Allosteric Modulators of Dopamine D2 Receptors for Fine-Tuning of Dopaminergic Neurotransmission in CNS Diseases: Overview, Pharmacology, Structural Aspects and Synthesis
Abstract
1. Introduction
2. Overview of Allosteric Modulators of Dopamine D2 Receptor
2.1. Small Molecules
2.2. Peptidomimetics
3. Computational Methods to Study Allostery in GPCRs—Dopamine Receptors as an Example
4. Synthesis of Allosteric Modulators of D2 Receptor
4.1. Small Molecules
4.2. Peptidomimetics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Usman, S.; Khawer, M.; Rafique, S.; Naz, Z.; Saleem, K. The Current Status of Anti GPCR Drugs against Different Cancers. J. Pharm. Anal. 2020, 10, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.C.S.; Li, Y.; Dahoun, T.; Vogel, H.; Yuan, S. New Binding Sites, New Opportunities for GPCR Drug Discovery. Trends Biochem. Sci. 2019, 44, 312–330. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.Forbes.Com/Sites/Simonking/2013/07/15/the-Best-Selling-Drugs-since-1996-Why-Abbvies-Humira-Is-Set-to-Eclipse-Pfizers-Lipitor (accessed on 15 November 2022).
- Jacobson, K.A. New Paradigms in GPCR Drug Discovery. Biochem. Pharmacol. 2015, 98, 541–555. [Google Scholar] [CrossRef] [PubMed]
- GPCRs, Desirable Therapeutic Targets in Oncology. Available online: https://www.Drugdiscoverytrends.Com/Gpcrs-Desirable-Therapeutic-Targets-in-Oncology/ (accessed on 15 November 2022).
- Lundstrom, K. An Overview on GPCRs and Drug Discovery: Structure-Based Drug Design and Structural Biology on GPCRs. Methods Mol. Biol. 2009, 552, 51–66. [Google Scholar] [CrossRef]
- Center Watch. Available online: http://www.Centerwatch.Com/Drug-Information/Fda-Approved-Drugs/Year/2015 (accessed on 15 November 2022).
- Blair, H.A. Lumateperone: First Approval. Drugs 2020, 80, 417–423. [Google Scholar] [CrossRef]
- Marti-Solano, M.; Kaczor, A.A.; Guixà-González, R.; Selent, J. Computational Strategies to Incorporate GPCR Complexity in Drug Design. In Frontiers in Computational Chemistry; Ul-Haq, Z., Madura, J.D., Eds.; Bentham Science Publishers: Soest, The Neatherlands, 2015; Chapter 1; pp. 3–43. ISBN 978-1-60805-865-5. [Google Scholar]
- Moro, S.; Hoffmann, C.; Jacobson, K.A. Role of the Extracellular Loops of G Protein-Coupled Receptors in Ligand Recognition: A Molecular Modeling Study of the Human P2Y1 Receptor. Biochemistry 1999, 38, 3498–3507. [Google Scholar] [CrossRef]
- Smith, N.J.; Bennett, K.A.; Milligan, G. When Simple Agonism Is Not Enough: Emerging Modalities of GPCR Ligands. Mol. Cell. Endocrinol. 2011, 331, 241–247. [Google Scholar] [CrossRef]
- Wu, Y.; Tong, J.; Ding, K.; Zhou, Q.; Zhao, S. GPCR Allosteric Modulator Discovery. In Protein Allostery in Drug Discovery; Zhang, J., Nussinov, R., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; pp. 225–251. ISBN 9789811387197. [Google Scholar]
- Persechino, M.; Hedderich, J.B.; Kolb, P.; Hilger, D. Allosteric Modulation of GPCRs: From Structural Insights to in silico Drug Discovery. Pharmacol. Ther. 2022, 237, 108242. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, G.; Li, S.; Liu, X.; Lu, S.; Chen, Z.; Song, K.; Yan, J.; Geng, L.; Huang, Z.; et al. ASD v3.0: Unraveling Allosteric Regulation with Structural Mechanisms and Biological Networks. Nucleic Acids Res. 2016, 44, D527–D535. [Google Scholar] [CrossRef]
- Brown, E.M. Clinical Utility of Calcimimetics Targeting the Extracellular Calcium-Sensing Receptor (CaSR). Biochem. Pharmacol. 2010, 80, 297–307. [Google Scholar] [CrossRef]
- Woollard, S.M.; Kanmogne, G.D. Maraviroc: A Review of Its Use in HIV Infection and Beyond. Drug Des. Dev. Ther. 2015, 9, 5447–5468. [Google Scholar] [CrossRef]
- De Clercq, E. Mozobil® (Plerixafor, AMD3100), 10 Years after Its Approval by the US Food and Drug Administration. Antivir. Chem. Chemother. 2019, 27, 2040206619829382. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, S.; Song, K.; Shen, Q.; Ni, D.; Li, Q.; He, X.; Zhang, H.; Wang, Q.; Chen, Y.; et al. Unraveling Allosteric Landscapes of Allosterome with ASD. Nucleic Acids Res. 2020, 48, D394–D401. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, A. Advances in G Protein-Coupled Receptor Allostery: From Function to Structure. Mol. Pharmacol. 2014, 86, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Langmead, C.J.; Christopoulos, A. Functional and Structural Perspectives on Allosteric Modulation of GPCRs. Curr. Opin. Cell Biol. 2014, 27, 94–101. [Google Scholar] [CrossRef]
- Bruder, M.; Polo, G.; Trivella, D.B.B. Natural Allosteric Modulators and Their Biological Targets: Molecular Signatures and Mechanisms. Nat. Prod. Rep. 2020, 37, 488–514. [Google Scholar] [CrossRef]
- Kumar, V.; Bonifazi, A.; Ellenberger, M.P.; Keck, T.M.; Pommier, E.; Rais, R.; Slusher, B.S.; Gardner, E.; You, Z.B.; Xi, Z.-X.; et al. Highly Selective Dopamine D3 Receptor (D3R) Antagonists and Partial Agonists Based on Eticlopride and the D3R Crystal Structure: New Leads for Opioid Dependence Treatment. J. Med. Chem. 2016, 59, 7634–7650. [Google Scholar] [CrossRef]
- Galaj, E.; Bi, G.H.; Klein, B.; Hempel, B.; Shaik, A.B.; Gogarnoiu, E.S.; Friedman, J.; Lam, J.; Rais, R.; Reed, J.F.; et al. A Highly D3R-Selective and Efficacious Partial Agonist (S)-ABS01-113 Compared to Its D3R-Selective Antagonist Enantiomer (R)-ABS01-113 as Potential Treatments for Opioid Use Disorder. Neuropsychopharmacology 2022, 47, 2309–2318. [Google Scholar] [CrossRef]
- Tetrahydroisoquinoline derivatives as modulators of dopamine D3 receptors. WO1998050364A1, 12 November 1998.
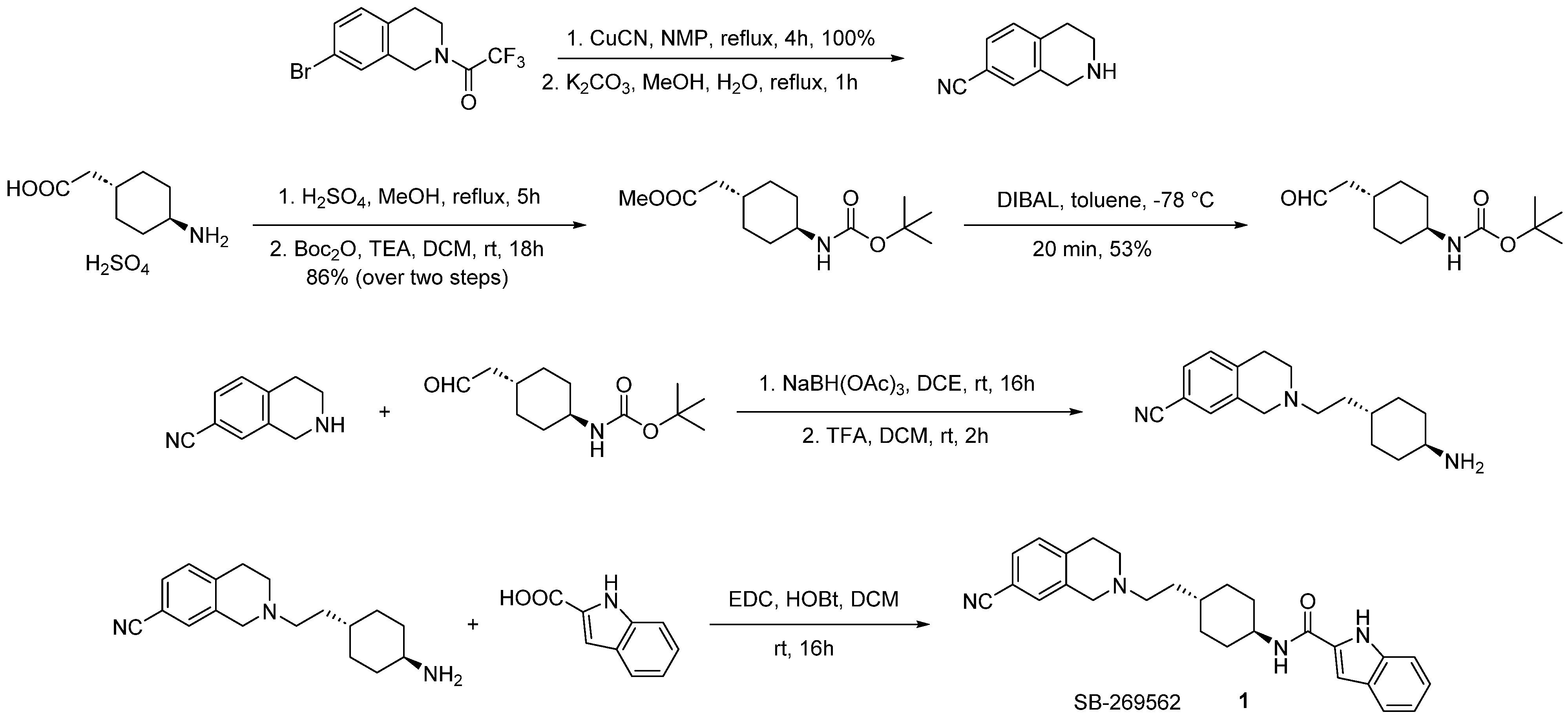
- Stemp, G.; Ashmeade, T.; Branch, C.L.; Hadley, M.S.; Hunter, A.J.; Johnson, C.N.; Nash, D.J.; Thewlis, K.M.; Vong, A.K.; Austin, N.E.; et al. Design and Synthesis of Trans-N-[4-[2-(6-Cyano-1,2,3, 4-Tetrahydroisoquinolin-2-Yl)Ethyl]Cyclohexyl]-4-Quinolinecarboxamide (SB-277011): A Potent and Selective Dopamine D3 Receptor Antagonist with High Oral Bioavailability and CNS Penetration in the Rat. J. Med. Chem. 2000, 43, 1878–1885. [Google Scholar] [CrossRef]
- Silvano, E.; Millan, M.J.; Mannoury la Cour, C.; Han, Y.; Duan, L.; Griffin, S.A.; Luedtke, R.R.; Aloisi, G.; Rossi, M.; Zazzeroni, F.; et al. The Tetrahydroisoquinoline Derivative SB269,652 Is an Allosteric Antagonist at Dopamine D3 and D2 Receptors. Mol. Pharmacol. 2010, 78, 925–934. [Google Scholar] [CrossRef]
