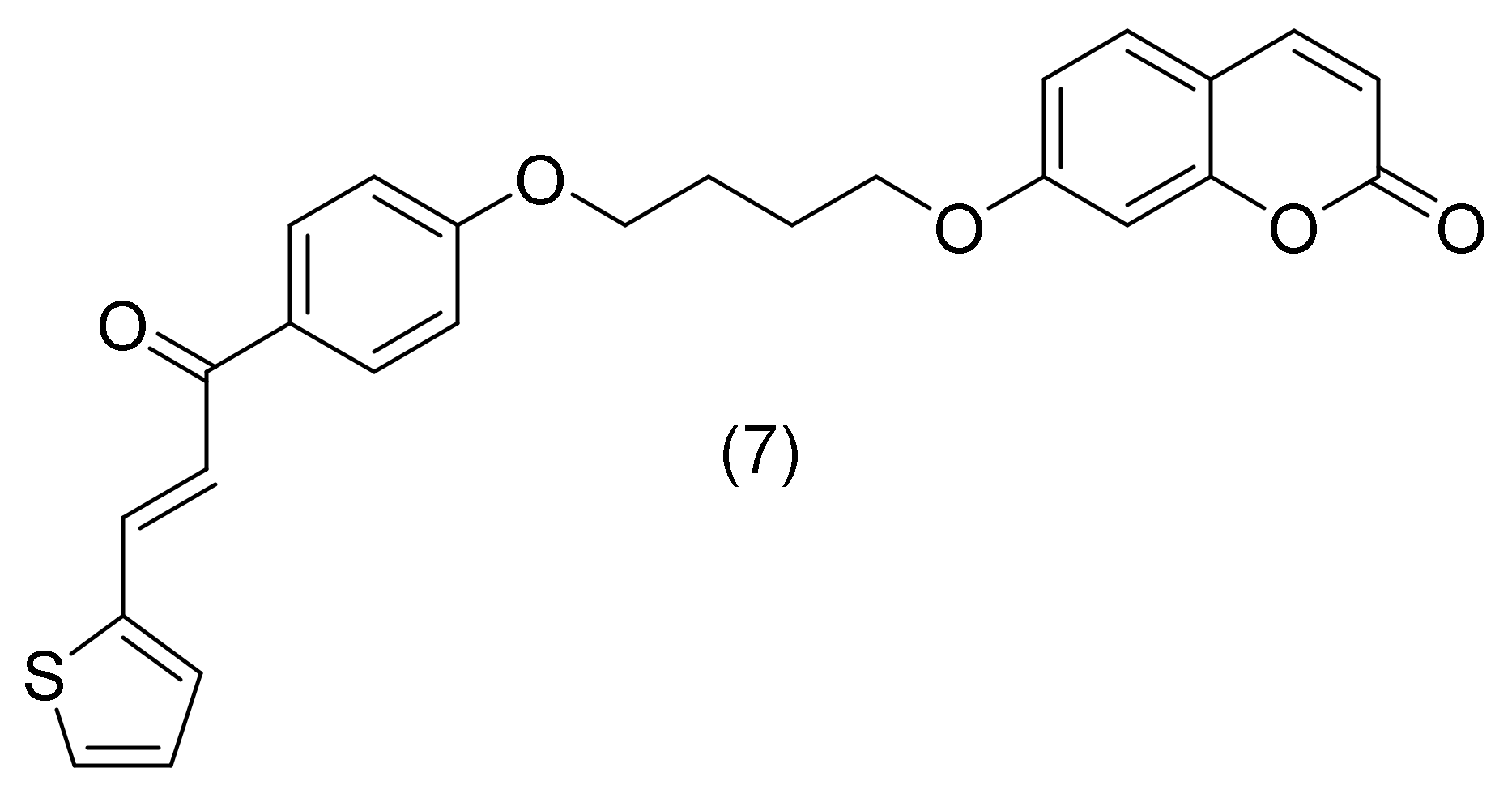
2.1.2. Thiophene Chalcone-Based Coumarin Analogues
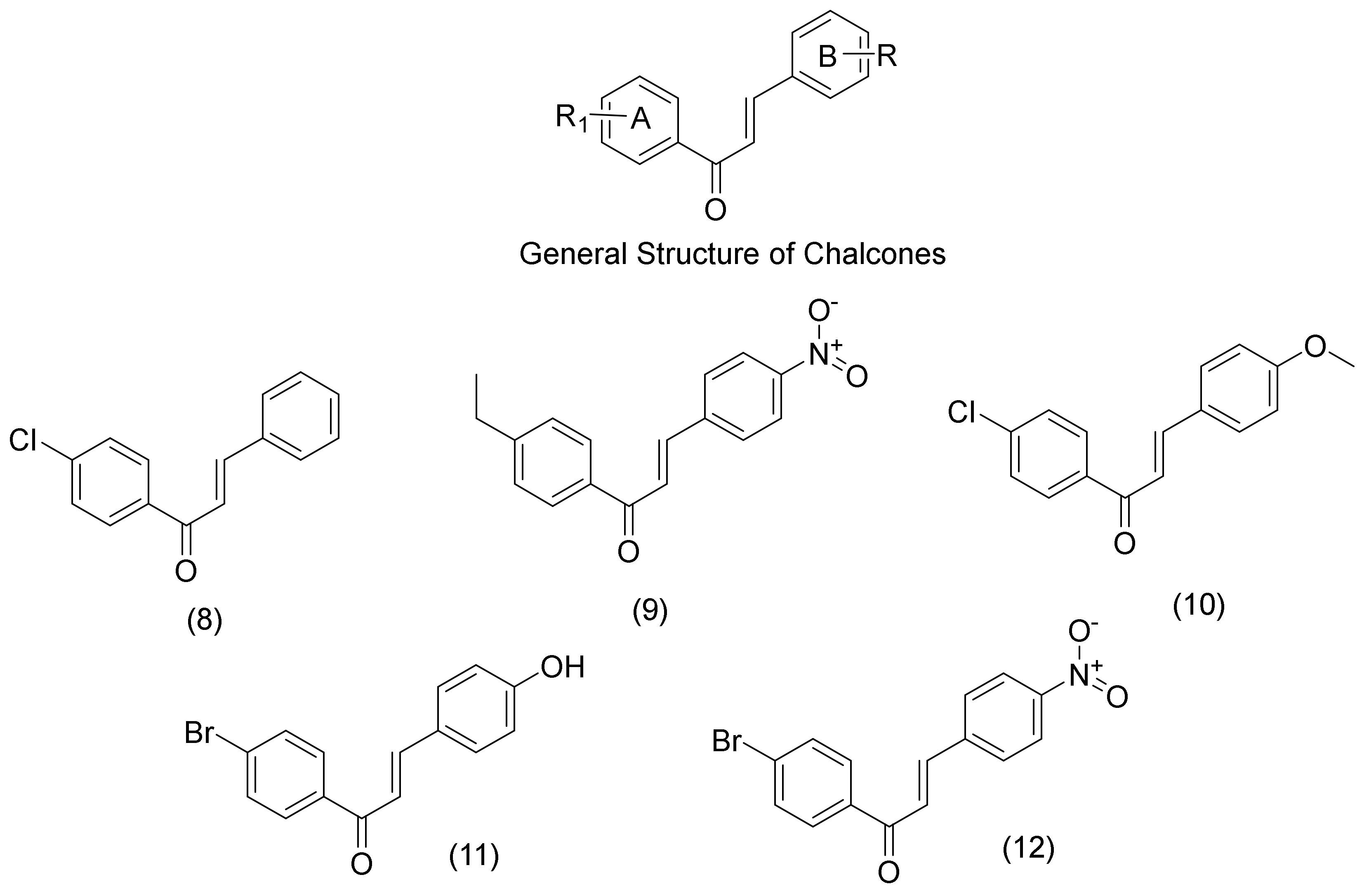
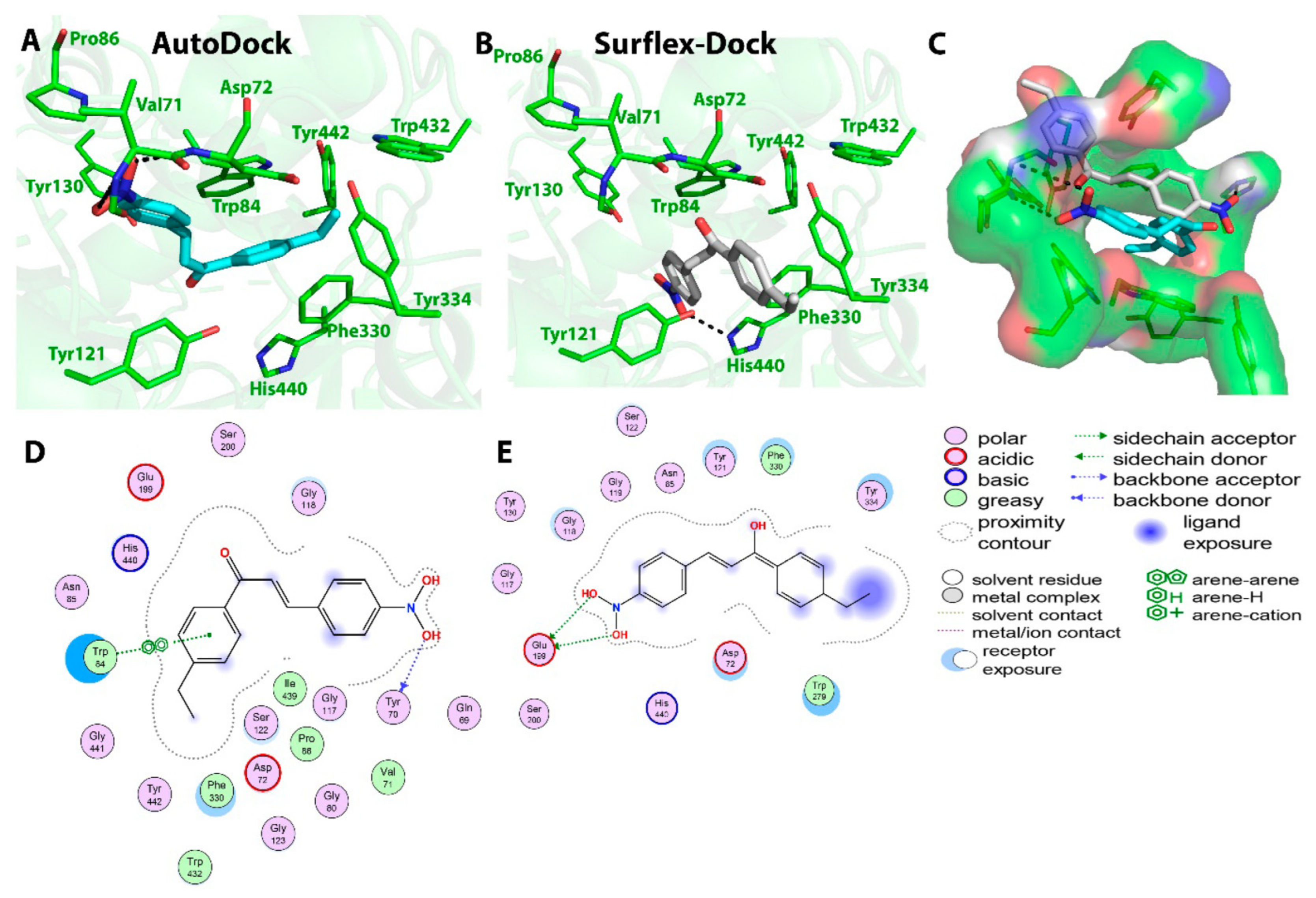
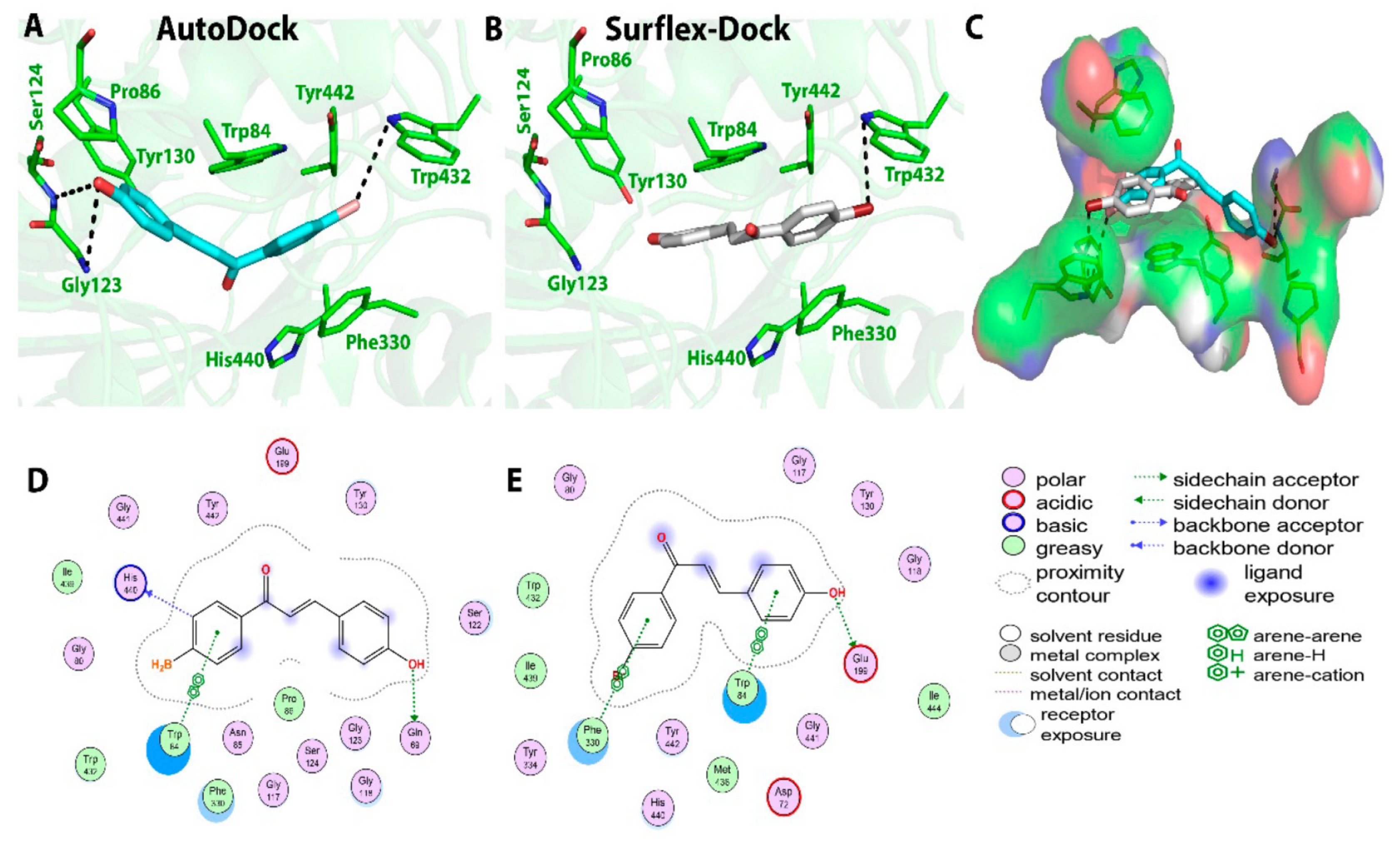
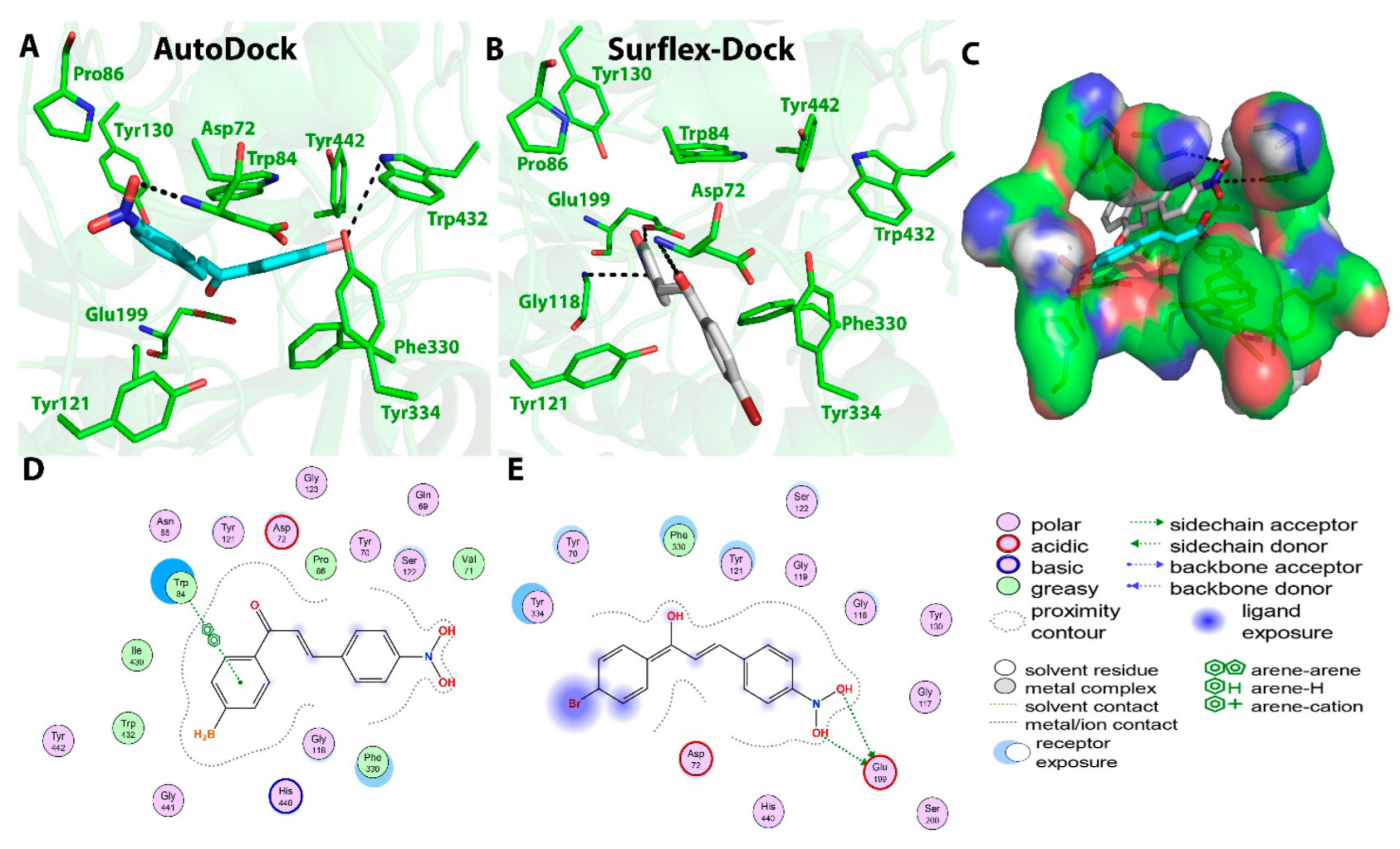
2.2. Chalcone Derivatives
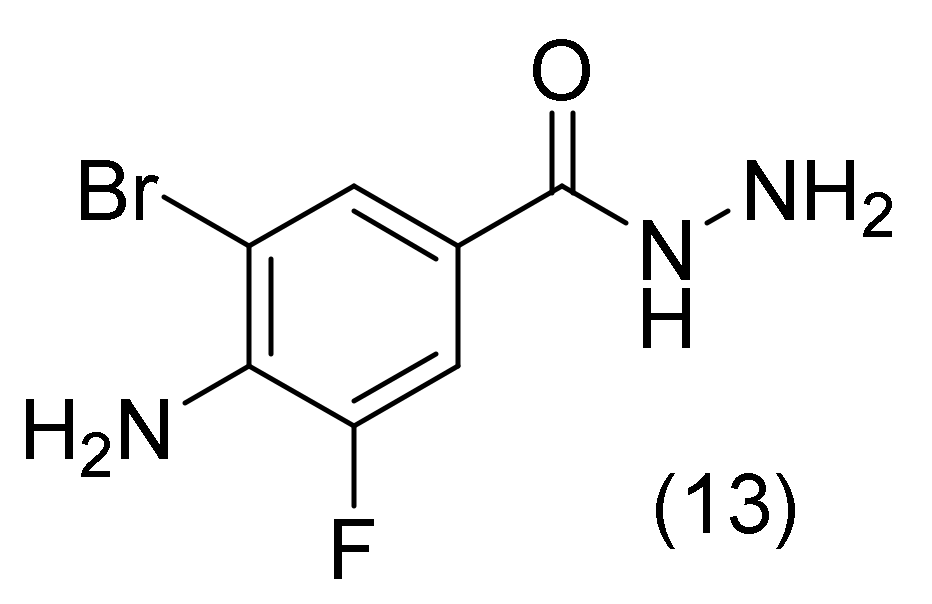
2.3. 4-Aminobenzohydrazide Derivative
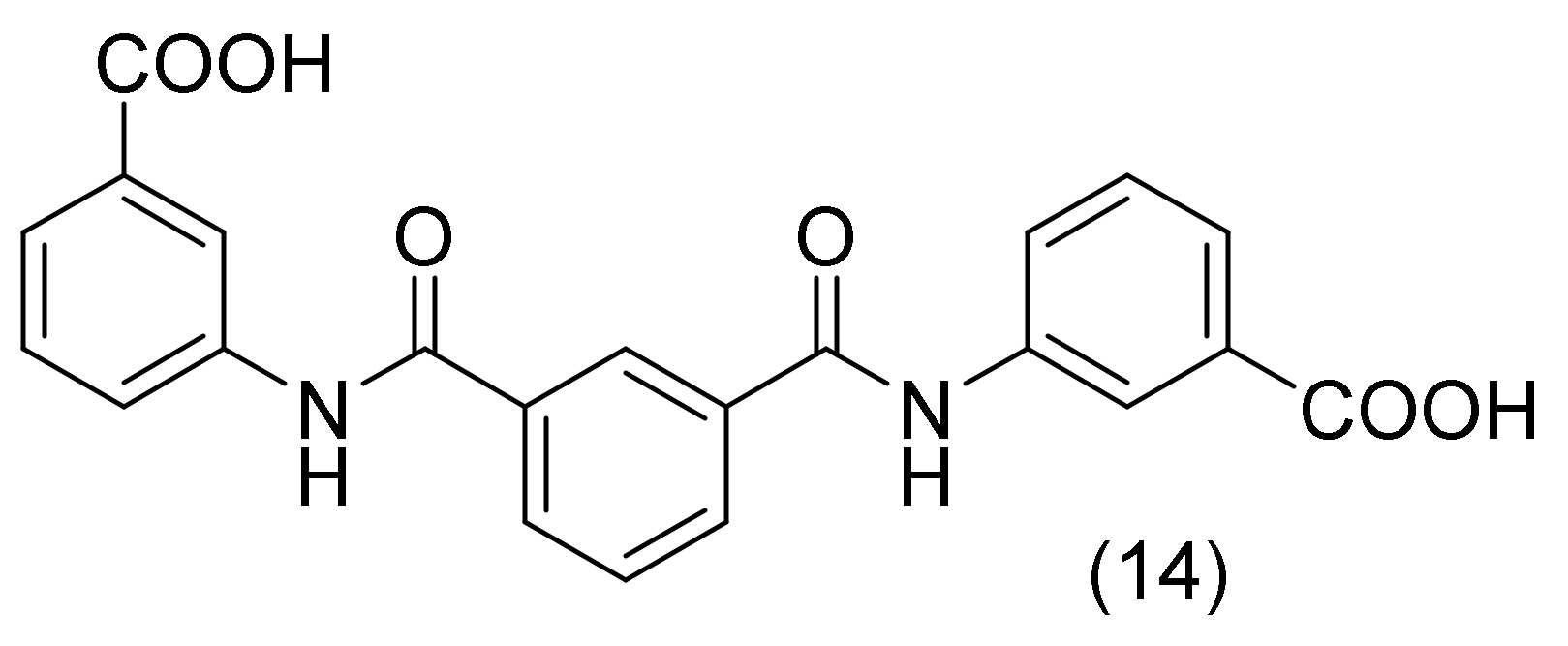
2.4. Aminobenzoic Acid Derivatives
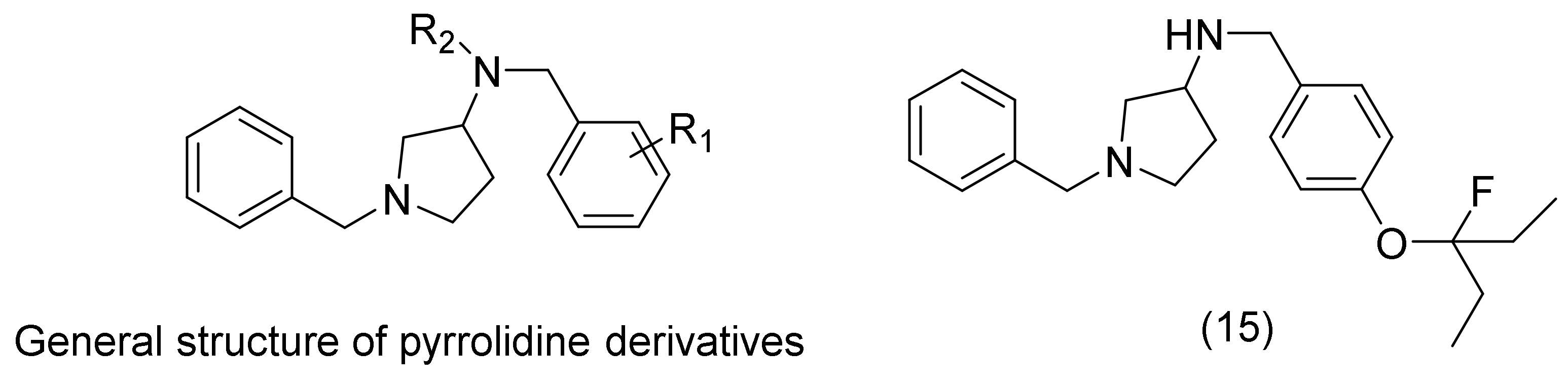
2.5. Pyrrolidine Derivatives
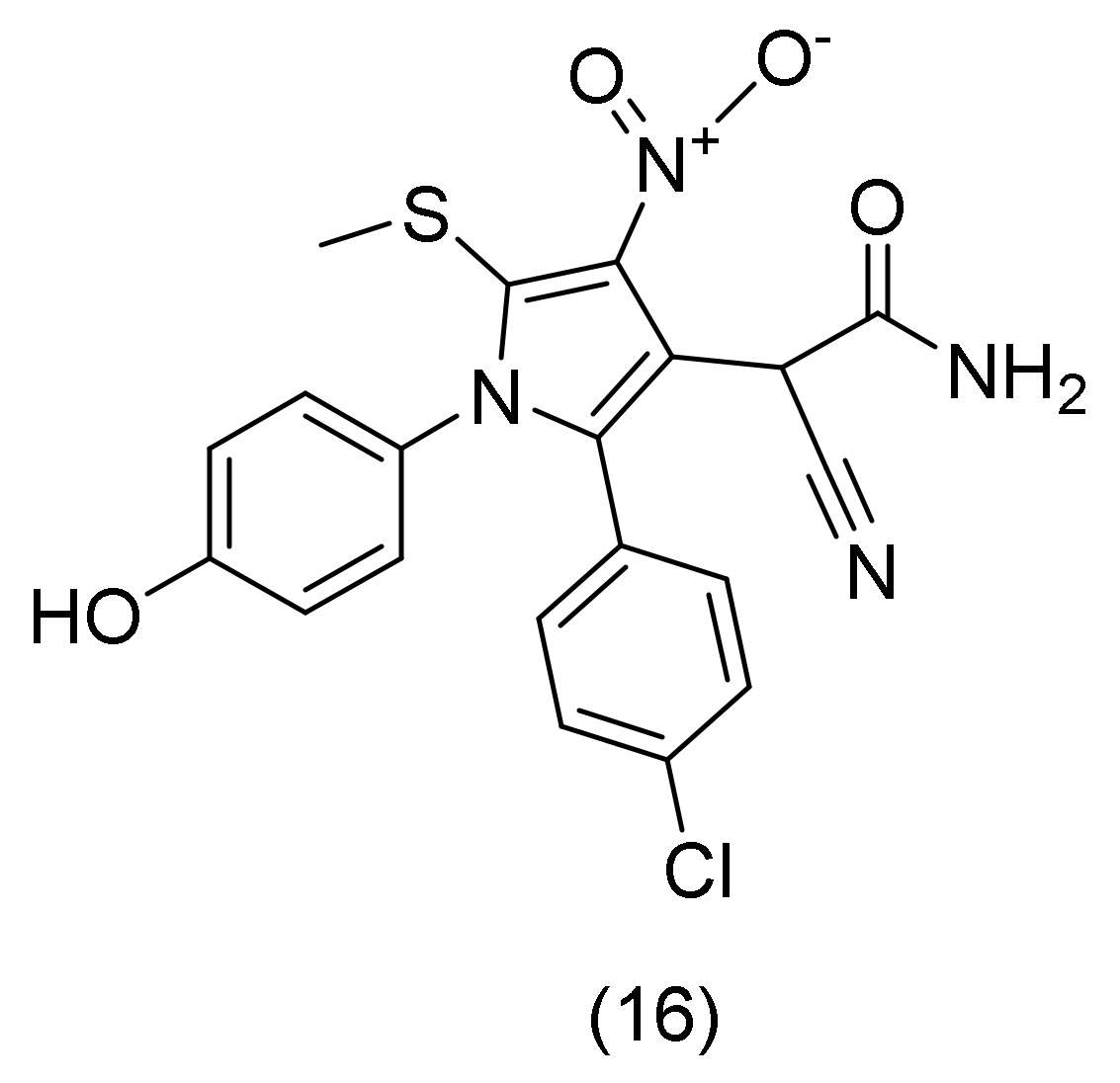
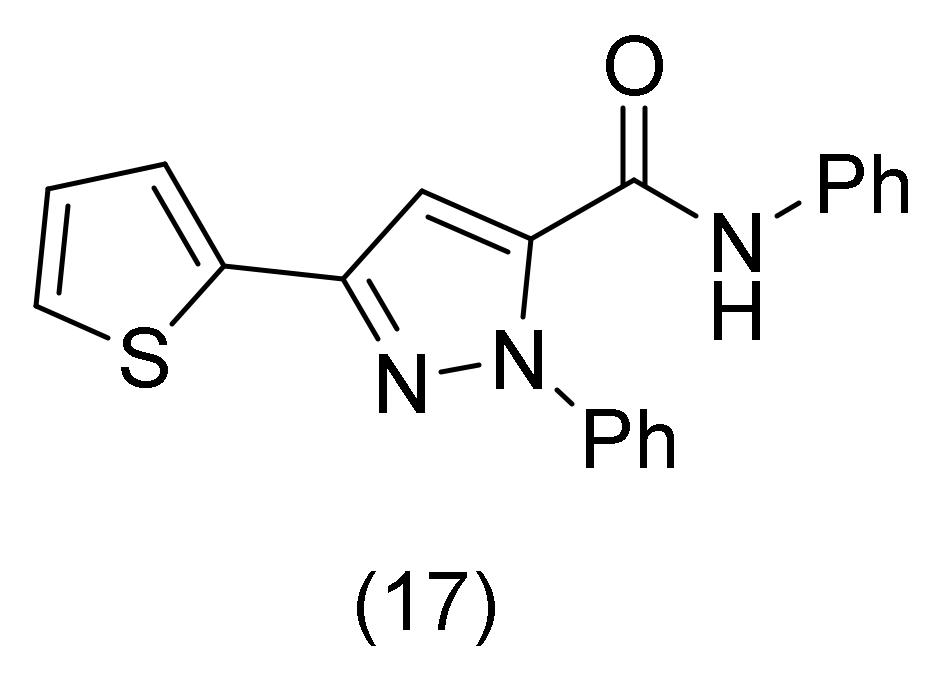
2.6. Pyrrole Derivatives
2.7. Pyrazole Derivatives
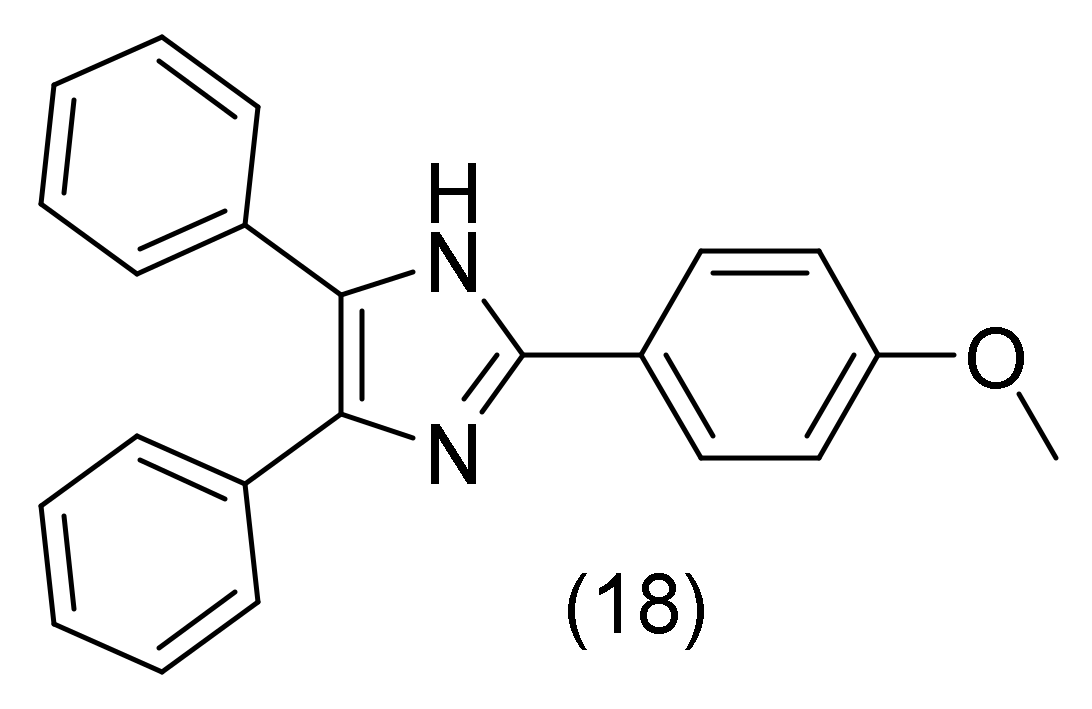
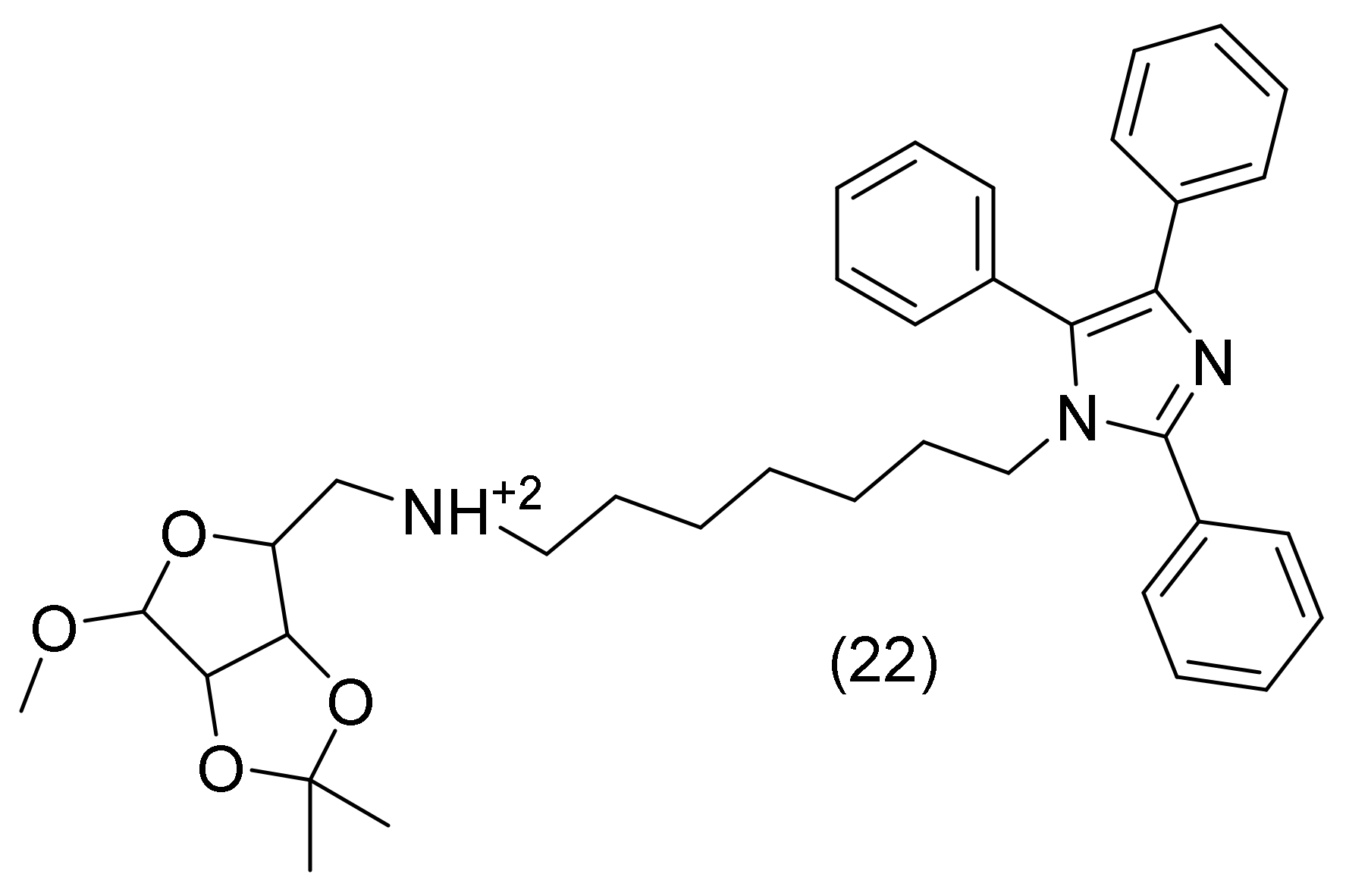
2.8. Imidazole Derivatives
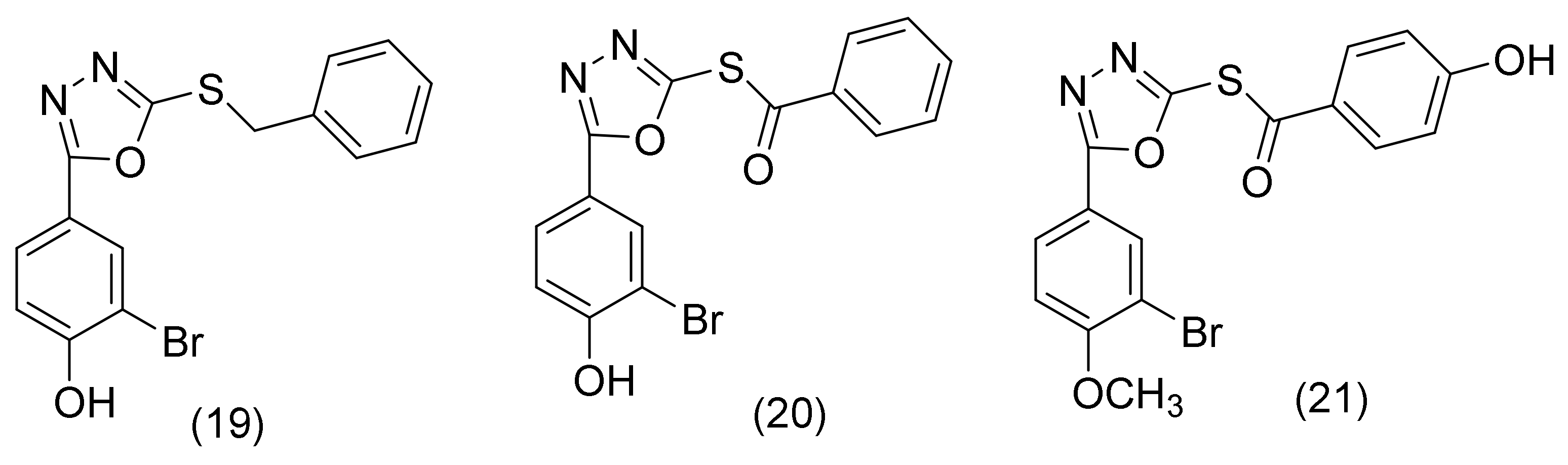
2.9. 1,3,4 Oxadiazole Derivatives
2.10. Lophine Derivatives
2.11. 1,2,4-Triazole Derivatives
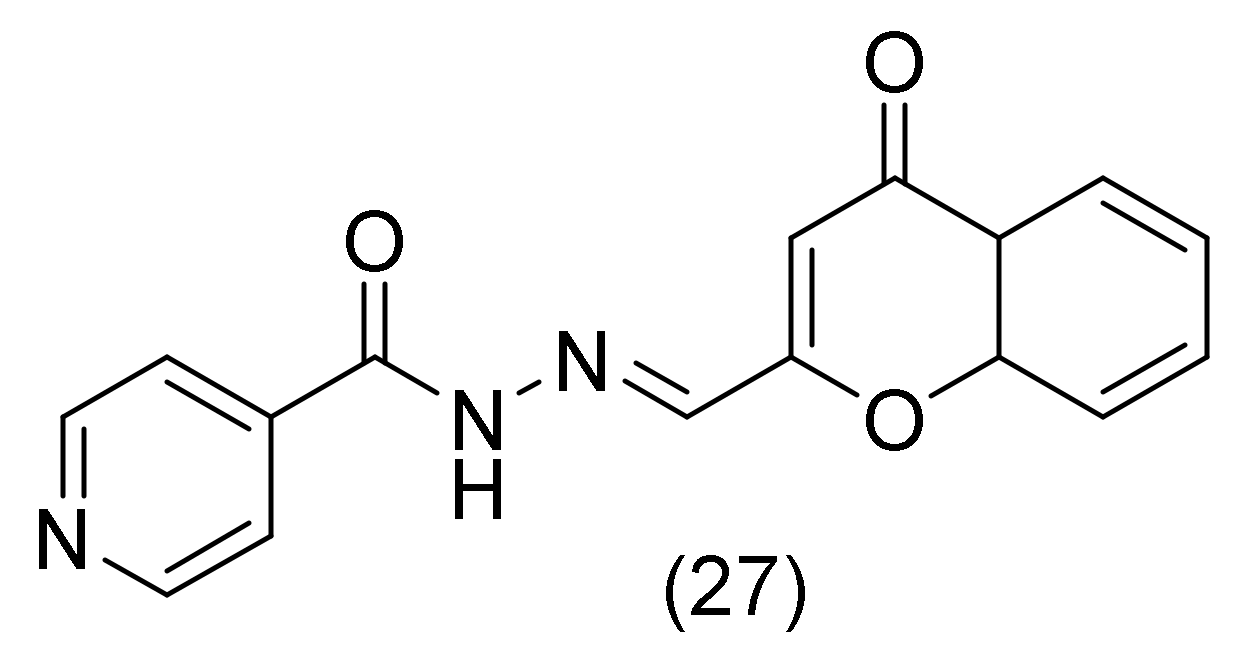
2.12. Isoniazid Derivatives
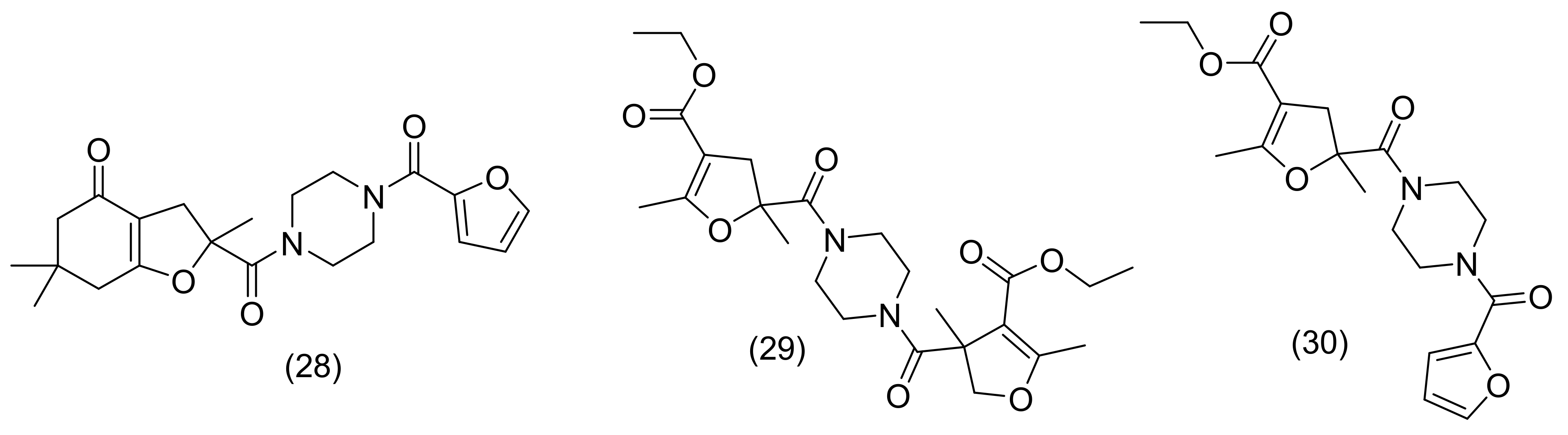
2.13. Piperazine Derivatives
2.14. Pyrimidire Derivatives
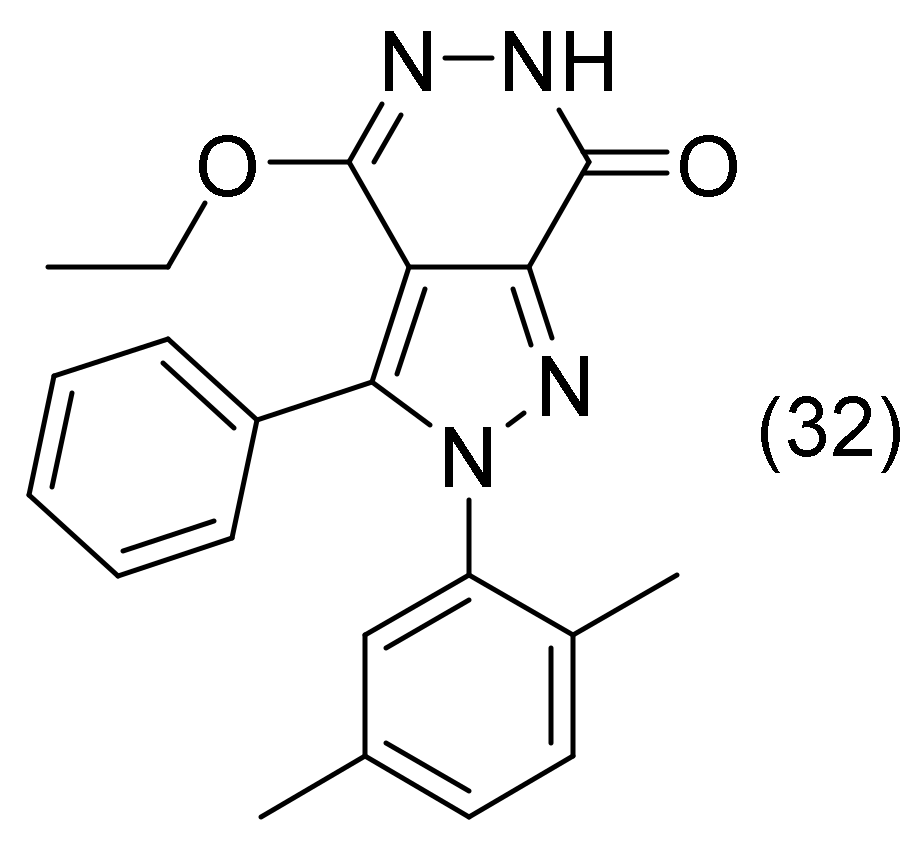
2.15. Pyrazole-Pyridazine Derivatives
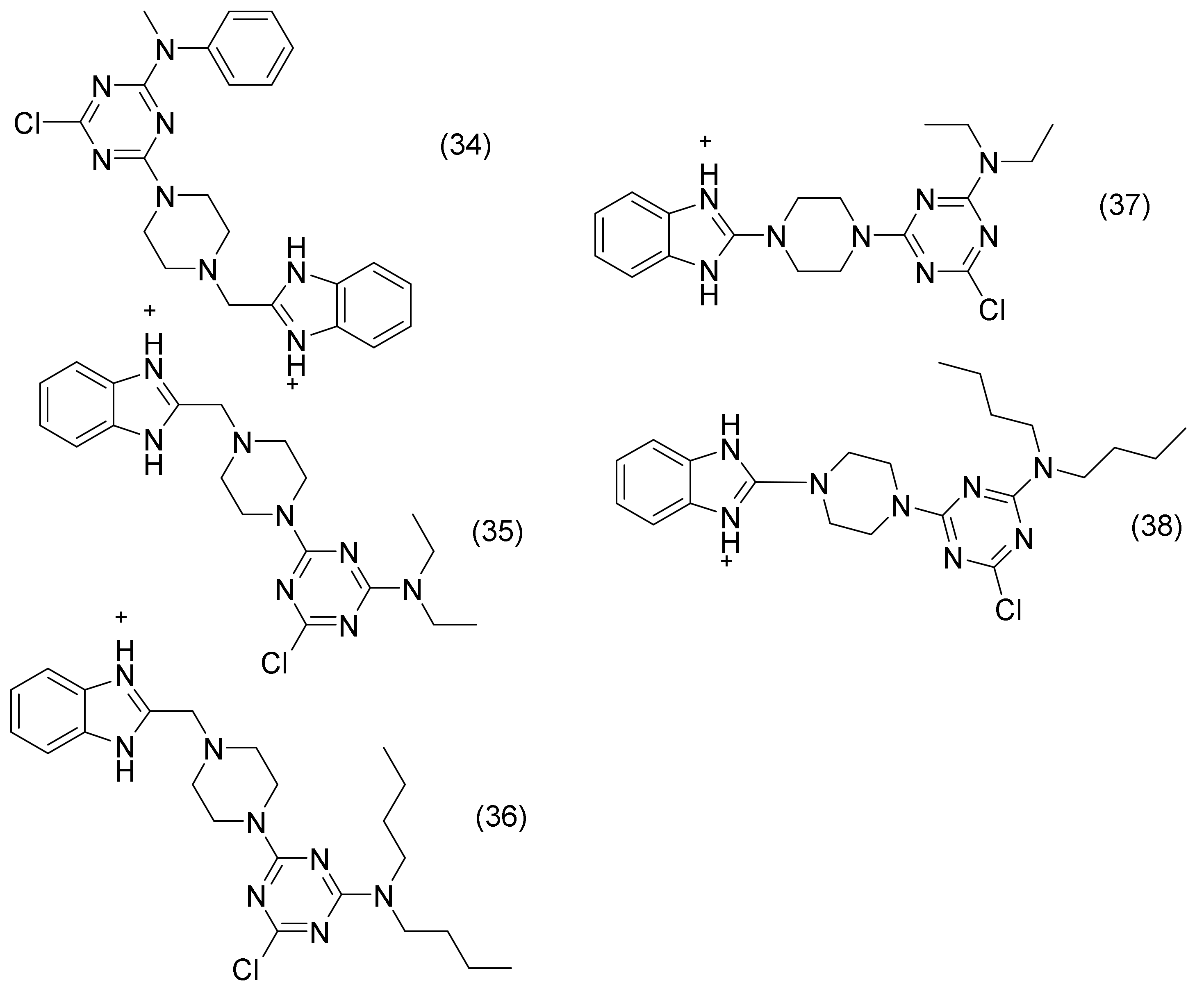
2.16. 1,3,5-Triazine Derivatives
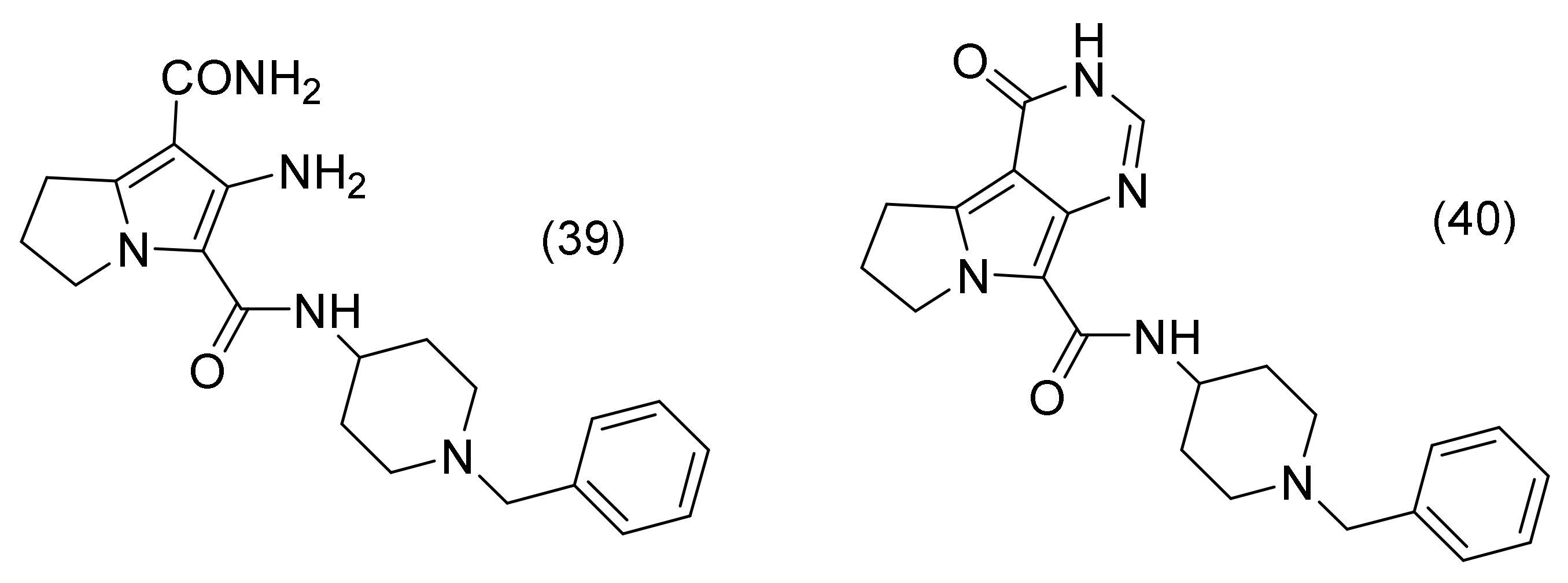
2.17. Pyrrolizine Derivatives
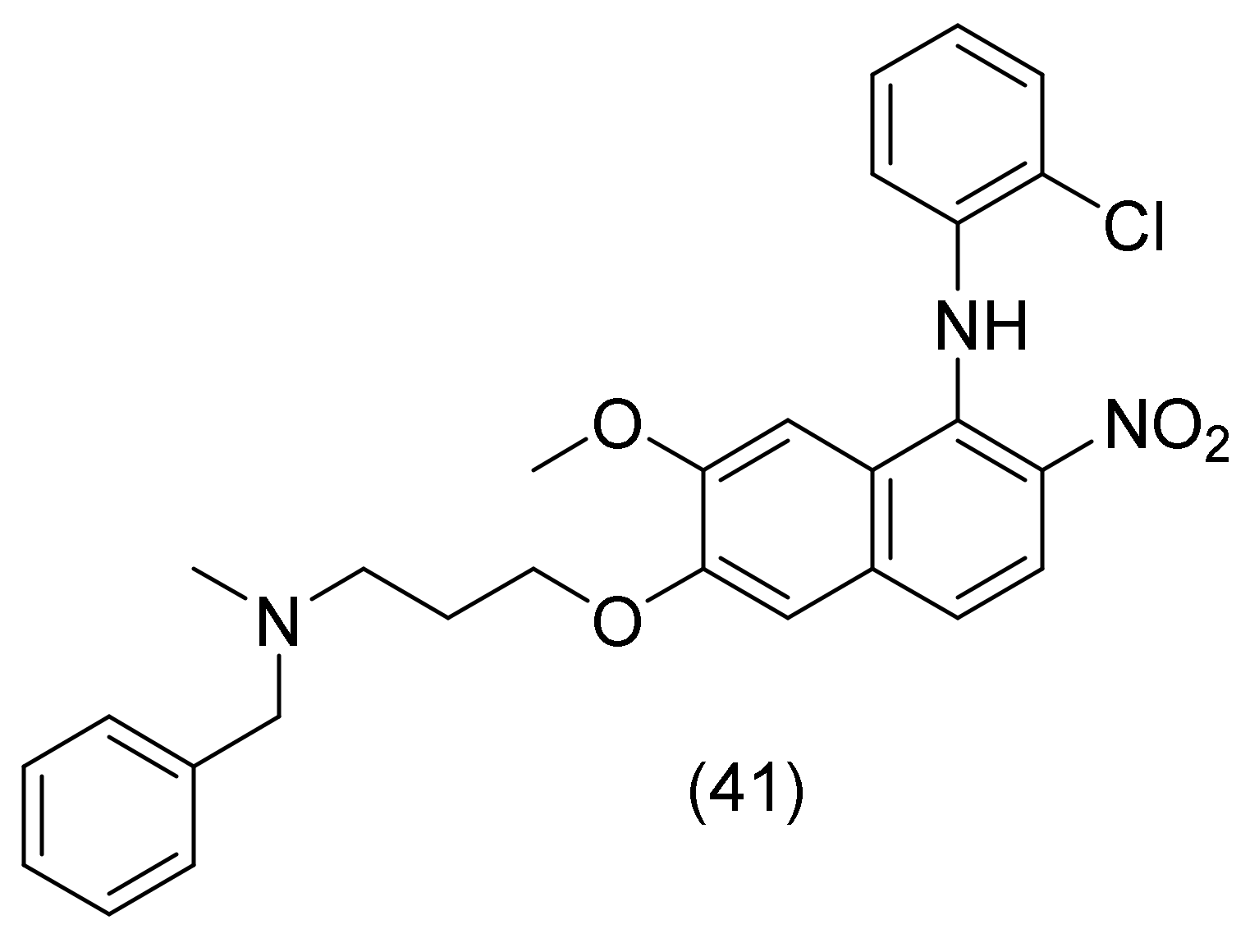
2.18. Quinoline Derivatives
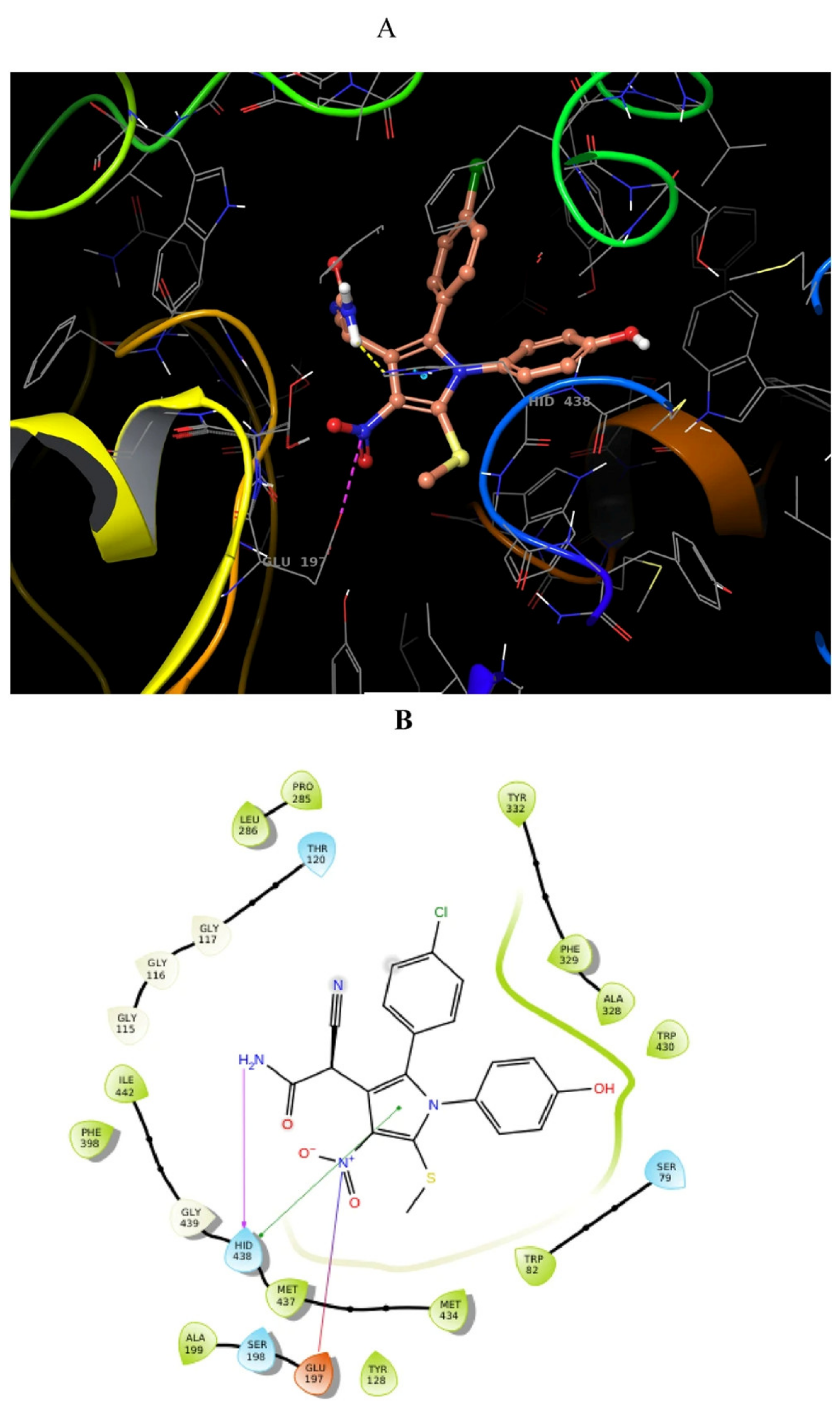
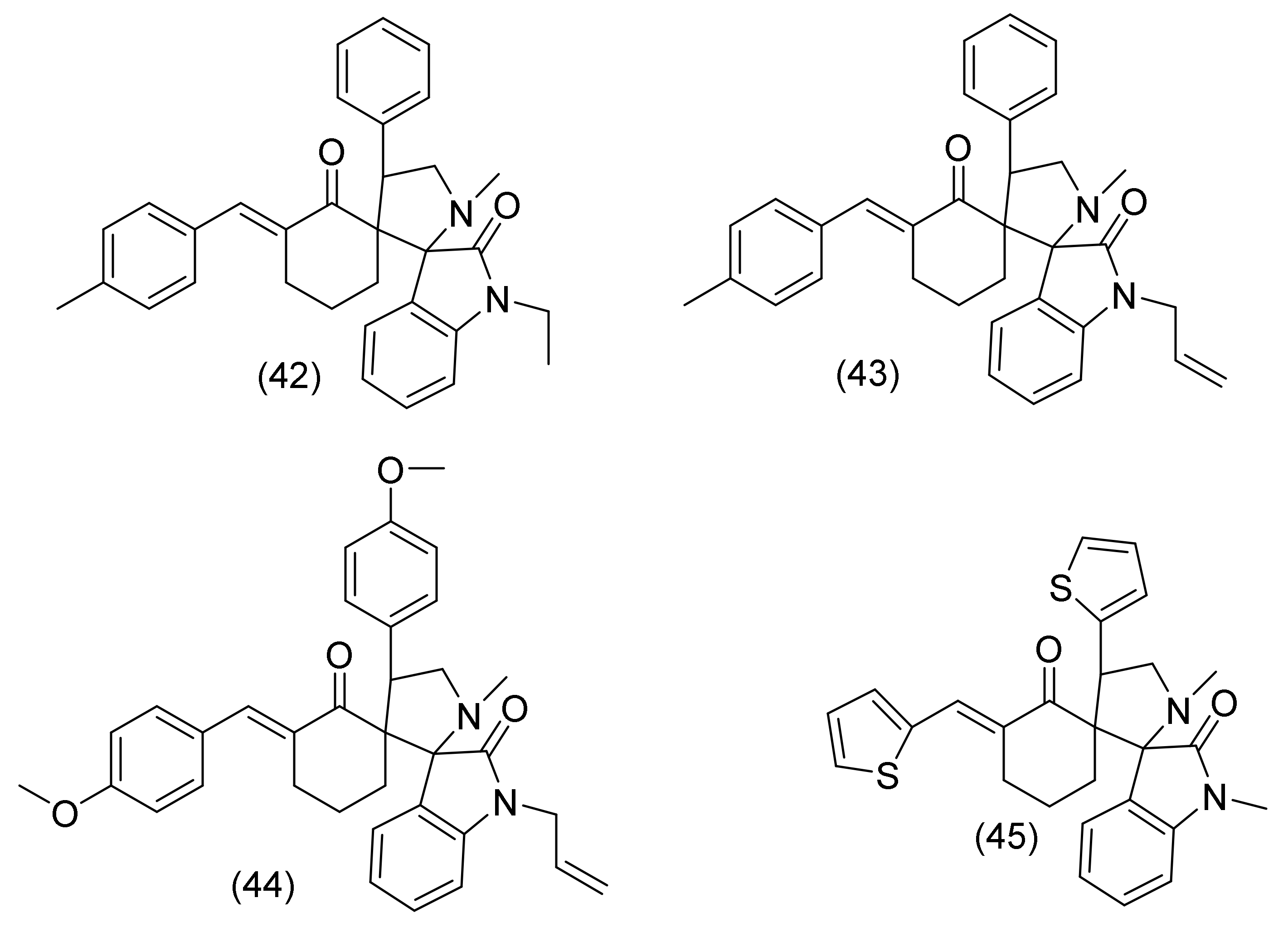
2.19. Oxindole Derivatives
2.20. Quinoxaline Derivatives
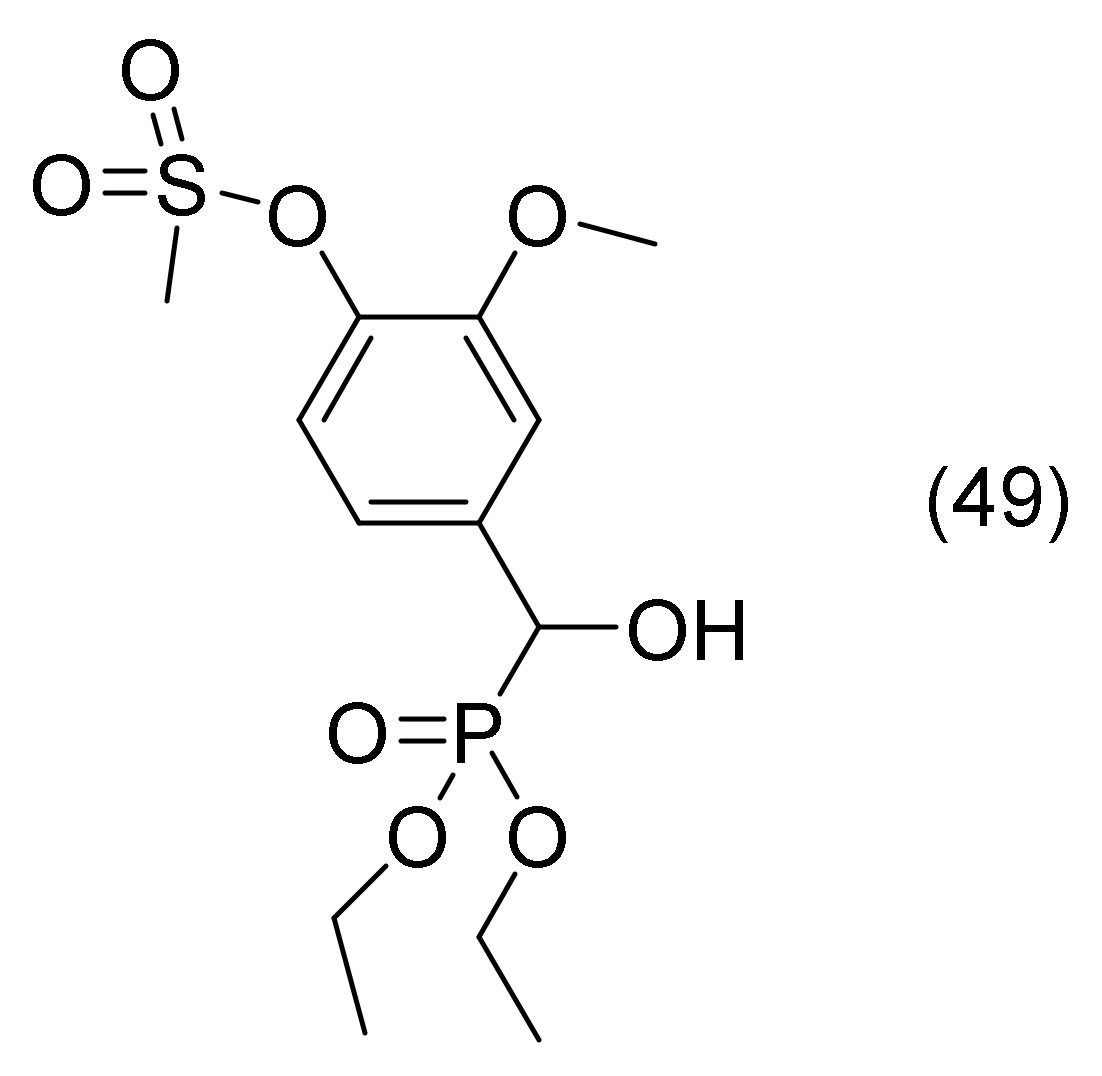
2.21. Sulfonate with Aryl α-Hydroxyphsphonate Group
2.22. Carbazole Based α-Aminophosphonate Derivatives
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kaur, S.; Bansal, Y. Design, Molecular Docking, Synthesis and Evaluation of Xanthoxylin Hybrids as Dual Inhibitors of IL-6 and Acetylcholinesterase for Alzheimer’s Disease. Bioorg. Chem. 2022, 121, 105670. [Google Scholar] [CrossRef] [PubMed]
- De Mendonça, L.J.C.; Ferrari, R.J. Alzheimer’s Disease Classification Based on Graph Kernel SVMs Constructed with 3D Texture Features Extracted from MR Images. Expert Syst. Appl. 2023, 211, 118633. [Google Scholar] [CrossRef]
- Lyketsos, C.G.; Carrillo, M.C.; Ryan, J.M.; Khachaturian, A.S.; Trzepacz, P.; Amatniek, J.; Cedarbaum, J.; Brashear, R.; Miller, D.S. Neuropsychiatric Symptoms in Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, K.S.; Waldorff, F.B.; Waldemar, G. O2-02-02: Physical Activity as a Predictor of Clinical Course in Mild Ad: The Danish Alzheimer’s Intervention Study (Daisy). Alzheimers Dement. 2014, 10, P165. [Google Scholar] [CrossRef]
- Berger, A. Positron Emission Tomography. BMJ 2003, 326, 1449. [Google Scholar] [CrossRef] [PubMed]
- Petrella, C.; Di Certo, M.G.; Barbato, C.; Gabanella, F.; Ralli, M.; Greco, A.; Possenti, R.; Severini, C. Neuropeptides in Alzheimer’s Disease: An Update. Curr. Alzheimer Res. 2019, 16, 544–558. [Google Scholar] [CrossRef]
- Iqbal, K.; Grundke-Iqbal, I. Neurofibrillary Pathology Leads to Synaptic Loss and Not the Other Way around in Alzheimer Disease. J. Alzheimers Dis. 2002, 4, 235–238. [Google Scholar] [CrossRef]
- Iqbal, K.; Liu, F.; Gong, C.-X. Tau and Neurodegenerative Disease: The Story so Far. Nat. Rev. Neurol. 2016, 12, 15–27. [Google Scholar] [CrossRef]
- Itagaki, S.; McGeer, P.L.; Akiyama, H.; Zhu, S.; Selkoe, D. Relationship of Microglia and Astrocytes to Amyloid Deposits of Alzheimer Disease. J. Neuroimmunol. 1989, 24, 173–182. [Google Scholar] [CrossRef]
