Multitarget Potential of Phytochemicals from Traditional Medicinal Tree, Terminalia arjuna (Roxb. ex DC.) Wight & Arnot as Potential Medicaments for Cardiovascular Disease: An In-Silico Approach
Abstract
1. Introduction
2. Results
2.1. Molecular Docking Analysis
2.2. MD Simulations Study
2.3. MM/GBSA Binding Free Energy Calculations
2.4. Assessment of Drug Likeness and Toxicity Prediction
3. Discussion
4. Materials and Methods
4.1. Retrieval of Proteins
4.2. Ligand Preparation
4.3. Molecular Docking
4.4. Molecular Dynamics Simulations
4.5. Binding Free Energy Calculations
4.6. Evaluation of Drug-Likeness and ADME/Toxicity Properties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thiriet, M. Vasculopathies: Behavioral, Chemical, Environmental, and Genetic Factors; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.E.; Hilgers, K.F.; Schlaich, M.P.; Schmidt, B.M. Renin-angiotensin system and cardiovascular risk. Lancet 2007, 369, 1208–1219. [Google Scholar] [CrossRef]
- Nasution, S.A. The use of ACE inhibitor in cardiovascular disease. Acta Med. Indones. 2006, 38, 60–64. [Google Scholar]
- Khera, A.V.; Kathhiresan, S. Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat. Rev. Genet. 2017, 18, 331–344. [Google Scholar] [CrossRef]
- Istvan, E.S.; Palnitkar, M.; Buchanan, S.K.; Deisenhofer, J. Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. EMBO J. 2000, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Hasimun, P.; Sulaeman, A.; Putra, H.M.; Lindasari, H. Inhibition of HMG CoA reductase and lipid peroxidation in the rats liver by selected zingiberaceae. Pharmacia 2018, 8, 232. [Google Scholar] [CrossRef]
- Hebert, P.R.; Gaziano, J.M.; Chan, K.S.; Hennekens, C.H. Cholesterol lowering with statin drugs, risk of stroke, and total mortality. J. Am. Med. Assoc. 1998, 42, 313–321. [Google Scholar] [CrossRef]
- Nematollahi, A.; Aminimoghadamfarouji, N.; Jalilvand, M.R.; Ali, S.A. Design and molecular studies of luteolin derivatives, from Biebersteinia multifida DC.; as novel HMG-CoA reductase inhibitors. Int. J. Chem. Tech. Res. 2012, 4, 733–738. [Google Scholar]
- Than, A.; Leow, M.K.; Chen, P. Control of adipogenesis by the autocrine interplays between angiotensin 1–7/Mas receptor and angiotensin II/AT1 receptor signaling pathways. J. Biol. Chem. 2013, 288, 15520–15531. [Google Scholar] [CrossRef]
- Braz, J.C.; Bueno, O.F.; Liang, Q.; Wilkins, B.J.; Dai, Y.S.; Parsons, S.; Braunwart, J.; Glascock, B.J.; Klevitsky, R.; Kimball, T.F.; et al. Targeted inhibition of p38 MAPK promotes hypertrophic cardiomyopathy through upregulation of calcineurin-NFAT signaling. J. Clin. Invest. 2003, 111, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Xu, Z.; Zhu, Q.; Thomas, C.; Kumar, R.; Feng, H.; Dostal, D.E.; White, M.F.; Baker, K.M.; Guo, S. Myocardial loss of IRS1 and IRS2 causes heart failure and is controlled by p38α MAPK during insulin resistance. Diabetes 2013, 62, 3887–3900. [Google Scholar] [CrossRef] [PubMed]