- Lane, J.R.; Donthamsetti, P.; Shonberg, J.; Draper-Joyce, C.J.; Dentry, S.; Michino, M.; Shi, L.; López, L.; Scammells, P.J.; Capuano, B.; et al. A New Mechanism of Allostery in a G Protein-Coupled Receptor Dimer. Nat. Chem. Biol. 2014, 10, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Fasciani, I.; Marampon, F.; Maggio, R.; Scarselli, M. The First Negative Allosteric Modulator for Dopamine D2 and D3 Receptors, SB269652 May Lead to a New Generation of Antipsychotic Drugs. Mol. Pharmacol. 2017, 91, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Draper-Joyce, C.J.; Verma, R.K.; Michino, M.; Shonberg, J.; Kopinathan, A.; Klein Herenbrink, C.; Scammells, P.J.; Capuano, B.; Abramyan, A.M.; Thal, D.M.; et al. The Action of a Negative Allosteric Modulator at the Dopamine D2 Receptor Is Dependent upon Sodium Ions. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ågren, R.; Sahlholm, K. Evidence for Two Modes of Binding of the Negative Allosteric Modulator SB269,652 to the Dopamine D2 Receptor. Biomedicines 2021, 10, 22. [Google Scholar] [CrossRef]
- Shonberg, J.; Draper-Joyce, C.; Mistry, S.N.; Christopoulos, A.; Scammells, P.J.; Lane, J.R.; Capuano, B. Structure-Activity Study of N-((Trans)-4-(2-(7-Cyano-3,4-Dihydroisoquinolin-2(1H)-Yl)Ethyl)Cyclohexyl)-1H-Indole-2-Carboxamide (SB269652), a Bitopic Ligand That Acts as a Negative Allosteric Modulator of the Dopamine D2 Receptor. J. Med. Chem. 2015, 58, 5287–5307. [Google Scholar] [CrossRef]
- Mistry, S.N.; Shonberg, J.; Draper-Joyce, C.J.; Klein Herenbrink, C.; Michino, M.; Shi, L.; Christopoulos, A.; Capuano, B.; Scammells, P.J.; Lane, J.R. Discovery of a Novel Class of Negative Allosteric Modulator of the Dopamine D2 Receptor Through Fragmentation of a Bitopic Ligand. J. Med. Chem. 2015, 58, 6819–6843. [Google Scholar] [CrossRef]
- Kopinathan, A.; Draper-Joyce, C.; Szabo, M.; Christopoulos, A.; Scammells, P.J.; Lane, J.R.; Capuano, B. Subtle Modifications to the Indole-2-Carboxamide Motif of the Negative Allosteric Modulator N-((Trans)-4-(2-(7-Cyano-3,4-Dihydroisoquinolin-2(1 H)-Yl)Ethyl)Cyclohexyl)-1 H-Indole-2-Carboxamide (SB269652) Yield Dramatic Changes in Pharmacological Activity at the Dopamine D2 Receptor. J. Med. Chem. 2019, 62, 371–377. [Google Scholar] [CrossRef]
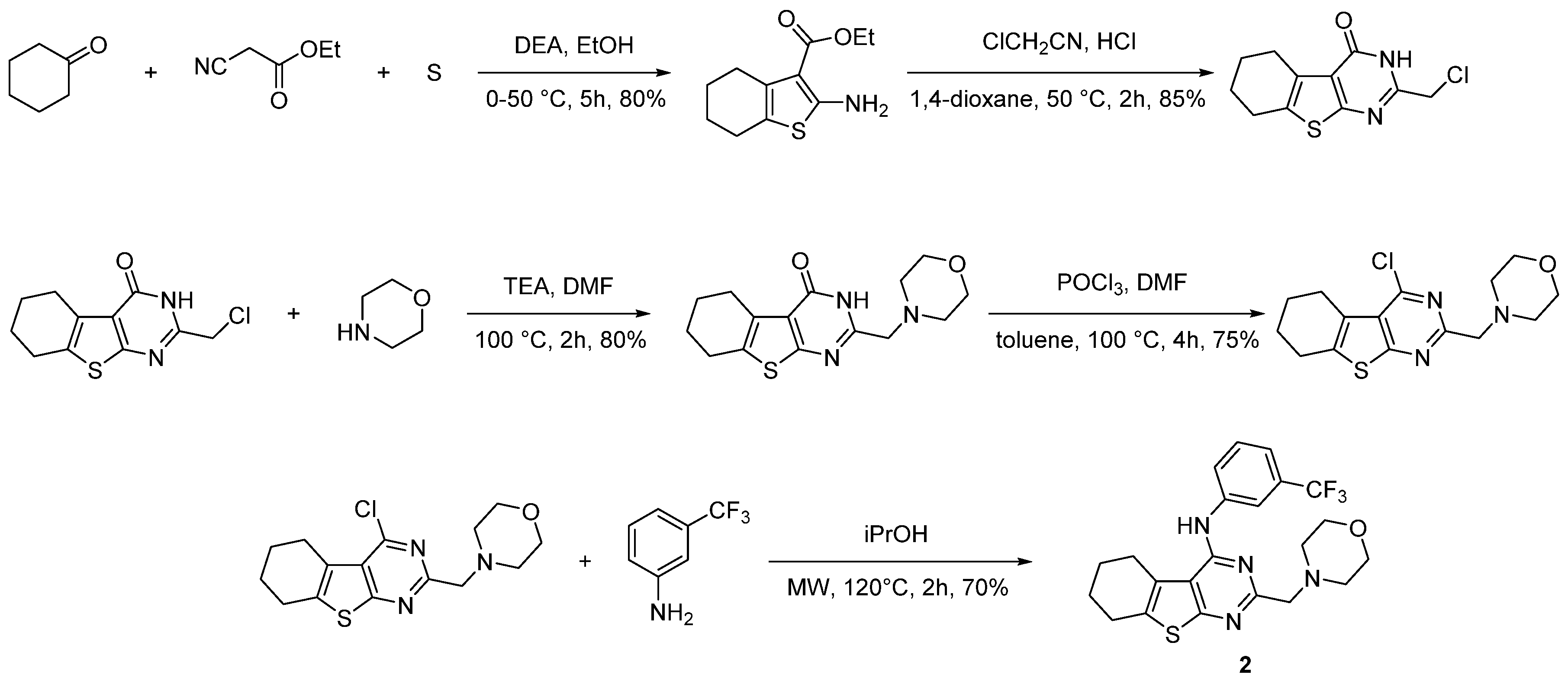
- Fyfe, T.J.; Zarzycka, B.; Lim, H.D.; Kellam, B.; Mistry, S.N.; Katrich, V.; Scammells, P.J.; Lane, J.R.; Capuano, B. A Thieno[2,3-d]Pyrimidine Scaffold Is a Novel Negative Allosteric Modulator of the Dopamine D2 Receptor. J. Med. Chem. 2019, 62, 174–206. [Google Scholar] [CrossRef]
- Fyfe, T.J.; Kellam, B.; Mistry, S.N.; Scammells, P.J.; Lane, J.R.; Capuano, B. Subtle Modifications to a Thieno[2,3-d] Pyrimidine Scaffold Yield Negative Allosteric Modulators and Agonists of the Dopamine D2 Receptor. Eur. J. Med. Chem. 2019, 168, 474–490. [Google Scholar] [CrossRef]
- Wood, M.; Ates, A.; Andre, V.M.; Michel, A.; Barnaby, R.; Gillard, M. In Vitro and in vivo Identification of Novel Positive Allosteric Modulators of the Human Dopamine D2 and D3 Receptor. Mol. Pharmacol. 2016, 89, 303–312. [Google Scholar] [CrossRef]
- Żuk, J.; Bartuzi, D.; Silva, A.G.; Pitucha, M.; Koszła, O.; Wróbel, T.M.; Matosiuk, D.; Castro, M.; Kaczor, A.A. Allosteric Modulation of Dopamine D2L Receptor in Complex with Gi1 and Gi2 Proteins: The Effect of Subtle Structural and Stereochemical Ligand Modifications. Pharmacol. Rep. 2022, 74, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Free, R.B.; Cuoco, C.A.; Xie, B.; Namkung, Y.; Prabhu, V.V.; Willette, B.K.A.; Day, M.M.; Sanchez-Soto, M.; Lane, J.R.; Laporte, S.A.; et al. Pharmacological Characterization of the Imipridone Anticancer Drug ONC201 Reveals a Negative Allosteric Mechanism of Action at the D2 Dopamine Receptor. Mol. Pharmacol. 2021, 100, 372–387. [Google Scholar] [CrossRef] [PubMed]
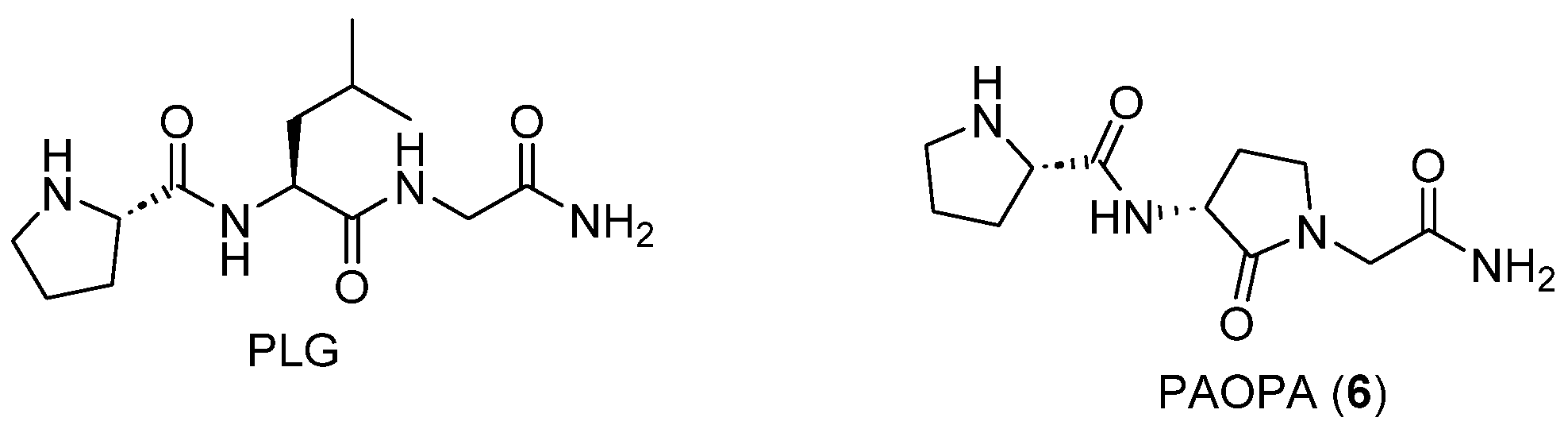
- Mishra, R.K.; Chiu, S.; Chiu, P.; Mishra, C.P. Pharmacology of L-Prolyl-L-Leucyl-Glycinamide (PLG): A Review. Methods Find. Exp. Clin. Pharmacol. 1983, 5, 203–233. [Google Scholar] [PubMed]
- Sampaio-Dias, I.E.; Reis-Mendes, A.; Costa, V.M.; García-Mera, X.; Brea, J.; Loza, M.I.; Pires-Lima, B.L.; Alcoholado, C.; Algarra, M.; Rodríguez-Borges, J.E. Discovery of New Potent Positive Allosteric Modulators of Dopamine D2 Receptors: Insights into the Bioisosteric Replacement of Proline to 3-Furoic Acid in the Melanostatin Neuropeptide. J. Med. Chem. 2021, 64, 6209–6220. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, L.K.; Bajwa, S.B.; Johnson, R.L.; Mishra, R.K. Interaction of L-Prolyl-l-Leucyl Glycinamide with Dopamine D2 Receptor: Evidence for Modulation of Agonist Affinity States in Bovine Striatal Membranes. J. Neurochem. 1988, 50, 960–968. [Google Scholar] [CrossRef]
- Bhargava, H.N. Effects of Prolyl-Leucyl-Glycinamide and Cyclo(Leucyl-Glycine) on the Supersensitivity of Dopamine Receptors in Brain Induced by Chronic Administration of Haloperidol to Rats. Neuropharmacology 1984, 23, 439–444. [Google Scholar] [CrossRef]
- Kostrzewa, R.M.; Kastin, A.J.; Sobrian, S.K. Potentiation of Apomorphine Action in Rats by L-Prolyl-l-Leucyl-Glycine Amide. Pharmacol. Biochem. Behav. 1978, 9, 375–378. [Google Scholar] [CrossRef]
- Sampaio-Dias, I.E.; Silva-Reis, S.C.; García-Mera, X.; Brea, J.; Loza, M.I.; Alves, C.S.; Algarra, M.; Rodríguez-Borges, J.E. Synthesis, Pharmacological, and Biological Evaluation of MIF-1 Picolinoyl Peptidomimetics as Positive Allosteric Modulators of D2R. ACS Chem. Neurosci. 2019, 10, 3690–3702. [Google Scholar] [CrossRef]
- Verma, V.; Mann, A.; Costain, W.; Pontoriero, G.; Castellano, J.M.; Skoblenick, K.; Gupta, S.K.; Pristupa, Z.; Niznik, H.B.; Johnson, R.L.; et al. Modulation of Agonist Binding to Human Dopamine Receptor Subtypes by L-Prolyl-L-Leucyl-Glycinamide and a Peptidomimetic Analog. J. Pharmacol. Exp. Ther. 2005, 315, 1228–1236. [Google Scholar] [CrossRef]
- Dyck, B.; Guest, K.; Sookram, C.; Basu, D.; Johnson, R.; Mishra, R.K. PAOPA, a Potent Analogue of Pro-Leu-Glycinamide and Allosteric Modulator of the Dopamine D2 Receptor, Prevents NMDA Receptor Antagonist (MK-801)-Induced Deficits in Social Interaction in the Rat: Implications for the Treatment of Negative Symptoms in Schizophrenia. Schizophr. Res. 2011, 125, 88–92. [Google Scholar] [CrossRef]
- Beyaert, M.G.R.; Daya, R.P.; Dyck, B.A.; Johnson, R.L.; Mishra, R.K. PAOPA, a Potent Dopamine D2 Receptor Allosteric Modulator, Prevents and Reverses Behavioral and Biochemical Abnormalities in an Amphetamine-Sensitized Preclinical Animal Model of Schizophrenia. Eur. Neuropsychopharmacol. 2013, 23, 253–262. [Google Scholar] [CrossRef]