- Choi, S.H.; Tanzi, R.E. Alzheimer’s Disease: Causes, Mechanisms, and Steps Toward Prevention. In The Oxford Handbook of the Neurobiology of Learning and Memory; Carew, T.J., Ed.; Oxford University Press: Oxford, UK, 2020; ISBN 978-0-19-006916-2. [Google Scholar]
- Xu, J.; Wang, F.; Guo, J.; Xu, C.; Cao, Y.; Fang, Z.; Wang, Q. Pharmacological Mechanisms Underlying the Neuroprotective Effects of Alpinia Oxyphylla Miq. on Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2071. [Google Scholar] [CrossRef]
- Hendy, G.N.; Guarnieri, V.; Canaff, L. Chapter 3 Calcium-Sensing Receptor and Associated Diseases. In Progress in Molecular Biology and Translational Science; Tao, Y.-X., Ed.; Academic Press: Cambridge, MA, USA, 2009; Volume 89, pp. 31–95. [Google Scholar]
- Bandyopadhyay, S.; Jeong, K.-H.; Hansen, J.T.; Vassilev, P.M.; Brown, E.M.; Chattopadhyay, N. Calcium-Sensing Receptor Stimulates Secretion of an Interferon-γ-Induced Monokine (CXCL10) and Monocyte Chemoattractant Protein-3 in Immortalized GnRH Neurons. J. Neurosci. Res. 2007, 85, 882–895. [Google Scholar] [CrossRef]
- Chiarini, A.; Pra, I.D.; Marconi, M.; Chakravarthy, B.; Whitfield, J.F.; Armato, U. Calcium-Sensing Receptor (CaSR) in Human Brain’s Pathophysiology: Roles in Late-Onset Alzheimer’s Disease (LOAD). Curr. Pharm. Biotechnol. 2009, 10, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, R.J.; Shen, J. Presenilin-1 Mutations and Alzheimer’s Disease. Proc. Natl. Acad. Sci. USA 2017, 114, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, C.; Sun, J.; Shen, H.-M.; Wang, J.; Yang, C. Impairment of the Autophagy–Lysosomal Pathway in Alzheimer’s Diseases: Pathogenic Mechanisms and Therapeutic Potential. Acta Pharm. Sin. B 2022, 12, 1019–1040. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, F.; Inestrosa, N. Interactions of AChE with Aβ Aggregates in Alzheimer’s Brain: Therapeutic Relevance of IDN 5706. Front. Mol. Neurosci. 2011, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Pitchai, A.; Rajaretinam, R.K.; Mani, R.; Nagarajan, N. Molecular Interaction of Human Acetylcholinesterase with Trans-Tephrostachin and Derivatives for Alzheimer’s Disease. Heliyon 2020, 6, e04930. [Google Scholar] [CrossRef]
- Myers, M.; Richmond, R.C.; Oakeshott, J.G. On the Origins of Esterases. Mol. Biol. Evol. 1988, 5, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Hormozi Jangi, S.R.; Akhond, M. Introducing a Covalent Thiol-Based Protected Immobilized Acetylcholinesterase with Enhanced Enzymatic Performances for Biosynthesis of Esters. Process Biochem. 2022, 120, 138–155. [Google Scholar] [CrossRef]
- Riaz, N.; Iftikhar, M.; Saleem, M.; Aziz-ur-Rehman; Hussain, S.; Rehmat, F.; Afzal, Z.; Khawar, S.; Ashraf, M.; al-Rashida, M. New Synthetic 1,2,4-Triazole Derivatives: Cholinesterase Inhibition and Molecular Docking Studies. Results Chem. 2020, 2, 100041. [Google Scholar] [CrossRef]
- Sussman, J.L.; Silman, I. Acetylcholinesterase: Structure and Use as a Model for Specific Cation—Protein Interactions. Curr. Opin. Struct. Biol. 1992, 2, 721–729. [Google Scholar] [CrossRef]
- Tripathi, A.; Srivastava, U.C. Acetylcholinesterase: A Versatile Enzyme of Nervous System. Ann. Neurosci. 2010, 15, 106–111. [Google Scholar] [CrossRef]
- Molecular Architecture and Biological Reactions. Chem. Eng. News Arch. 1946, 24, 1375–1377. [CrossRef]