- Bisoendial, R.J.; Boekholdt, S.M.; Vergeer, M.; Stroes, E.S.; Kastelein, J.J. C-reactive protein is a mediator of cardiovascular disease. Eur. Heart J. 2010, 31, 2087–2091. [Google Scholar] [CrossRef] [PubMed]
- Kaptoge, S.; Di Angelantonio, E.; Lowe, G.; Pepys, M.B.; Thompson, S.G.; Collins, R.; Danesh, J. Emerging Risk Factors Collaboration: C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: An individual participant meta-analysis. Lancet 2010, 375, 132–140. [Google Scholar] [PubMed]
- Reid, J.L.; MacFadyen, R.J.; Squire, I.B.; Lees, K.R. Blood pressure response to the first dose of angiotensin-converting enzyme inhibitors in congestive heart failure. Am. J. Cardiol. 1993, 71, 57E–60E. [Google Scholar] [CrossRef]
- Bezalel, S.; Mahlab-Guri, K.; Asher, I.; Werner, B.; Sthoeger, Z.M. Angiotensin-converting enzyme inhibitor-induced angioedema. Am. J. Med. 2015, 128, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Slater, E.E.; Merrill, D.D.; Guess, H.A.; Roylance, P.J.; Cooper, W.D.; Inman, W.H.; Ewan, P.W. Clinical profile of angioedema associated with angiotensin converting-enzyme inhibition. JAMA 1988, 260, 967–970. [Google Scholar] [CrossRef]
- Ramkumar, S.; Raghunath, A.; Raghunath, S. Statin therapy: Review of safety and potential side effects. Acta Cardiol. Sin. 2016, 32, 631. [Google Scholar]
- Sen, S.; Chakraborty, R. Revival, modernization and integration of Indian traditional herbal medicine in clinical practice: Importance, challenges and future. J. Tradit. Complement. Med. 2017, 7, 234–244. [Google Scholar] [CrossRef]
- Dwivedi, S. Terminalia arjuna Wight & Arn.—A useful drug for cardiovascular disorders. J. Ethnopharmacol. 2007, 114, 114–129. [Google Scholar]
- Kapoor, D.; Vijayvergiya, R.; Dhawan, V. Terminalia arjuna in coronary artery disease: Ethnopharmacology, pre-clinical, clinical & safety evaluation. J. Ethnopharmacol. 2014, 155, 1029–1045. [Google Scholar] [PubMed]
- Amalraj, A.; Gopi, S. Medicinal properties of Terminalia arjuna (Roxb.) Wight & Arn.: A review. J. Tradit. Complement. Med. 2017, 7, 65–78. J. Tradit. Complement. Med. 2017, 7, 65–78. [Google Scholar] [PubMed]
- Gauthaman, K.; Maulik, M.; Manchanda, S.C.; Maulik, S.K. Antioxidant effect of chronic treatment with Terminalia arjuna bark on ischaemic reperfusion injury in rat heart. Ind. J. Pharmacol. 1999, 31, 78. [Google Scholar]