- Daya, R.P.; Bhandari, J.; Kooner, S.K.; Ho, J.; Rowley, C.D.; Bock, N.A.; Farncombe, T.; Mishra, R.K. The Dopamine Allosteric Agent, PAOPA, Demonstrates Therapeutic Potential in the Phencyclidine NMDA Pre-Clinical Rat Model of Schizophrenia. Front. Behav. Neurosci. 2018, 12, 302. [Google Scholar] [CrossRef]
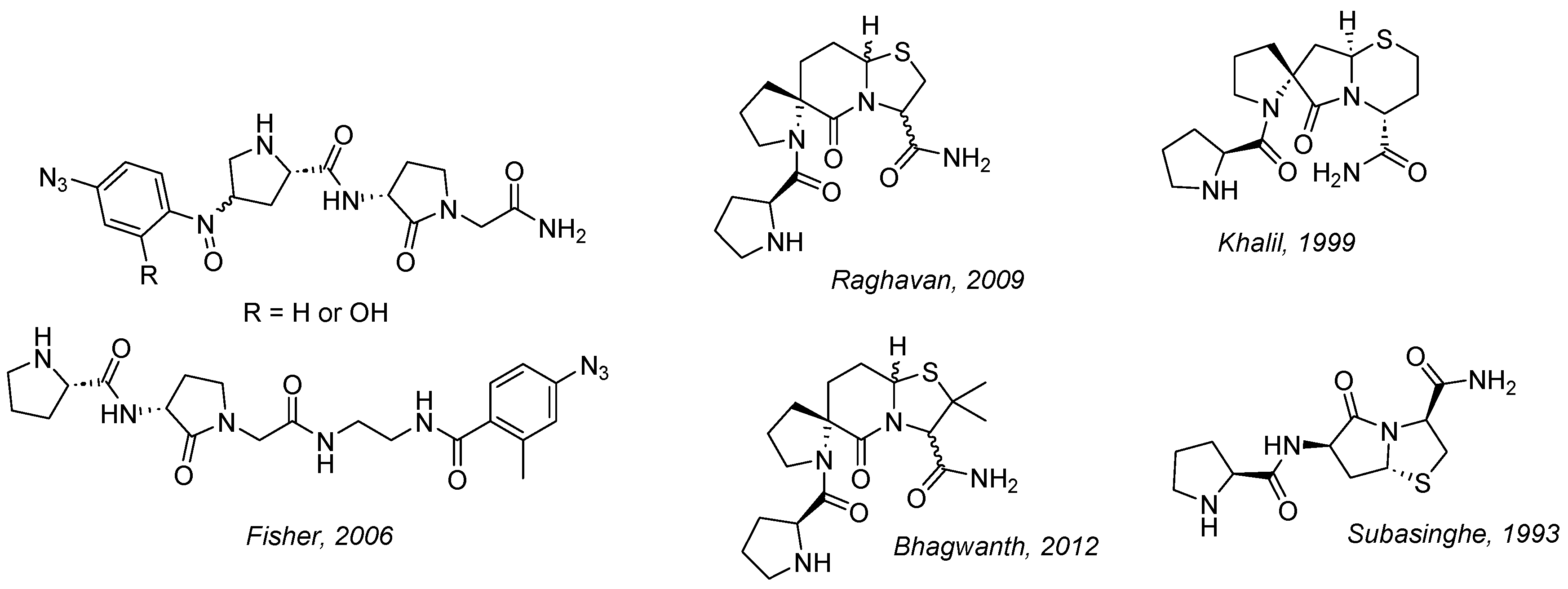
- Fisher, A.; Mann, A.; Verma, V.; Thomas, N.; Mishra, R.K.; Johnson, R.L. Design and Synthesis of Photoaffinity-Labeling Ligands of the L-Prolyl-L-Leucylglycinamide Binding Site Involved in the Allosteric Modulation of the Dopamine Receptor. J. Med. Chem. 2006, 49, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, B.; Skoblenick, K.J.; Bhagwanth, S.; Argintaru, N.; Mishra, R.K.; Johnson, R.L. Allosteric Modulation of the Dopamine D2 Receptor by Pro-Leu-Gly-NH2 Peptidomimetics Constrained in Either a Polyproline II Helix or a Type II Beta-Turn Conformation. J. Med. Chem. 2009, 52, 2043–2051. [Google Scholar] [CrossRef]
- Bhagwanth, S.; Mishra, S.; Daya, R.; Mah, J.; Mishra, R.K.; Johnson, R.L. Transformation of Pro-Leu-Gly-NH2 Peptidomimetic Positive Allosteric Modulators of the Dopamine D2 Receptor into Negative Modulators. ACS Chem. Neurosci. 2012, 3, 274–284. [Google Scholar] [CrossRef]
- Subasinghe, N.L.; Bontems, R.J.; McIntee, E.; Mishra, R.K.; Johnson, R.L. Bicyclic Thiazolidine Lactam Peptidomimetics of the Dopamine Receptor Modulating Peptide Pro-Leu-Gly-NH2. J. Med. Chem. 1993, 36, 2356–2361. [Google Scholar] [CrossRef] [PubMed]
- Khalil, E.M.; Ojala, W.H.; Pradhan, A.; Nair, V.D.; Gleason, W.B.; Mishra, R.K.; Johnson, R.L. Design, Synthesis, and Dopamine Receptor Modulating Activity of Spiro Bicyclic Peptidomimetics of L-Prolyl-L-Leucyl-Glycinamide. J. Med. Chem. 1999, 42, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Bhagwanth, S.; Mishra, R.K.; Johnson, R.L. Development of Peptidomimetic Ligands of Pro-Leu-Gly-NH(2) as Allosteric Modulators of the Dopamine D2 Receptor. Beilstein J. Org. Chem. 2013, 9, 204–214. [Google Scholar] [CrossRef]
- Ferreira da Costa, J.; Caamaño, O.; Fernández, F.; García-Mera, X.; Sampaio-Dias, I.E.; Brea, J.M.; Cadavid, M.I. Synthesis and Allosteric Modulation of the Dopamine Receptor by Peptide Analogs of L-Prolyl-L-Leucyl-Glycinamide (PLG) Modified in the L-Proline or L-Proline and L-Leucine Scaffolds. Eur. J. Med. Chem. 2013, 69, 146–158. [Google Scholar] [CrossRef]
- Oliver, M.; Gadais, C.; García-Pindado, J.; Teixidó, M.; Lensen, N.; Chaume, G.; Brigaud, T. Trifluoromethylated Proline Analogues as Efficient Tools to Enhance the Hydrophobicity and to Promote Passive Diffusion Transport of the L-Prolyl-L-Leucyl Glycinamide (PLG) Tripeptide. RSC Adv. 2018, 8, 14597–14602. [Google Scholar] [CrossRef]
- Sampaio-Dias, I.E.; Sousa, C.A.D.; García-Mera, X.; Ferreira da Costa, J.; Caamaño, O.; Rodríguez-Borges, J.E. Novel L-Prolyl-L-Leucylglycinamide (PLG) Tripeptidomimetics Based on a 2-Azanorbornane Scaffold as Positive Allosteric Modulators of the D2R. Org. Biomol. Chem. 2016, 14, 11065–11069. [Google Scholar] [CrossRef] [PubMed]
- Deflorian, F.; Mason, J.S.; Bortolato, A.; Tehan, B.G. Impact of Recently Determined Crystallographic Structures of GPCRs on Drug Discovery. In Structural Biology in Drug Discovery; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 449–477. ISBN 978-1-118-68112-1. [Google Scholar]
- Livingston, K.E.; Stanczyk, M.A.; Burford, N.T.; Alt, A.; Canals, M.; Traynor, J.R. Pharmacologic Evidence for a Putative Conserved Allosteric Site on Opioid Receptors. Mol. Pharmacol. 2018, 93, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; del Sol, A.; Nussinov, R. Allostery: Absence of a Change in Shape Does Not Imply That Allostery Is Not at Play. J. Mol. Biol. 2008, 378, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Tsai, C.J. Allostery without a Conformational Change? Revisiting the Paradigm. Curr. Opin. Struct. Biol. 2015, 30, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The Ensemble Nature of Allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef]
- Gusach, A.; Maslov, I.; Luginina, A.; Borshchevskiy, V.; Mishin, A.; Cherezov, V. Beyond Structure: Emerging Approaches to Study GPCR Dynamics. Curr. Opin. Struct. Biol. 2020, 63, 18–25. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Chen, K.Y.M.; Keri, D.; Barth, P. Computational Design of G Protein-Coupled Receptor Allosteric Signal Transductions. Nat. Chem. Biol. 2020, 16, 77–86. [Google Scholar] [CrossRef]
- Greener, J.G.; Sternberg, M.J. Structure-Based Prediction of Protein Allostery. Curr. Opin. Struct. Biol. 2018, 50, 1–8. [Google Scholar] [CrossRef]
- Sheik Amamuddy, O.; Veldman, W.; Manyumwa, C.; Khairallah, A.; Agajanian, S.; Oluyemi, O.; Verkhivker, G.; Tastan Bishop, O. Integrated Computational Approaches and Tools ForAllosteric Drug Discovery. Int. J. Mol. Sci. 2020, 21, 847. [Google Scholar] [CrossRef]
- Bowerman, S.; Wereszczynski, J. Detecting Allosteric Networks Using Molecular Dynamics Simulation. Methods Enzymol. 2016, 578, 429–447. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Opportunities and Challenges in the Discovery of Allosteric Modulators of GPCRs. In Computational Methods for GPCR Drug Discovery; Heifetz, A., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 297–319. ISBN 978-1-4939-7465-8. [Google Scholar]
- Dokholyan, N.V. Controlling Allosteric Networks in Proteins. Chem. Rev. 2016, 116, 6463–6487. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, D.; Kaczor, A.A.; Targowska-Duda, K.M.; Matosiuk, D. Recent Advances and Applications of Molecular Docking to G Protein-Coupled Receptors. Molecules 2017, 22, 340. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Lee, C.T.; Durrant, J.D.; Malmstrom, R.D.; Feher, V.A.; Amaro, R.E. Emerging Computational Methods for the Rational Discovery of Allosteric Drugs. Chem. Rev. 2016, 116, 6370–6390. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.M.; Wilkins, A.D.; Rodriguez, G.J.; Wensel, T.G.; Lichtarge, O. Intramolecular Allosteric Communication in Dopamine D2 Receptor Revealed by Evolutionary Amino Acid Covariation. Proc. Natl. Acad. Sci. USA 2016, 113, 3539–3544. [Google Scholar] [CrossRef] [PubMed]
- Panjkovich, A.; Daura, X. PARS: A Web Server for the Prediction of Protein Allosteric and Regulatory Sites. Bioinformatics 2014, 30, 1314–1315. [Google Scholar] [CrossRef]
- Goncearenco, A.; Mitternacht, S.; Yong, T.; Eisenhaber, B.; Eisenhaber, F.; Berezovsky, I.N. SPACER: Server for Predicting Allosteric Communication and Effects of Regulation. Nucleic Acids Res. 2013, 41, W266–W272. [Google Scholar] [CrossRef]