- El-Sayed, N.A.-E.; Farag, A.E.-S.; Ezzat, M.A.F.; Akincioglu, H.; Gülçin, İ.; Abou-Seri, S.M. Design, Synthesis, in Vitro and in Vivo Evaluation of Novel Pyrrolizine-Based Compounds with Potential Activity as Cholinesterase Inhibitors and Anti-Alzheimer’s Agents. Bioorg. Chem. 2019, 93, 103312. [Google Scholar] [CrossRef]
- Wang, L.; Moraleda, I.; Iriepa, I.; Romero, A.; López-Muñoz, F.; Chioua, M.; Inokuchi, T.; Bartolini, M.; Marco-Contelles, J. 5-Methyl- N -(8-(5,6,7,8-Tetrahydroacridin-9-Ylamino)Octyl)-5 H -Indolo[2,3- b ]Quinolin-11-Amine: A Highly Potent Human Cholinesterase Inhibitor. MedChemComm 2017, 8, 1307–1317. [Google Scholar] [CrossRef]
- Kuzu, B.; Tan, M.; Taslimi, P.; Gülçin, İ.; Taşpınar, M.; Menges, N. Mono- or Di-Substituted Imidazole Derivatives for Inhibition of Acetylcholine and Butyrylcholine Esterases. Bioorg. Chem. 2019, 86, 187–196. [Google Scholar] [CrossRef] [PubMed]
- McGleenon, B.M.; Dynan, K.B.; Passmore, A.P. Acetylcholinesterase Inhibitors in Alzheimer’s Disease. Br. J. Clin. Pharm. 1999, 48, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Kurt, B.Z.; Ozten Kandas, N.; Dag, A.; Sonmez, F.; Kucukislamoglu, M. Synthesis and Biological Evaluation of Novel Coumarin-Chalcone Derivatives Containing Urea Moiety as Potential Anticancer Agents. Arab. J. Chem. 2020, 13, 1120–1129. [Google Scholar] [CrossRef]
- Amin, K.M.; Abdel Rahman, D.E.; Abdelrasheed Allam, H.; El-Zoheiry, H.H. Design and Synthesis of Novel Coumarin Derivatives as Potential Acetylcholinesterase Inhibitors for Alzheimer’s Disease. Bioorg. Chem. 2021, 110, 104792. [Google Scholar] [CrossRef]
- Benazzouz-Touami, A.; Chouh, A.; Halit, S.; Terrachet-Bouaziz, S.; Makhloufi-Chebli, M.; Ighil-Ahriz, K.; Silva, A.M.S. New Coumarin-Pyrazole Hybrids: Synthesis, Docking Studies and Biological Evaluation as Potential Cholinesterase Inhibitors. J. Mol. Struct. 2022, 1249, 131591. [Google Scholar] [CrossRef]
- Hasan, A.H.; Murugesan, S.; Amran, S.I.; Chander, S.; Alanazi, M.M.; Hadda, T.B.; Shakya, S.; Pratama, M.R.F.; Das, B.; Biswas, S.; et al. Novel Thiophene Chalcones-Coumarin as Acetylcholinesterase Inhibitors: Design, Synthesis, Biological Evaluation, Molecular Docking, ADMET Prediction and Molecular Dynamics Simulation. Bioorg. Chem. 2022, 119, 105572. [Google Scholar] [CrossRef]
- Matos, M.J.; Vazquez-Rodriguez, S.; Uriarte, E.; Santana, L. Potential Pharmacological Uses of Chalcones: A Patent Review (from June 2011–2014). Expert Opin. Ther. Pat. 2015, 25, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Medhi, B.; Sehgal, R. Chalcones as an Emerging Lead Molecule for Antimalarial Therapy: A Review. J. Mod. Med. Chem. 2013, 1, 64–77. [Google Scholar] [CrossRef]
- Malik, Y.A.; Awad, T.A.; Abdalla, M.; Yagi, S.; Alhazmi, H.A.; Ahsan, W.; Albratty, M.; Najmi, A.; Muhammad, S.; Khalid, A. Chalcone Scaffolds Exhibiting Acetylcholinesterase Enzyme Inhibition: Mechanistic and Computational Investigations. Molecules 2022, 27, 3181. [Google Scholar] [CrossRef] [PubMed]
- Almaz, Z.; Oztekin, A.; Tan, A.; Ozdemir, H. Biological Evaluation and Molecular Docking Studies of 4-Aminobenzohydrazide Derivatives as Cholinesterase Inhibitors. J. Mol. Struct. 2021, 1244, 130918. [Google Scholar] [CrossRef]