- Gauthaman, K.; Maulik, M.; Kumari, R.; Manchanda, S.C.; Dinda, A.K.; Maulik, S.K. Effect of chronic treatment with bark of Terminalia arjuna: A study on the isolated ischemic reperfused rat heart. J. Ethnopharmacol. 2001, 75, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, K.; Bai, B.S.; Gauthaman, K.; Sathish, K.S.; Devaraj, S.N. Cardioprotective effect of the alcoholic extract of Terminalia arjuna bark in an in vivo model of myocardial ischemic reperfusion injury. Life Sci. 2003, 73, 2727–2739. [Google Scholar] [CrossRef]
- Parveen, A.; Babbar, R.; Agarwal, S.; Kotwani, A.; Fahim, M. Terminalia arjuna enhances baroreflex sensitivity and myocardial function in isoproterenol-induced chronic heart failure rats. J. Cardiovasc. Pharmacol. Ther. 2012, 17, 199–220. [Google Scholar] [CrossRef]
- Kokkiripati, P.K.; Kamsala, R.V.; Bashyam, L.; Manthapuram, N.; Bitla, P.; Peddada, V.; Raghavendra, A.S.; Tetali, S.D. Stem-bark of Terminalia arjuna attenuates human monocytic (THP-1) and aortic endothelial cell activation. J. Ethnopharmacol. 2013, 146, 456–464. [Google Scholar] [CrossRef]
- Mythili, P.; Parameswari, C.S.; Dayana, J. Phytochemical analysis of the bark extract of Terminalia arjuna and its cardioprotective effect. Ind. J. Innov. Dev. 2012, 1, 40–42. [Google Scholar]
- Loza-Mejía, M.A.; Salazar, J.R.; Sánchez-Tejeda, J.F. In Silico studies on compounds derived from Calceolaria: Phenylethanoid glycosides as potential multitarget inhibitors for the development of pesticides. Biomolecules 2018, 8, 121. [Google Scholar] [CrossRef]
- Rallabandi, H.R.; Mekapogu, M.; Natesan, K.; Saindane, M.; Dhupal, M.; Swamy, M.K.; Vasamsetti, B.M. Computational Methods Used in Phytocompound-Based Drug Discovery. In Plant-derived Bioactives; Springer: Singapore, 2020; pp. 549–573. [Google Scholar]
- Lee, K.; Kim, D. In-silico molecular binding prediction for human drug targets using deep neural multi-task learning. Genes 2019, 10, 906. [Google Scholar] [CrossRef]
- Kazmi, S.R.; Jun, R.; Yu, M.S.; Jung, C.; Na, D. In silico approaches and tools for the prediction of drug metabolism and fate: A review. Comput. Biol. Med. 2019, 106, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Lounnas, V.; Ritschel, T.; Kelder, J.; McGuire, R.; Bywater, R.P.; Foloppe, N. Current progress in structure-based rational drug design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 2013, 5, e201302011. [Google Scholar] [CrossRef] [PubMed]