- Greener, J.G.; Sternberg, M.J.E. AlloPred: Prediction of Allosteric Pockets on Proteins Using Normal Mode Perturbation Analysis. BMC Bioinform. 2015, 16, 335. [Google Scholar] [CrossRef]
- Salmas, R.E.; Stein, M.; Yurtsever, M.; Seeman, P.; Erol, I.; Mestanoglu, M.; Durdagi, S. The Signaling Pathway of Dopamine D2 Receptor (D2R) Activation Using Normal Mode Analysis (NMA) and the Construction of Pharmacophore Models for D2R Ligands. J. Biomol. Struct. Dyn. 2017, 35, 2040–2048. [Google Scholar] [CrossRef]
- Kaya, C.; Armutlulu, A.; Ekesan, S.; Haliloglu, T. MCPath: Monte Carlo Path Generation Approach to Predict Likely Allosteric Pathways and Functional Residues. Nucleic Acids Res. 2013, 41, W249–W255. [Google Scholar] [CrossRef]
- McClendon, C.L.; Friedland, G.; Mobley, D.L.; Amirkhani, H.; Jacobson, M.P. Quantifying Correlations Between Allosteric Sites in Thermodynamic Ensembles. J. Chem. Theory Comput. 2009, 5, 2486–2502. [Google Scholar] [CrossRef] [PubMed]
- Van Wart, A.T.; Durrant, J.; Votapka, L.; Amaro, R.E. Weighted Implementation of Suboptimal Paths (WISP): An Optimized Algorithm and Tool for Dynamical Network Analysis. J. Chem. Theory Comput. 2014, 10, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Molecular Mechanisms of Allosteric Probe Dependence in μ Opioid Receptor. J. Biomol. Struct. Dyn. 2019, 37, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Bruzzese, A.; Dalton, J.A.R.; Giraldo, J. Insights into Adenosine A2A Receptor Activation through Cooperative Modulation of Agonist and Allosteric Lipid Interactions. PLoS Comput. Biol. 2020, 16, e1007818. [Google Scholar] [CrossRef]
- Ng, H.W.; Laughton, C.A.; Doughty, S.W. Molecular Dynamics Simulations of the Adenosine A2a Receptor in POPC and POPE Lipid Bilayers: Effects of Membrane on Protein Behavior. J. Chem. Inf. Model. 2014, 54, 573–581. [Google Scholar] [CrossRef]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Activation and Allosteric Modulation of Human μ Opioid Receptor in Molecular Dynamics. J. Chem. Inf. Model. 2015, 55, 2421–2434. [Google Scholar] [CrossRef]
- Dror, R.O.; Green, H.F.; Valant, C.; Borhani, D.W.; Valcourt, J.R.; Pan, A.C.; Arlow, D.H.; Canals, M.; Lane, J.R.; Rahmani, R.; et al. Structural Basis for Modulation of a G-Protein-Coupled Receptor by Allosteric Drugs. Nature 2013, 503, 295–299. [Google Scholar] [CrossRef]
- Shaw, D.E.; Dror, R.O.; Salmon, J.K.; Grossman, J.P.; Mackenzie, K.M.; Bank, J.A.; Young, C.; Deneroff, M.M.; Batson, B.; Bowers, K.J.; et al. Millisecond-Scale Molecular Dynamics Simulations on Anton. In Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis; Association for Computing Machinery: Portland, OR, USA, 2009; pp. 1–11. [Google Scholar]
- Saleh, N.; Ibrahim, P.; Saladino, G.; Gervasio, F.L.; Clark, T. An Efficient Metadynamics-Based Protocol to Model the Binding Affinity and the Transition State Ensemble of G-Protein-Coupled Receptor Ligands. J. Chem. Inf. Model. 2017, 57, 1210–1217. [Google Scholar] [CrossRef]
- Söldner, C.A.; Horn, A.H.C.; Sticht, H. A Metadynamics-Based Protocol for the Determination of GPCR-Ligand Binding Modes. Int. J. Mol. Sci. 2019, 20, 1970. [Google Scholar] [CrossRef]
- Schneider, S.; Provasi, D.; Filizola, M. The Dynamic Process of Drug-GPCR Binding at Either Orthosteric or Allosteric Sites Evaluated by Metadynamics. Methods Mol. Biol. 2015, 1335, 277–294. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yeatman, H.R.; Provasi, D.; Alt, A.; Christopoulos, A.; Canals, M.; Filizola, M. Proposed Mode of Binding and Action of Positive Allosteric Modulators at Opioid Receptors. ACS Chem. Biol. 2016, 11, 1220–1229. [Google Scholar] [CrossRef]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Interplay between Two Allosteric Sites and Their Influence on Agonist Binding in Human μ Opioid Receptor. J. Chem. Inf. Model. 2016, 56, 563–570. [Google Scholar] [CrossRef]
- Lane, J.R.; Chubukov, P.; Liu, W.; Canals, M.; Cherezov, V.; Abagyan, R.; Stevens, R.C.; Katritch, V. Structure-Based Ligand Discovery Targeting Orthosteric and Allosteric Pockets of Dopamine Receptors. Mol. Pharmacol. 2013, 84, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [PubMed]
- Pelé, J.; Abdi, H.; Moreau, M.; Thybert, D.; Chabbert, M. Multidimensional Scaling Reveals the Main Evolutionary Pathways of Class A G-Protein-Coupled Receptors. PLoS ONE 2011, 6, e19094. [Google Scholar] [CrossRef] [PubMed]
- Selent, J.; Sanz, F.; Pastor, M.; de Fabritiis, G. Induced Effects of Sodium Ions on Dopaminergic G-Protein Coupled Receptors. PLoS Comput. Biol. 2010, 6, e1000884. [Google Scholar] [CrossRef]
- Harvey, M.J.; Giupponi, G.; Fabritiis, G.D. ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J. Chem. Theory Comput. 2009, 5, 1632–1639. [Google Scholar] [CrossRef]
- Neve, K.A.; Cumbay, M.G.; Thompson, K.R.; Yang, R.; Buck, D.C.; Watts, V.J.; DuRand, C.J.; Teeter, M.M. Modeling and Mutational Analysis of a Putative Sodium-Binding Pocket on the Dopamine D2 Receptor. Mol. Pharmacol. 2001, 60, 373–381. [Google Scholar] [CrossRef]
- Fenalti, G.; Giguere, P.M.; Katritch, V.; Huang, X.P.; Thompson, A.A.; Cherezov, V.; Roth, B.L.; Stevens, R.C. Molecular Control of δ-Opioid Receptor Signalling. Nature 2014, 506, 191–196. [Google Scholar] [CrossRef]
- Żuk, J.; Bartuzi, D.; Matosiuk, D.; Kaczor, A.A. Preferential Coupling of Dopamine D2S and D2L Receptor Isoforms with Gi1 and Gi2 Proteins-In Silico Study. Int. J. Mol. Sci. 2020, 21, 436. [Google Scholar] [CrossRef]
- Żuk, J.; Bartuzi, D.; Miszta, P.; Kaczor, A.A. The Role of Lipids in Allosteric Modulation of Dopamine D2 Receptor-In Silico Study. Molecules 2022, 27, 1335. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shimon, A.; Niv, M.Y. AnchorDock: Blind and Flexible Anchor-Driven Peptide Docking. Structure 2015, 23, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.M.; Alhossary, A.A.; Mu, Y.; Kwoh, C.K. Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration. Sci. Rep. 2017, 7, 15451. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xu, X.; Zou, X. Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. Structure 2016, 24, 1842–1853. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the Molecular Mechanics/Poisson Boltzmann Surface Area and Molecular Mechanics/Generalized Born Surface Area Methods. II. The Accuracy of Ranking Poses Generated from Docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef]
- Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2017. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2017. Available online: https://www.schrodinger.com/products/desmond (accessed on 15 November 2022).