- Iftikhar, K.; Murtaza, S.; Kausar, N.; Abbas, A.; Tahir, M.N. Aminobenzoic Acid Derivatives as Antioxidants and Cholinesterase Inhibitors; Synthesis, Biological Evaluation and Molecular Docking Studies. Acta Pol. Pharm. Drug Res. 2018, 75, 385–396. [Google Scholar]
- El Khatabi, K.; El-Mernissi, R.; Aanouz, I.; Ajana, M.A.; Lakhlifi, T.; Khan, A.; Wei, D.-Q.; Bouachrine, M. Identification of Novel Acetylcholinesterase Inhibitors through 3D-QSAR, Molecular Docking, and Molecular Dynamics Simulation Targeting Alzheimer’s Disease. J. Mol. Model. 2021, 27, 302. [Google Scholar] [CrossRef]
- Marth, G.; Anderson, R.J.; Thompson, B.G.; Ashton, M.; Groundwater, P.W. Chemo- and Regioselectivity in the Reactions of Polyfunctional Pyrroles. Tetrahedron 2010, 66, 6113–6120. [Google Scholar] [CrossRef]
- Pourtaher, H.; Hasaninejad, A.; Iraji, A. Design, Synthesis, in Silico and Biological Evaluations of Novel Polysubstituted Pyrroles as Selective Acetylcholinesterase Inhibitors against Alzheimer’s Disease. Sci. Rep. 2022, 12, 15236. [Google Scholar] [CrossRef]
- Dias, I.M.; Junior, H.C.S.; Costa, S.C.; Cardoso, C.M.; Cruz, A.G.B.; Santos, C.E.R.; Candela, D.R.S.; Soriano, S.; Marques, M.M.; Ferreira, G.B.; et al. Mononuclear Coordination Compounds Containing a Pyrazole-Based Ligand: Syntheses, Magnetism and Acetylcholinesterase Inhibition Assays. J. Mol. Struct. 2020, 1205, 127564. [Google Scholar] [CrossRef]
- Cetin, A.; Türkan, F.; Bursal, E.; Murahari, M. Synthesis, Characterization, Enzyme Inhibitory Activity, and Molecular Docking Analysis of a New Series of Thiophene-Based Heterocyclic Compounds. Russ. J. Org. Chem. 2021, 57, 598–604. [Google Scholar] [CrossRef]
- Lin, N.-H.; Wang, L.; Cohen, J.; Gu, W.-Z.; Frost, D.; Zhang, H.; Rosenberg, S.; Sham, H. Synthesis and Biological Evaluation of 4-[3-Biphenyl-2-Yl-1-Hydroxy-1-(3-Methyl-3H-Imidazol-4-Yl)-Prop-2-Ynyl]-1-Yl-Benzonitrile as Novel Farnesyltransferase Inhibitor. Bioorg. Med. Chem. Lett. 2003, 13, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Niculescu-Duvaz, D.; Niculescu-Duvaz, I.; Suijkerbuijk, B.M.J.M.; Ménard, D.; Zambon, A.; Davies, L.; Pons, J.-F.; Whittaker, S.; Marais, R.; Springer, C.J. Potent BRAF Kinase Inhibitors Based on 2,4,5-Trisubstituted Imidazole with Naphthyl and Benzothiophene 4-Substituents. Bioorg. Med. Chem. 2013, 21, 1284–1304. [Google Scholar] [CrossRef] [PubMed]
- Mano, T.; Stevens, R.W.; Ando, K.; Nakao, K.; Okumura, Y.; Sakakibara, M.; Okumura, T.; Tamura, T.; Miyamoto, K. Novel Imidazole Compounds as a New Series of Potent, Orally Active Inhibitors of 5-Lipoxygenase. Bioorg. Med. Chem. 2003, 11, 3879–3887. [Google Scholar] [CrossRef] [PubMed]
- Organocatalyzed Solvent Free and Efficient Synthesis of 2,4,5-Trisubstituted Imidazoles as Potential Acetylcholinesterase Inhibitors for Alzheimer’s Disease—Pervaiz—2020—Chemistry & Biodiversity—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1002/cbdv.201900493 (accessed on 29 November 2022).
- Mishra, P.; Sharma, P.; Tripathi, P.N.; Gupta, S.K.; Srivastava, P.; Seth, A.; Tripathi, A.; Krishnamurthy, S.; Shrivastava, S.K. Design and Development of 1,3,4-Oxadiazole Derivatives as Potential Inhibitors of Acetylcholinesterase to Ameliorate Scopolamine-Induced Cognitive Dysfunctions. Bioorg. Chem. 2019, 89, 103025. [Google Scholar] [CrossRef]