- Yuriev, E.; Ramsland, P.A. Latest developments in molecular docking: 2010–2011 in review. J. Mol. Recognit. 2013, 26, 215–239. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Tahir, R.A.; Bashir, A.; Yousaf, M.N.; Ahmed, A.; Dali, Y.; Khan, S.; Sehgal, S.A. In Silico identification of angiotensin-converting enzyme inhibitory peptides from MRJP1. PloS ONE 2020, 15, e0228265. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.A.; Fatima, N. In silico analysis and molecular docking studies of potential angiotensin-converting enzyme inhibitor using quercetin glycosides. Phcog. Mag. 2015, 11, S123. [Google Scholar] [CrossRef]
- Ahmad, I. Isolation, elucidation, and molecular docking studies of active compounds from Phyllanthus niruri with angiotensin-converting enzyme inhibition. Phcog. Mag. 2018, 14, 604–610. [Google Scholar]
- Krushna, G.S.; Shivaranjani, V.L.; Umamaheswari, J.; Srinivasulu, C.; Hussain, S.A.; Kareem, M.A.; Reddy, V.D.; Ali, D.; Lokhande, K.B.; Swamy, K.V.; et al. In vivo and molecular docking studies using whole extract and phytocompounds of Aegle marmelos fruit protective effects against Isoproterenol-induced myocardial infarction in rats. Biomed. Pharmacother. 2017, 91, 880–889. [Google Scholar] [CrossRef]
- Jasmine, J.M.; Vanaja, R. In silico analysis of phytochemical compounds for optimizing the inhibitors of HMG CoA reductase. J. Appl. Pharma. Sci. 2013, 3, 43. [Google Scholar]
- Talapatra, S.N.; Talukdar, P.; Swarnakar, S. Interaction between C-Reactive Protein and Phytochemical (s) from Calotropis procera: An Approach on Molecular Docking. Int. Lett. Nat. Sci. 2017, 61, 43–55. [Google Scholar] [CrossRef]
- Shahbazi, S.; Sahrawat, T.R.; Ray, M.; Dash, S.; Kar, D.; Singh, S. Drug targets for cardiovascular-safe anti-inflammatory: In silico rational drug studies. PloS ONE 2016, 11, e0156156. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.N. Structure based docking studies towards exploring potential angiotensin-I receptor blockers of selected Alangium salvifolium phytochemicals against protective vascular remodeling. Int J. Adv. Res. 2018, 6, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fang, L.; Min, W.; Liu, J.; Li, H. Exploration of the molecular interactions between angiotensin-I-converting enzyme (ACE) and the inhibitory peptides derived from hazelnut (Corylus heterophylla Fisch.). Food Chem. 2018, 245, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Sellappan, M.; Pandian, M.R.; Krishnamoorthi, A.; Senthilnathan, S. Computational studies of plant derived natural compounds with HMG-CoA enzyme through in silico analysis. IOSR J. Pharma Biol. Sci. 2019, 14, 21–25. [Google Scholar]
- Gaikwad, D.T.; Jadhav, N.R. Discovery of potential inhibitors for phosphodiesterase 5A, sodium-potassium pump and beta-adrenergic receptor from Terminalia arjuna: In silico approach. J. Biomol. Struct. Dyn. 2020, 39, 1754–1765. [Google Scholar] [CrossRef]