- Newman, A.H.; Beuming, T.; Banala, A.K.; Donthamsetti, P.; Pongetti, K.; LaBounty, A.; Levy, B.; Cao, J.; Michino, M.; Luedtke, R.R.; et al. Molecular Determinants of Selectivity and Efficacy at the Dopamine D3 Receptor. J. Med. Chem. 2012, 55, 6689–6699. [Google Scholar] [CrossRef]
- Draper-Joyce, C.J.; Michino, M.; Verma, R.K.; Klein Herenbrink, C.; Shonberg, J.; Kopinathan, A.; Scammells, P.J.; Capuano, B.; Thal, D.M.; Javitch, J.A.; et al. The Structural Determinants of the Bitopic Binding Mode of a Negative Allosteric Modulator of the Dopamine D2 Receptor. Biochem. Pharmacol. 2018, 148, 315–328. [Google Scholar] [CrossRef]
- Verma, R.K.; Abramyan, A.M.; Michino, M.; Free, R.B.; Sibley, D.R.; Javitch, J.A.; Lane, J.R.; Shi, L. The E2.65 A Mutation Disrupts Dynamic Binding Poses of SB269652 at the Dopamine D2 and D3 Receptors. PLoS Comput. Biol. 2018, 14, e1005948. [Google Scholar] [CrossRef]
- Wang, X.; Heinz, B.A.; Qian, Y.W.; Carter, J.H.; Gadski, R.A.; Beavers, L.S.; Little, S.P.; Yang, C.R.; Beck, J.P.; Hao, J.; et al. Intracellular Binding Site for a Positive Allosteric Modulator of the Dopamine D1 Receptor. Mol. Pharmacol. 2018, 94, 1232–1245. [Google Scholar] [CrossRef]
- Männel, B.; Jaiteh, M.; Zeifman, A.; Randakova, A.; Möller, D.; Hübner, H.; Gmeiner, P.; Carlsson, J. Structure-Guided Screening for Functionally Selective D2 Dopamine Receptor Ligands from a Virtual Chemical Library. ACS Chem. Biol. 2017, 12, 2652–2661. [Google Scholar] [CrossRef] [PubMed]
- Katritch, V.; Rueda, M.; Abagyan, R. Ligand-Guided Receptor Optimization. Methods Mol. Biol. 2012, 857, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.A.C.; Totrov, M.; Abagyan, R. Docking and Scoring with ICM: The Benchmarking Results and Strategies for Improvement. J. Comput. Aided Mol. Des. 2012, 26, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Shoichet, B.K. Rapid Context-Dependent Ligand Desolvation in Molecular Docking. J. Chem. Inf. Model. 2010, 50, 1561–1573. [Google Scholar] [CrossRef]