- Tariq, S.; Mutahir, S.; Khan, M.A.; Mutahir, Z.; Hussain, S.; Ashraf, M.; Bao, X.; Zhou, B.; Stark, C.B.W.; Khan, I.U. Synthesis, in Vitro Cholinesterase Inhibition, Molecular Docking, DFT, and ADME Studies of Novel 1,3,4-Oxadiazole-2-Thiol Derivatives. Chem. Biodivers 2022, 19, e202200157. [Google Scholar] [CrossRef]
- Yanover, D.; Kaftory, M. Lophine (2,4,5-Triphenyl-1H-Imidazole). Acta Cryst. E 2009, 65, o711. [Google Scholar] [CrossRef]
- Bizarro Lopes, J.P.; Silva, L.; Antonio Ceschi, M.; Seibert Lüdtke, D.; Rigon Zimmer, A.; Carine Ruaro, T.; Ferreira Dantas, R.; de Salles, C.M.C.; Paes Silva-Jr, F.; Roberto Senger, M.; et al. Synthesis of New Lophine–Carbohydrate Hybrids as Cholinesterase Inhibitors: Cytotoxicity Evaluation and Molecular Modeling. MedChemComm 2019, 10, 2089–2101. [Google Scholar] [CrossRef]
- Sahu, J.K.; Ganguly, S.; Kaushik, A. Triazoles: A Valuable Insight into Recent Developments and Biological Activities. Chin. J. Nat. Med. 2013, 11, 456–465. [Google Scholar] [CrossRef]
- Holm, S.C.; Straub, B.F. Synthesis of N-Substituted 1,2,4-Triazoles. A Review. Org. Prep. Proced. Int. 2011, 43, 319–347. [Google Scholar] [CrossRef]
- Maddila, S.; Pagadala, R.; Jonnalagadda, S. 1,2,4-Triazoles: A Review of Synthetic Approaches and the Biological Activity. LOC 2013, 10, 693–714. [Google Scholar] [CrossRef]
- Sztanke, K.; Tuzimski, T.; Rzymowska, J.; Pasternak, K.; Kandefer-Szerszeń, M. Synthesis, Determination of the Lipophilicity, Anticancer and Antimicrobial Properties of Some Fused 1,2,4-Triazole Derivatives. Eur. J. Med. Chem. 2008, 43, 404–419. [Google Scholar] [CrossRef]
- Stillings, M.R.; Welbourn, A.P.; Walter, D.S. Substituted 1,3,4-Thiadiazoles with Anticonvulsant Activity. 2. Aminoalkyl Derivatives. J. Med. Chem. 1986, 29, 2280–2284. [Google Scholar] [CrossRef]
- Hunashal, R.D.; Satyanarayana, D. One Pot Synthesis Of 3-(Substituted Phenoxymethyl)-6- Phenyl/Substituted Phenoxymethyl-1,2,4-Triazolo[3,4-B][1,3,4] Thiadiazole Derivatives As Antimicrobial Agents. Int. J. Pharm. Biol. Sci. 2012, 3, 183–192. [Google Scholar]
- Bulut, N.; Kocyigit, U.M.; Gecibesler, I.H.; Dastan, T.; Karci, H.; Taslimi, P.; Durna Dastan, S.; Gulcin, I.; Cetin, A. Synthesis of Some Novel Pyridine Compounds Containing Bis-1,2,4-Triazole/Thiosemicarbazide Moiety and Investigation of Their Antioxidant Properties, Carbonic Anhydrase, and Acetylcholinesterase Enzymes Inhibition Profiles. J. Biochem. Mol. Toxicol. 2018, 32, e22006. [Google Scholar] [CrossRef] [PubMed]
- Adlard, P.A.; Cherny, R.A.; Finkelstein, D.I.; Gautier, E.; Robb, E.; Cortes, M.; Volitakis, I.; Liu, X.; Smith, J.P.; Perez, K.; et al. Rapid Restoration of Cognition in Alzheimer’s Transgenic Mice with 8-Hydroxy Quinoline Analogs Is Associated with Decreased Interstitial Aβ. Neuron 2008, 59, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Lannfelt, L.; Blennow, K.; Zetterberg, H.; Batsman, S.; Ames, D.; Harrison, J.; Masters, C.L.; Targum, S.; Bush, A.I.; Murdoch, R.; et al. Safety, Efficacy, and Biomarker Findings of PBT2 in Targeting Aβ as a Modifying Therapy for Alzheimer’s Disease: A Phase IIa, Double-Blind, Randomised, Placebo-Controlled Trial. Lancet Neurol. 2008, 7, 779–786. [Google Scholar] [CrossRef]
- De Freitas, L.V.; da Silva, C.C.P.; Ellena, J.; Costa, L.A.S.; Rey, N.A. Structural and Vibrational Study of 8-Hydroxyquinoline-2-Carboxaldehyde Isonicotinoyl Hydrazone—A Potential Metal–Protein Attenuating Compound (MPAC) for the Treatment of Alzheimer’s Disease. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 116, 41–48. [Google Scholar] [CrossRef]