- Trzaskowski, B.; Latek, D.; Yuan, S.; Ghoshdastider, U.; Debinski, A.; Filipek, S. Action of Molecular Switches in GPCRs--Theoretical and Experimental Studies. Curr. Med. Chem. 2012, 19, 1090–1109. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Salomon-Ferrer, R.; Lee, S.; Vaidehi, N. Conserved Mechanism of Conformational Stability and Dynamics in G-Protein-Coupled Receptors. J. Chem. Theory Comput. 2016, 12, 5575–5584. [Google Scholar] [CrossRef]
- Vaidehi, N.; Bhattacharya, S. Allosteric Communication Pipelines in G-Protein-Coupled Receptors. Curr. Opin. Pharmacol. 2016, 30, 76–83. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Vaidehi, N. Differences in Allosteric Communication Pipelines in the Inactive and Active States of a GPCR. Biophys. J. 2014, 107, 422–434. [Google Scholar] [CrossRef]
- Luderman, K.D.; Conroy, J.L.; Free, R.B.; Southall, N.; Ferrer, M.; Sanchez-Soto, M.; Moritz, A.E.; Willette, B.K.A.; Fyfe, T.J.; Jain, P.; et al. Identification of Positive Allosteric Modulators of the D1 Dopamine Receptor That Act at Diverse Binding Sites. Mol. Pharmacol. 2018, 94, 1197–1209. [Google Scholar] [CrossRef]
- Bonifazi, A.; Yano, H.; Guerrero, A.M.; Kumar, V.; Hoffman, A.F.; Lupica, C.R.; Shi, L.; Newman, A.H. Novel and Potent Dopamine D2 Receptor Go-Protein Biased Agonists. ACS Pharmacol. Transl. Sci. 2019, 2, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Czuczi, T.; Murányi, J.; Bárány, P.; Móra, I.; Borbély, A.; Csala, M.; Csámpai, A. Synthesis and Antiproliferative Activity of Novel Imipridone–Ferrocene Hybrids with Triazole and Alkyne Linkers. Pharmaceuticals 2022, 15, 468. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.L.; Rajakumar, G.; Srivastava, L.K.; Mishra, R.K.; Johnson, R.L. Dopamine Receptor Modulation by Conformationally Constrained Analogues of Pro-Leu-Gly-NH2. J. Med. Chem. 1988, 31, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