- Santos, D.C.; Henriques, R.R.; Junior, M.A.D.A.L.; Farias, A.B.; do Nogueira, T.L.C.; Quimas, J.V.F.; Romeiro, N.C.; da Silva, L.L.; Souza, A.L.F. de Acylhydrazones as Isoniazid Derivatives with Multi-Target Profiles for the Treatment of Alzheimer’s Disease: Radical Scavenging, Myeloperoxidase/Acetylcholinesterase Inhibition and Biometal Chelation. Bioorg. Med. Chem. 2020, 28, 115470. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-H.; Wu, J.W.; Liu, H.-L.; Zhao, J.-H.; Liu, K.-T.; Chuang, C.-K.; Lin, H.-Y.; Tsai, W.-B.; Ho, Y. The Discovery of Potential Acetylcholinesterase Inhibitors: A Combination of Pharmacophore Modeling, Virtual Screening, and Molecular Docking Studies. J. Biomed. Sci. 2011, 18, 8. [Google Scholar] [CrossRef]
- Szabo, M.; Klein Herenbrink, C.; Christopoulos, A.; Lane, J.R.; Capuano, B. Structure–Activity Relationships of Privileged Structures Lead to the Discovery of Novel Biased Ligands at the Dopamine D2 Receptor. J. Med. Chem. 2014, 57, 4924–4939. [Google Scholar] [CrossRef]
- Chaudhary, P.; Kumar, R.; Verma, A.K.; Singh, D.; Yadav, V.; Chhillar, A.K.; Sharma, G.L.; Chandra, R. Synthesis and Antimicrobial Activity of N-Alkyl and N-Aryl Piperazine Derivatives. Bioorg. Med. Chem. 2006, 14, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kadow, J.F.; Zhang, Z.; Yin, Z.; Gao, Q.; Wu, D.; Parker, D.D.; Yang, Z.; Zadjura, L.; Robinson, B.A.; et al. Inhibitors of HIV-1 Attachment. Part 4: A Study of the Effect of Piperazine Substitution Patterns on Antiviral Potency in the Context of Indole-Based Derivatives. Bioorg. Med. Chem. Lett. 2009, 19, 5140–5145. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.-C.; Ma, Y.-C.; Zhang, E.; Shi, X.-J.; Wang, M.-M.; Ye, X.-W.; Liu, H.-M. Design and Synthesis of Novel 1,2,3-Triazole-Dithiocarbamate Hybrids as Potential Anticancer Agents. Eur. J. Med. Chem. 2013, 62, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Piemontese, L.; Tomás, D.; Hiremathad, A.; Capriati, V.; Candeias, E.; Cardoso, S.M.; Chaves, S.; Santos, M.A. Donepezil Structure-Based Hybrids as Potential Multifunctional Anti-Alzheimer’s Drug Candidates. J. Enzym. Inhib. Med. Chem. 2018, 33, 1212–1224. [Google Scholar] [CrossRef]
- Sari, S.; Yilmaz, M. Synthesis, Characterization, Acetylcholinesterase Inhibition, and Molecular Docking Studies of New Piperazine Substituted Dihydrofuran Compounds. Med. Chem. Res. 2020, 29, 1804–1818. [Google Scholar] [CrossRef]
- Merugu, R.; Garimella, S.; Balla, D.; Sambaru, K. Synthesis and Biological Activities of Pyrimidines: A Review. Int. J. PharmTech Res. 2015, 8, 88–93. [Google Scholar] [CrossRef]
- Duran, H.E. Pyrimidines: Molecular Docking and Inhibition Studies on Carbonic Anhydrase and Cholinesterases. Biotechnol. Appl. Biochem. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Taslimi, P.; Türkan, F.; Cetin, A.; Burhan, H.; Karaman, M.; Bildirici, I.; Gulçin, İ.; Şen, F. Pyrazole[3,4-d]Pyridazine Derivatives: Molecular Docking and Explore of Acetylcholinesterase and Carbonic Anhydrase Enzymes Inhibitors as Anticholinergics Potentials. Bioorg. Chem. 2019, 92, 103213. [Google Scholar] [CrossRef]
- Singla, P.; Luxami, V.; Paul, K. Synthesis and in Vitro Evaluation of Novel Triazine Analogues as Anticancer Agents and Their Interaction Studies with Bovine Serum Albumin. Eur. J. Med. Chem. 2016, 117, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Makowska, A.; Sączewski, F.; Bednarski, P.J.; Sączewski, J.; Balewski, Ł. Hybrid Molecules Composed of 2,4-Diamino-1,3,5-Triazines and 2-Imino-Coumarins and Coumarins. Synthesis and Cytotoxic Properties. Molecules 2018, 23, 1616. [Google Scholar] [CrossRef]
- Alpan, A.S.; Parlar, S.; Carlino, L.; Tarikogullari, A.H.; Alptüzün, V.; Güneş, H.S. Synthesis, Biological Activity and Molecular Modeling Studies on 1H-Benzimidazole Derivatives as Acetylcholinesterase Inhibitors. Bioorg. Med. Chem. 2013, 21, 4928–4937. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, M.; Manral, A.; Jameel, E.; Kumar, J.; Saini, V.; Shandilya, A.; Tiwari, M.; Hoda, N.; Jayaram, B. Development of Cyanopyridine–Triazine Hybrids as Lead Multitarget Anti-Alzheimer Agents. Bioorg. Med. Chem. 2016, 24, 2777–2788. [Google Scholar] [CrossRef] [PubMed]
- Lolak, N.; Akocak, S.; Türkeş, C.; Taslimi, P.; Işık, M.; Beydemir, Ş.; Gülçin, İ.; Durgun, M. Synthesis, Characterization, Inhibition Effects, and Molecular Docking Studies as Acetylcholinesterase, α-Glycosidase, and Carbonic Anhydrase Inhibitors of Novel Benzenesulfonamides Incorporating 1,3,5-Triazine Structural Motifs. Bioorg. Chem. 2020, 100, 103897. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-L.; Wen, Z.-Y.; Qian, J.-J.; Zou, J.-P.; Liu, S.-M.; Yang, S.; Qin, T.; Yang, Q.; Liu, Y.-H.; Liu, W.-W.; et al. Design, Synthesis, Characterization and Evaluation of 1,3,5-Triazine-Benzimidazole Hybrids as Multifunctional Acetylcholinesterases Inhibitors. J. Mol. Struct. 2022, 1257, 132498. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, L.-N.; Cai, R.; Geng, S.-Q.; Dong, Y.-F.; Liu, Y.-M. Design, Synthesis, Evaluation and Molecular Modeling Study of 4-N-Phenylaminoquinolines for Alzheimer Disease Treatment. Bioorg. Med. Chem. Lett. 2019, 29, 1325–1329. [Google Scholar] [CrossRef]
- Ahles, T.A.; Saykin, A.J. Candidate Mechanisms for Chemotherapy-Induced Cognitive Changes. Nat. Rev. Cancer 2007, 7, 192–201. [Google Scholar] [CrossRef]
- Vardy, J.; Rourke, S.; Tannock, I.F. Evaluation of Cognitive Function Associated With Chemotherapy: A Review of Published Studies and Recommendations for Future Research. J. Clin. Oncol. 2007, 25, 2455–2463. [Google Scholar] [CrossRef]
- Kesler, S.R.; Rao, V.; Ray, W.J.; Rao, A. Probability of Alzheimer’s Disease in Breast Cancer Survivors Based on Gray-Matter Structural Network Efficiency. Alzheimers Dement. Diagn. Assess. Dis. Monit. 2017, 9, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Srour, A.M.; Dawood, D.H.; Nossier, E.S.; El-Shiekh, R.A.; Mahmoud, A.E.; Hussien, A.G.; Omran, M.M.; Ali, M.M. Design, Synthesis and Molecular Docking Simulation of Oxindole-Based Derivatives with Dual VEGFR-2 and Cholinesterase Inhibitory Activities. J. Mol. Struct. 2023, 1271, 134130. [Google Scholar] [CrossRef]
- Suwanhom, P.; Saetang, J.; Khongkow, P.; Nualnoi, T.; Tipmanee, V.; Lomlim, L. Synthesis, Biological Evaluation, and In Silico Studies of New Acetylcholinesterase Inhibitors Based on Quinoxaline Scaffold. Molecules 2021, 26, 4895. [Google Scholar] [CrossRef]
- Christensen, N.D.; Reed, C.A.; Culp, T.D.; Hermonat, P.L.; Howett, M.K.; Anderson, R.A.; Zaneveld, L.J.D. Papillomavirus Microbicidal Activities of High-Molecular-Weight Cellulose Sulfate, Dextran Sulfate, and Polystyrene Sulfonate. Antimicrob. Agents Chemother. 2001, 45, 3427–3432. [Google Scholar] [CrossRef] [PubMed]