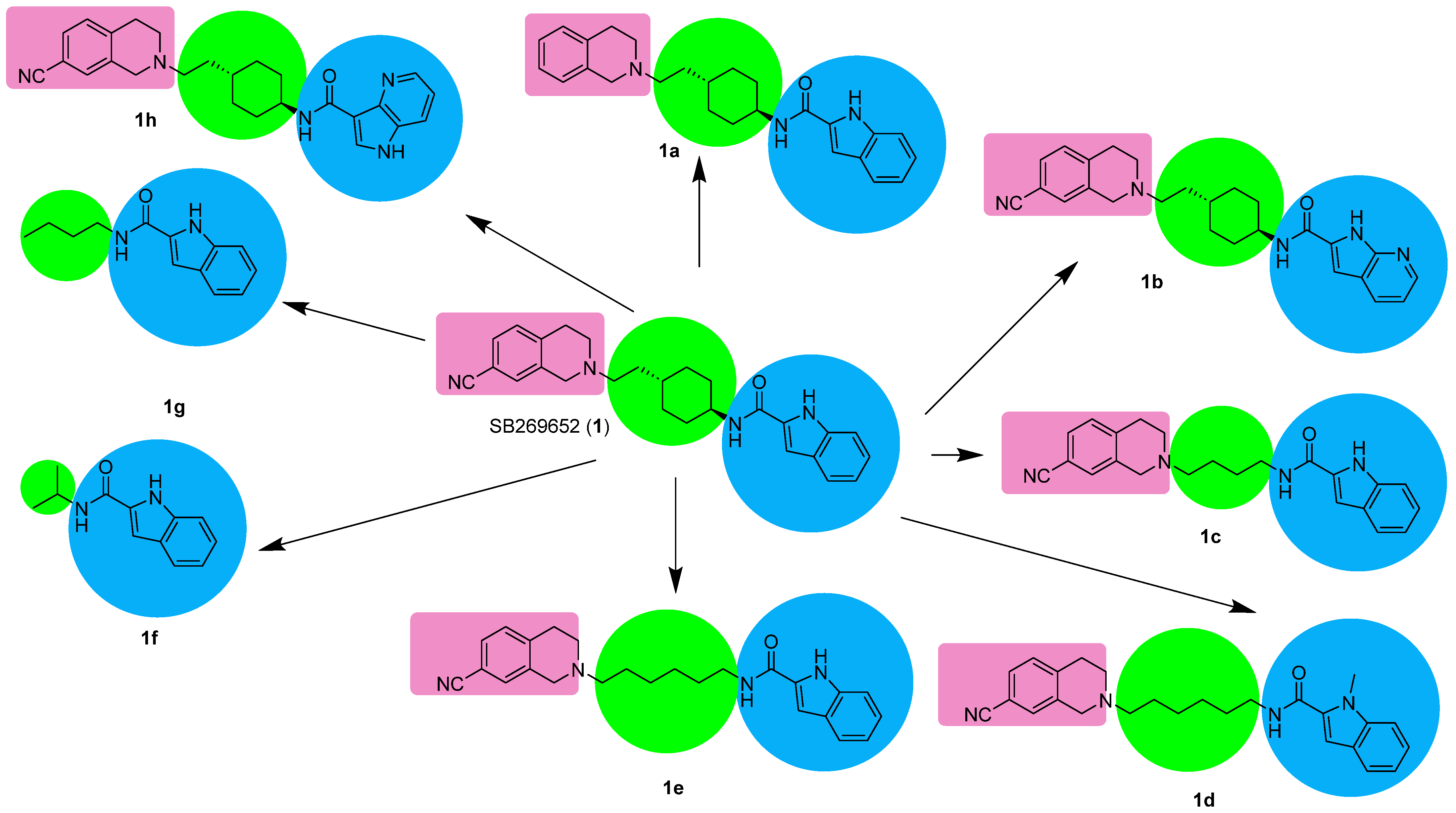
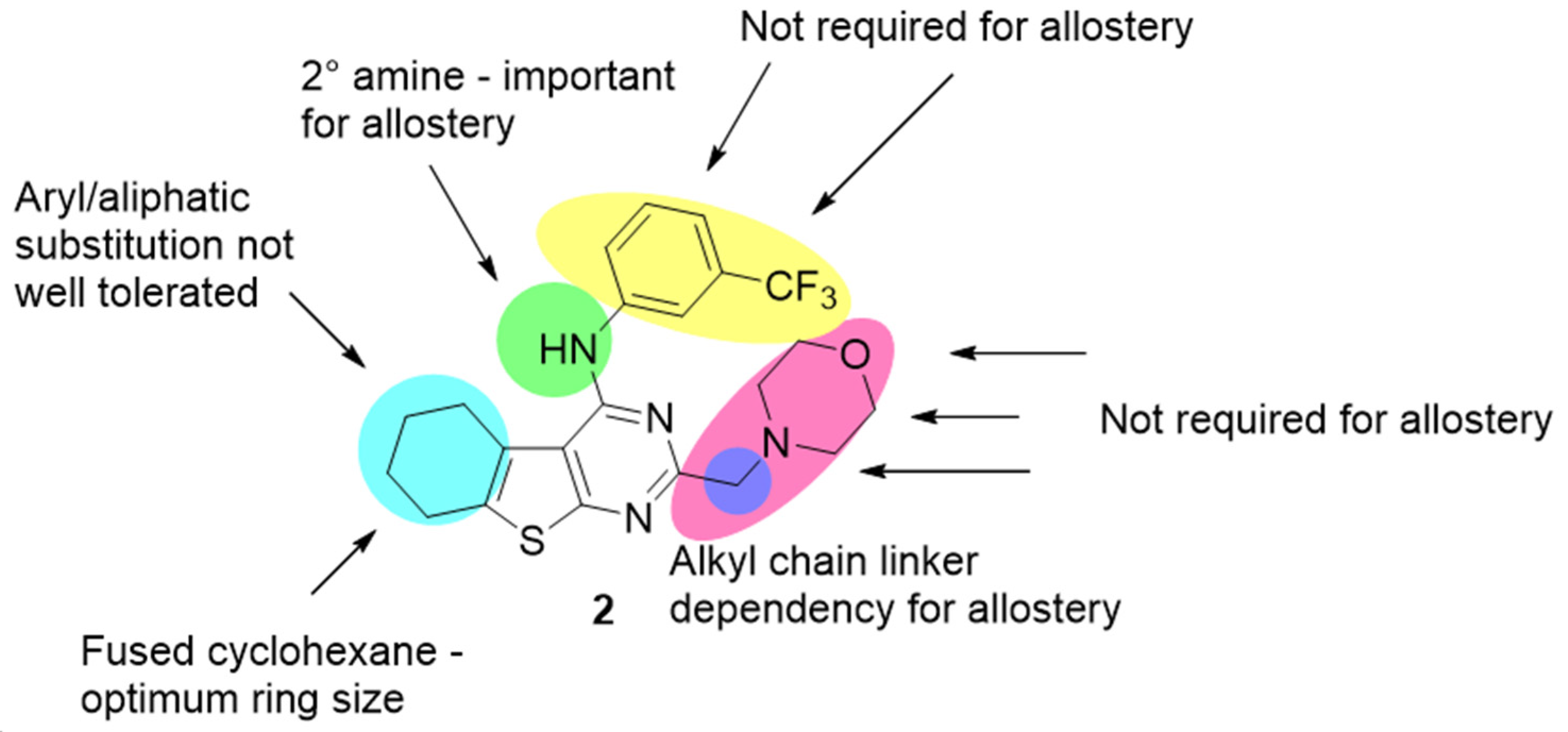

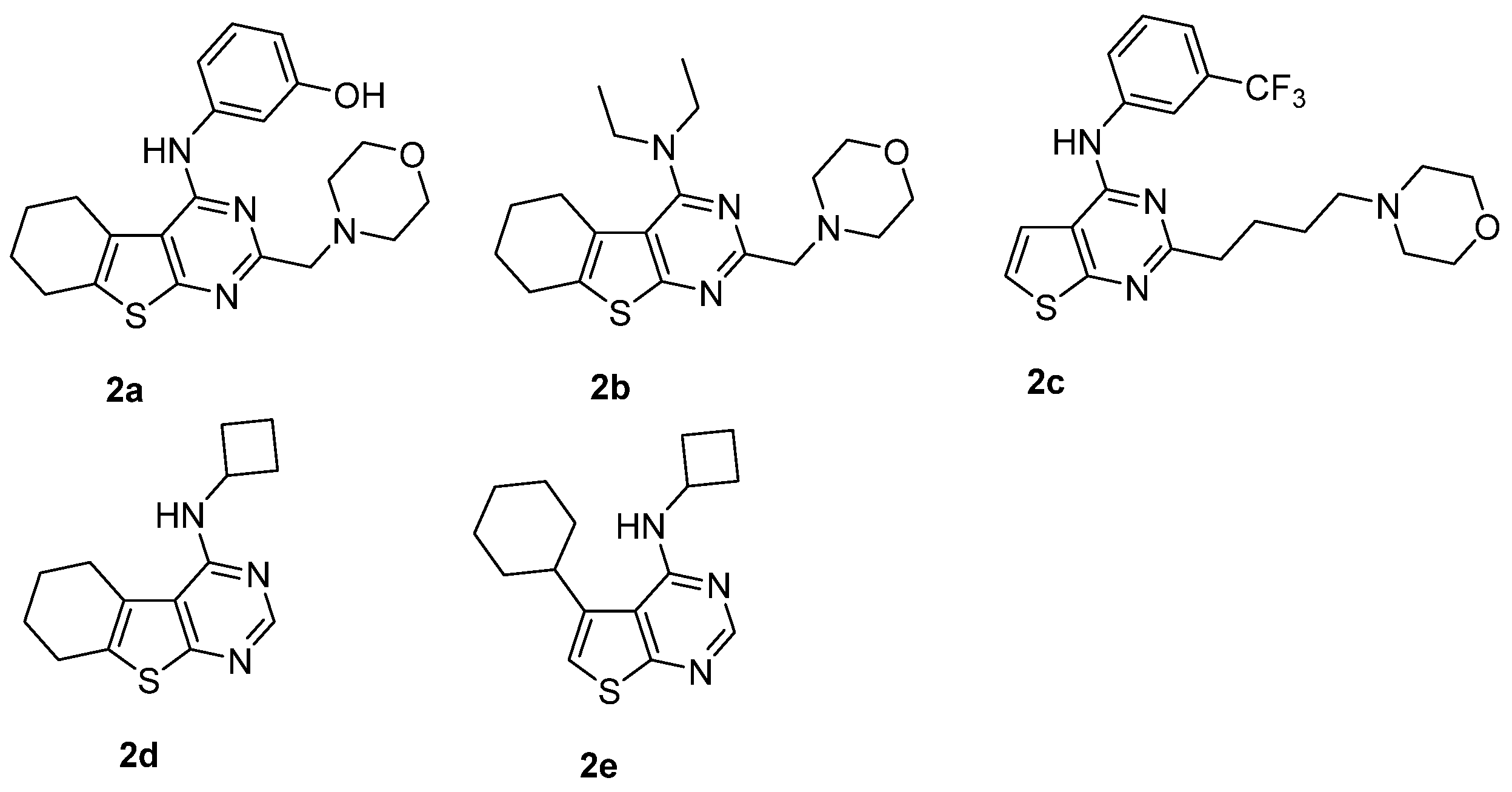
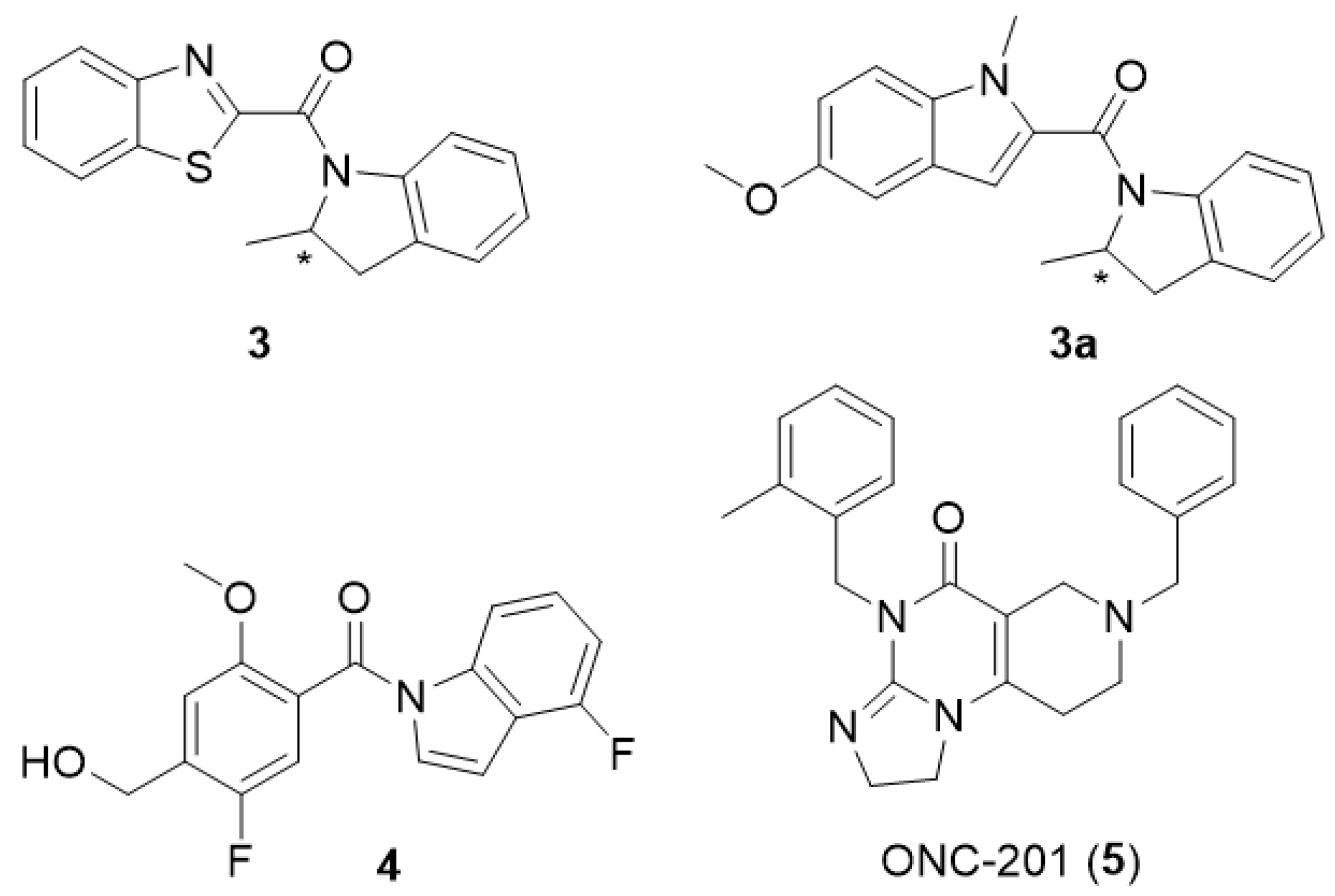

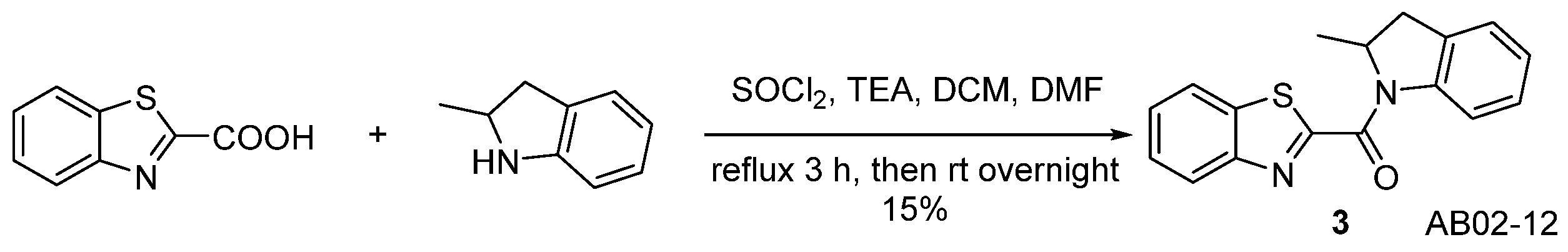
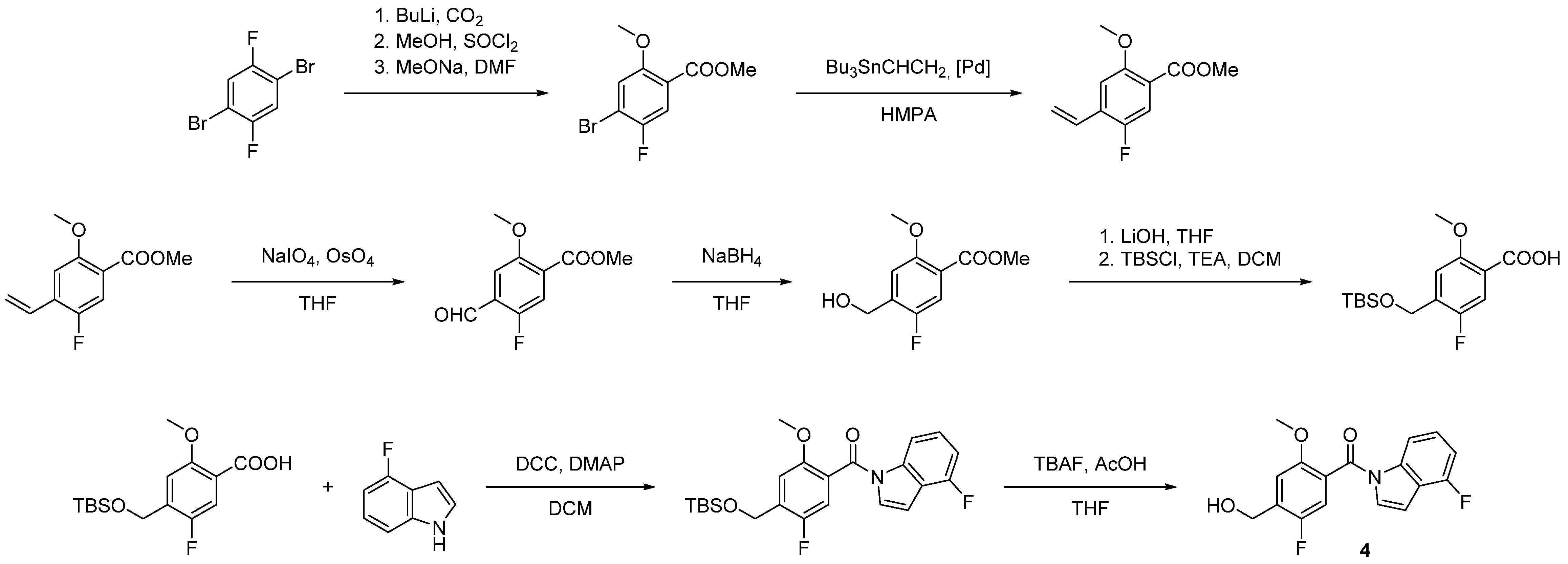
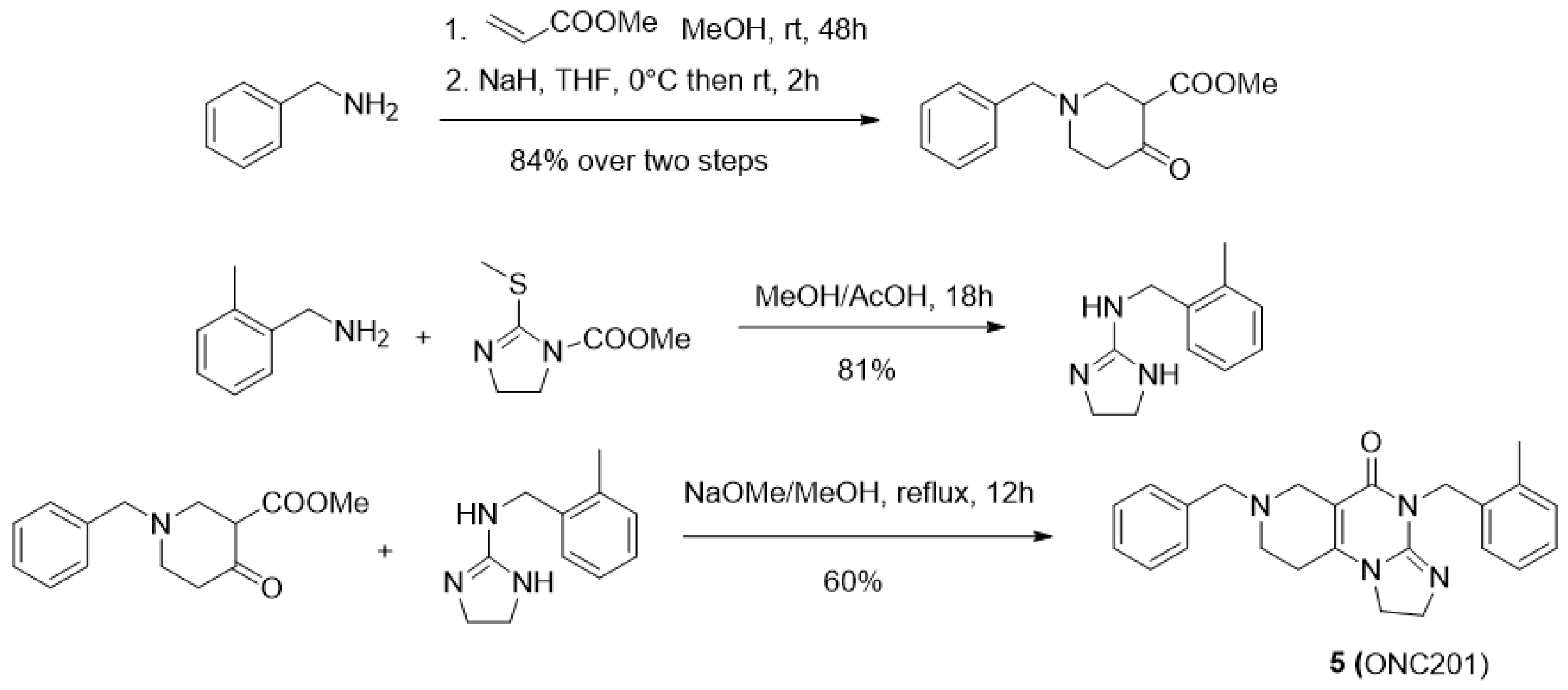
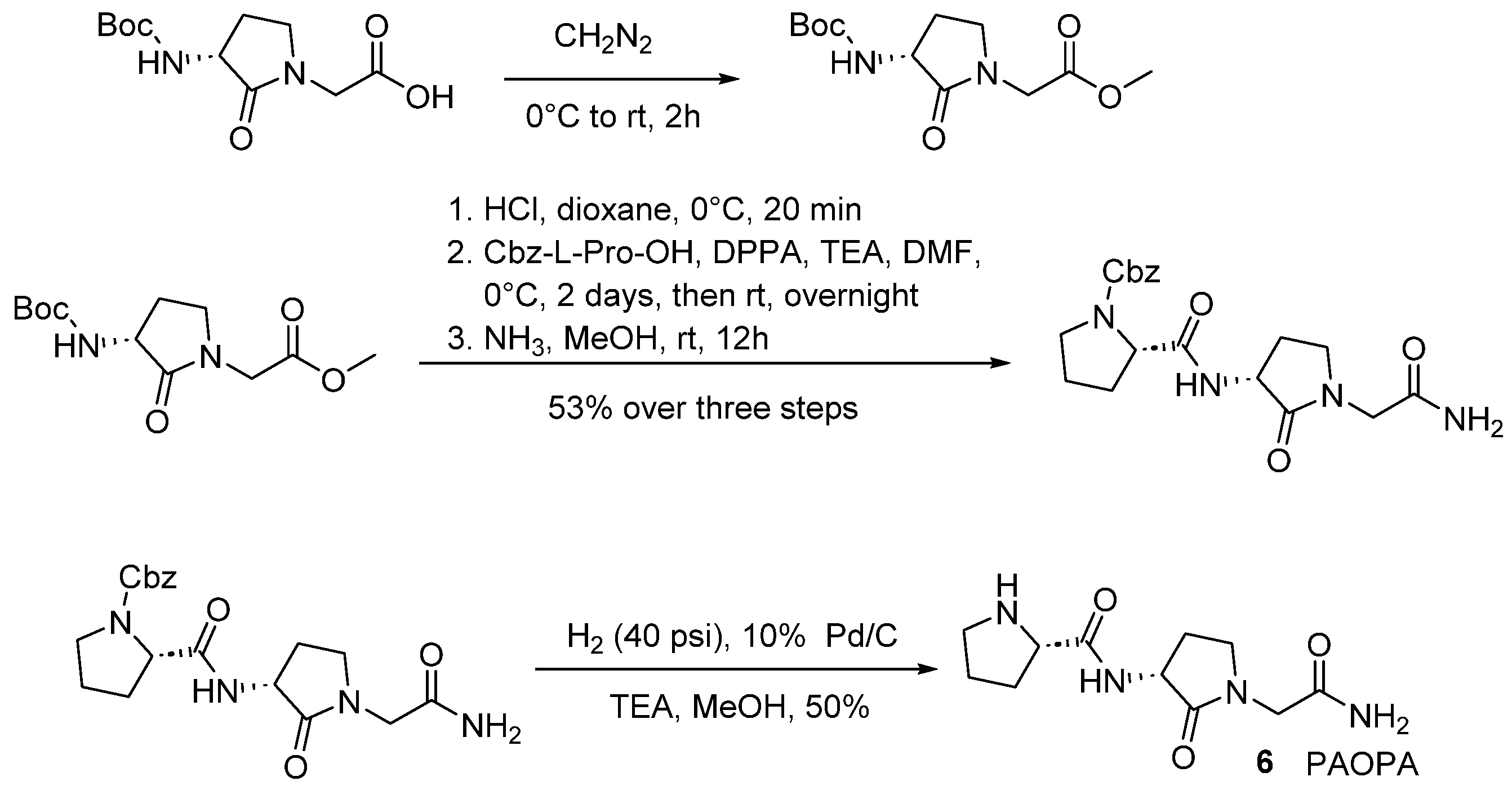
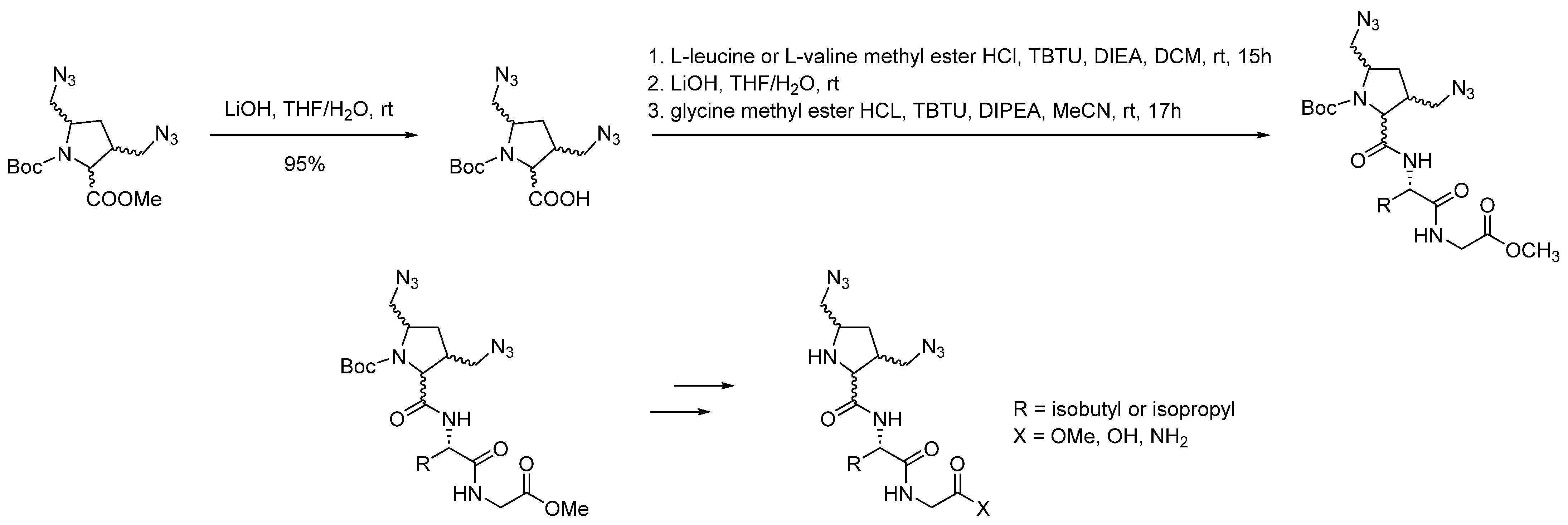
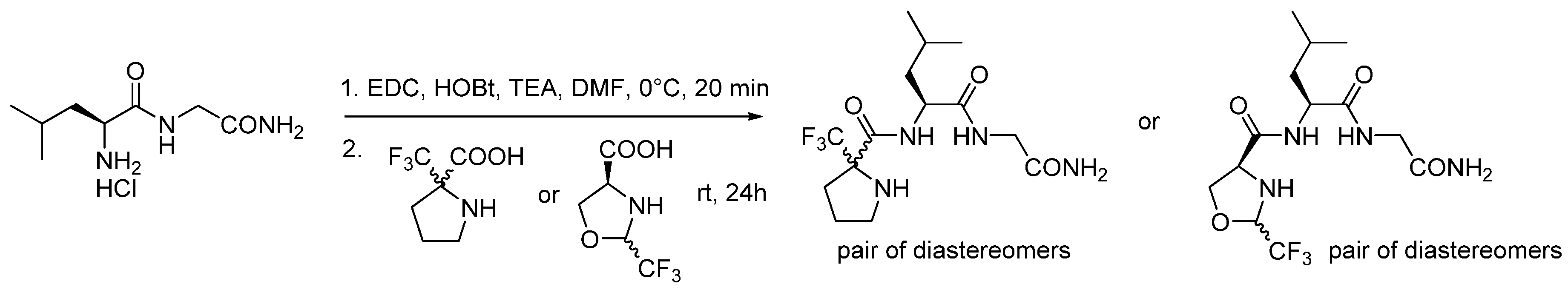

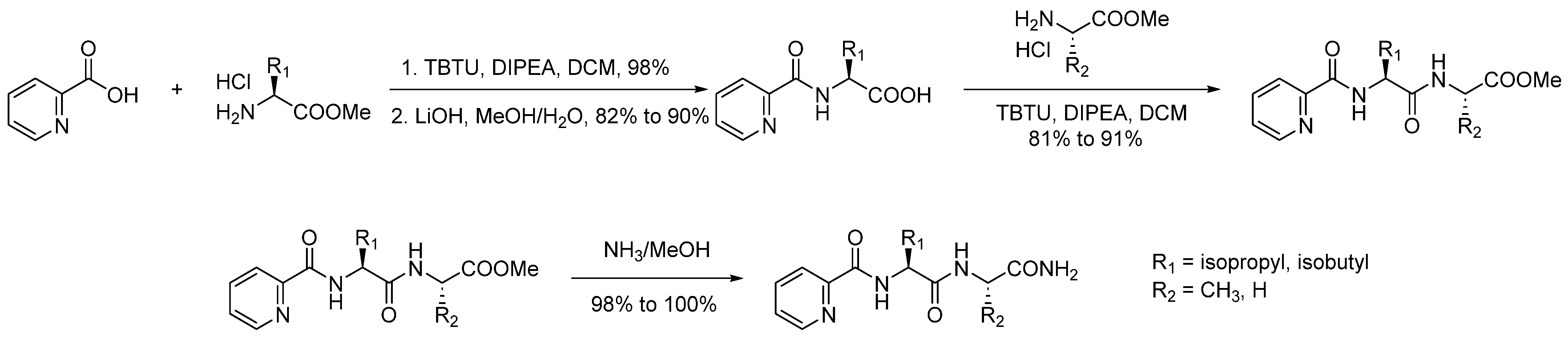
| Compound | KB a, nM | αβ b |
|---|---|---|
| SB-269562 (1) [27] | 776 | 0.06 |
| 1a [31] | 87 | 0.091 |
| 1b [31] | 23.4 | 0.04 |
| 1c [32] | 81 | 0.05 |
| 1d [31] | 72.4 | NA |
| 1e [31] | 30 | 0.021 |
| 1f [32] | 4600 | 0.08 |
| 1g [32] | 6000 | 0.02 |
| 1h [33] | 0.148 | α = 0.048 Β = 0.16 |
| Compound | pKB a (KB) [μM] | Logα b (α) | Logβ c (β) |
|---|---|---|---|
| 2 [34] | 5.41 ± 0.22 (3.87) | 0 | −0.55 ± 0.08 (0.28) |
| 2a [34] | 5.55 ± 0.12 (2.81) | −1.22 ± 0.16 (0.06) | −3.0 |
| 2b [34] | 6.18 ± 0.16 (0.662) | −0.17 ± 0.17 (0.68) | −1.10 ± 0.10 (0.08) |
| 2c [34] | 5.58 ± 0.10 (1.4) | −0.88 ± 0.16 (0.13) | −1.21 ± 0.16 (0.06) |
| 2d [35] | 6.25 ± 0.12 (0.57) | −0.72 ± 0.12 (0.19) | −0.65 ± 0.07 (0.22) |
| 2e [35] | 5.53 ± 0.13 (2.92) | 0 | −1.55 ± 0.19 (0.03) |
| Parameter | +R Isomer of 3 (10 μM) | +S Isomer of 3 (10 μM) | +Compound 4 (10 μM) |
|---|---|---|---|
| Kd [nM] | 13 ± 3 | 17 ± 6 | 11 ± 4 |
| Bmax [%control] | 137 ± 10 | 82 ± 16 | 278 ± 51 |
| Assay | logτA a | logτB b | pKA c | pKB d | Logα e | Logβ f | Slope g |
|---|---|---|---|---|---|---|---|
| β-arrestin | 1.57 [0.73–2.40] | −3.00 | 5.26 [4.39–6.14] | 5.53 5.42–5.64] (3 μM) | −0.70 [−1.59–0.20] (0.2) | −3.00 | 1.21 [1.06–1.35] |