- Baron, A.D. Hemodynamic Actions of Insulin. Am. J. Physiol. Endocrinol. Metab. 1994, 267, E187–E202. [Google Scholar] [CrossRef] [PubMed]
- Lenartowicz, P.; Kafarski, P.; Lipok, J. The Overproduction of 2,4-DTBP Accompanying to the Lack of Available Form of Phosphorus during the Biodegradative Utilization of Aminophosphonates by Aspergillus Terreus. Biodegradation 2015, 26, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Şahin, İ.; Bingöl, Z.; Onur, S.; Güngör, S.A.; Köse, M.; Gülçin, İ.; Tümer, F. Enzyme Inhibition Properties and Molecular Docking Studies of 4-Sulfonate Containing Aryl α-Hydroxyphosphonates Based Hybrid Molecules. Chem. Biodivers 2022, 19, e202100787. [Google Scholar] [CrossRef] [PubMed]
- Choubdar, N.; Golshani, M.; Jalili-Baleh, L.; Nadri, H.; Küçükkilinç, T.T.; Ayazgök, B.; Moradi, A.; Moghadam, F.H.; Abdolahi, Z.; Ameri, A.; et al. New Classes of Carbazoles as Potential Multi-Functional Anti-Alzheimer’s Agents. Bioorg. Chem. 2019, 91, 103164. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.-H.; Min, W.; Song, M.; Si, X.-X.; Li, M.-C.; Zhang, Z.; Liu, Y.-W.; Liu, W.-W. Synthesis, Characterization, Crystal Structure and Evaluation of Four Carbazole-Coumarin Hybrids as Multifunctional Agents for the Treatment of Alzheimer’s Disease. J. Mol. Struct. 2020, 1209, 127897. [Google Scholar] [CrossRef]
- Bachurin, S.O.; Shevtsova, E.F.; Makhaeva, G.F.; Grigoriev, V.V.; Boltneva, N.P.; Kovaleva, N.V.; Lushchekina, S.V.; Shevtsov, P.N.; Neganova, M.E.; Redkozubova, O.M.; et al. Novel Conjugates of Aminoadamantanes with Carbazole Derivatives as Potential Multitarget Agents for AD Treatment. Sci. Rep. 2017, 7, 45627. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Wang, S.; Chen, Q.; Tu, Y.; Yang, X.; Chen, J.; Yan, J.; Pi, R.; Wang, Y. Discovery of a Novel Multifunctional Carbazole–Aminoquinoline Dimer for Alzheimer’s Disease: Copper Selective Chelation, Anti-Amyloid Aggregation, and Neuroprotection. Med. Chem. Res. 2018, 27, 777–784. [Google Scholar] [CrossRef]
- Shaikh, S.; Dhavan, P.; Singh, P.; Uparkar, J.; Vaidya, S.P.; Jadhav, B.L.; Ramana, M.V. Synthesis of Carbazole Based α-Aminophosphonate Derivatives: Design, Molecular Docking and in Vitro Cholinesterase Activity. J. Biomol. Struct. Dyn. 2022, 40, 4801–4814. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peitzika, S.-C.; Pontiki, E. A Review on Recent Approaches on Molecular Docking Studies of Novel Compounds Targeting Acetylcholinesterase in Alzheimer Disease. Molecules 2023, 28, 1084. https://doi.org/10.3390/molecules28031084
Peitzika S-C, Pontiki E. A Review on Recent Approaches on Molecular Docking Studies of Novel Compounds Targeting Acetylcholinesterase in Alzheimer Disease. Molecules. 2023; 28(3):1084. https://doi.org/10.3390/molecules28031084
Chicago/Turabian StylePeitzika, Stergiani-Chrysovalanti, and Eleni Pontiki. 2023. "A Review on Recent Approaches on Molecular Docking Studies of Novel Compounds Targeting Acetylcholinesterase in Alzheimer Disease" Molecules 28, no. 3: 1084. https://doi.org/10.3390/molecules28031084
APA StylePeitzika, S.-C., & Pontiki, E. (2023). A Review on Recent Approaches on Molecular Docking Studies of Novel Compounds Targeting Acetylcholinesterase in Alzheimer Disease. Molecules, 28(3), 1084. https://doi.org/10.3390/molecules28031084