| cAMP | 1.68 [1.21–2.23] | −3.00 | 5.62 [5.05–6.15] | 5.64 [5.37–5.92] (2.3 μM) | −0.28 [−0.85–0.34] (0.5) | −3.00 | 0.88 [0.77–1.02] |
| Condition | Binding Parameter | ||||
|---|---|---|---|---|---|
| KH [nM] | KL [nM] | RH [%] | RL [%] | RH/RL | |
| PLG-treated-Gpp(NH)p | 0.020 ± 0.001 | 78.0 ± 5.0 | 73.0 ± 6.0 | 27.0 ± 3.0 | 2.70 ± 0.28 |
| PLG-treated+Gpp(NH)p | 0.040 ± 0.002 | 92.0 ± 8.0 | 34.0 ± 2.5 | 66.0 ± 11 | 0.55 ± 0.040 |
| PAOPA-treated-Gpp(NH)p | 0.030 ± 0.002 | 63.0 ± 4.0 | 74.0 ± 8.0a | 26.0 ± 3.0 | 2.80 ± 0.19 |
| PAOPA-treated+Gpp(NH)p | 0.050 ± 0.003 | 88.0 ± 9.0 | 44.0 ± 6.0 | 56.0 ± 7.0 | 0.78 ± 0.050 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaczor, A.A.; Wróbel, T.M.; Bartuzi, D. Allosteric Modulators of Dopamine D2 Receptors for Fine-Tuning of Dopaminergic Neurotransmission in CNS Diseases: Overview, Pharmacology, Structural Aspects and Synthesis. Molecules 2023, 28, 178. https://doi.org/10.3390/molecules28010178
Kaczor AA, Wróbel TM, Bartuzi D. Allosteric Modulators of Dopamine D2 Receptors for Fine-Tuning of Dopaminergic Neurotransmission in CNS Diseases: Overview, Pharmacology, Structural Aspects and Synthesis. Molecules. 2023; 28(1):178. https://doi.org/10.3390/molecules28010178
Chicago/Turabian StyleKaczor, Agnieszka A., Tomasz M. Wróbel, and Damian Bartuzi. 2023. "Allosteric Modulators of Dopamine D2 Receptors for Fine-Tuning of Dopaminergic Neurotransmission in CNS Diseases: Overview, Pharmacology, Structural Aspects and Synthesis" Molecules 28, no. 1: 178. https://doi.org/10.3390/molecules28010178
APA StyleKaczor, A. A., Wróbel, T. M., & Bartuzi, D. (2023). Allosteric Modulators of Dopamine D2 Receptors for Fine-Tuning of Dopaminergic Neurotransmission in CNS Diseases: Overview, Pharmacology, Structural Aspects and Synthesis. Molecules, 28(1), 178. https://doi.org/10.3390/molecules28010178