- Murad, H.A.; Alqurashi, T.M.; Hussien, M.A. Interactions of selected cardiovascular active natural compounds with CXCR4 and CXCR7 receptors: A molecular docking, molecular dynamics, and pharmacokinetic/toxicity prediction study. BMC Complement. Med. Ther. 2022, 22, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC’06, Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; IEEE: New York, NY, USA, 2006; p. 43. [Google Scholar]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef]
- Patel, C.N.; Georrge, J.J.; Modi, K.M.; Narechania, M.B.; Patel, D.P.; Gonzalez, F.J.; Pandya, H.A. Pharmacophore-based virtual screening of catechol-o-methyltransferase (COMT) inhibitors to combat Alzheimer’s disease. J. Biomol. Struct. Dyn. 2018, 36, 3938–3957. [Google Scholar] [CrossRef]
- Patel, C.N.; Kumar, S.P.; Modi, K.M.; Soni, M.N.; Modi, N.R.; Pandya, H.A. Cardiotonic steroids as potential Na+/K+-ATPase inhibitors–a computational study. J. Recept. Signal. Transduct. 2019, 39, 226–234. [Google Scholar] [CrossRef]
- Reddy, S.V.G.; Reddy, K.T.; Kumari, V.V.; Basha, S.H. Molecular docking and dynamic simulation studies evidenced plausible immunotherapeutic anticancer property by Withaferin A targeting indoleamine 2, 3-dioxygenase. J. Biomol. Struct. Dyn. 2015, 33, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Panchal, U.; Modi, K.; Dey, S.; Prajapati, U.; Patel, C.; Jain, V.K. A resorcinarene-based “turn-off” fluorescence sensor for 4-nitrotoluene: Insights from fluorescence and 1H NMR titration with computational approach. J. Lumin. 2017, 184, 74–82. [Google Scholar] [CrossRef]
- Mall, S.; Srivastava, R.; Sharma, N.; Patel, C.N.; Rolta, R.; Sourirajan, A.; Dev, K.; Ghosh, A.; Kumar, V. Antihypertensive activity of phytocompounds from selected medicinal plants via inhibition of angiotensin-converting enzyme (ACE) protein: An in-silico approach. Nat. Prod. Res. 2021, 36, 4526–4529. [Google Scholar] [CrossRef]
- Sharma, N.; Gupta, N.; Orfali, R.; Kumar, V.; Patel, C.N.; Peng, J.; Perveen, S. Evaluation of the Antifungal, Antioxidant, and Anti-Diabetic Potential of the Essential Oil of Curcuma longa Leaves from the North-Western Himalayas by In Vitro and In Silico Analysis. Molecules 2022, 27, 7664. [Google Scholar]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Patel, C.N.; Goswami, D.; Jaiswal, D.G.; Parmar, R.M.; Solanki, H.A.; Pandya, H.A. Pinpointing the potential hits for hindering interaction of SARS-CoV-2 S-protein with ACE2 from the pool of antiviral phytochemicals utilizing molecular docking and molecular dynamics (MD) simulations. J. Mol. Graph. Mod. 2021, 105, 107874. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, 257–263. [Google Scholar] [CrossRef]

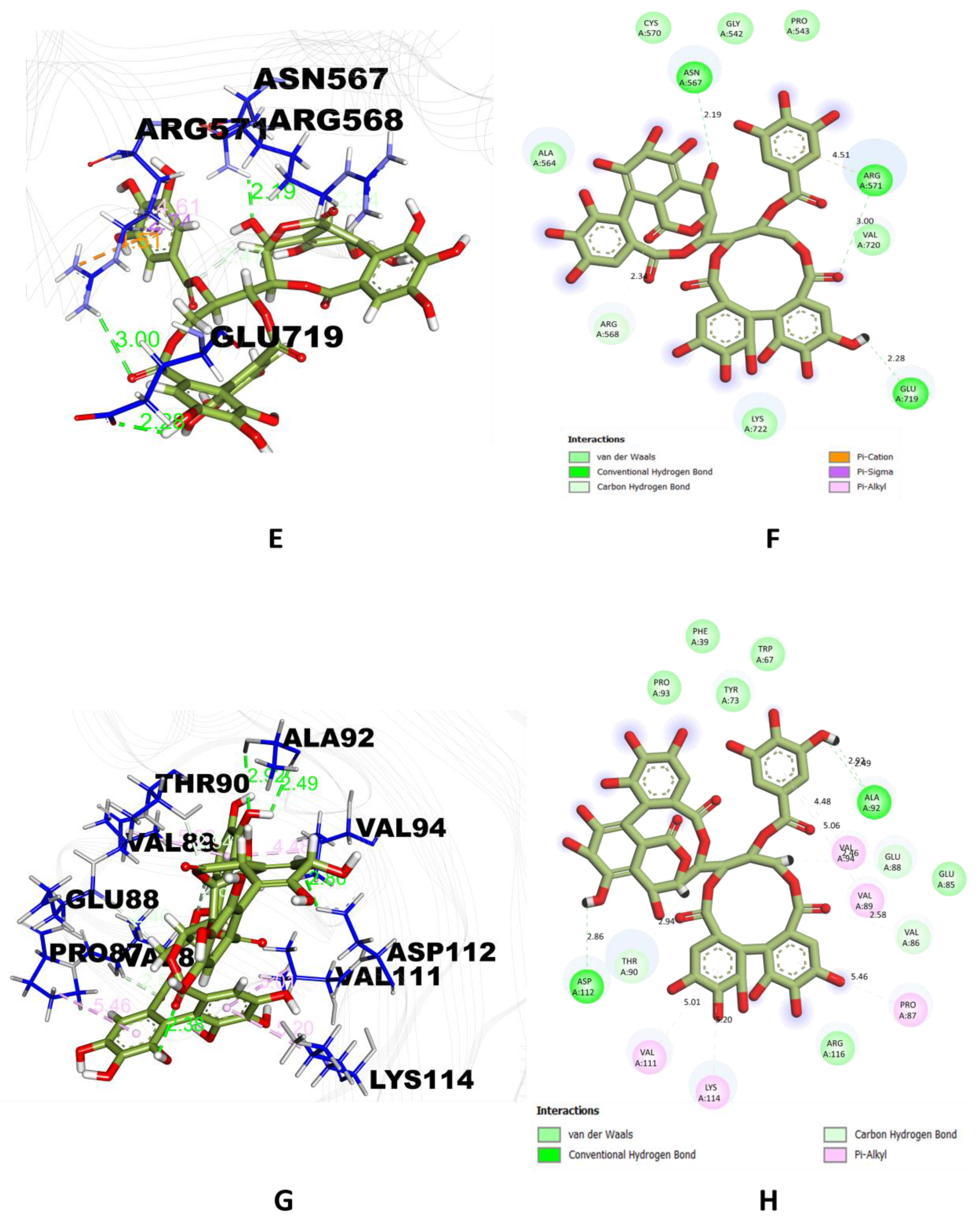
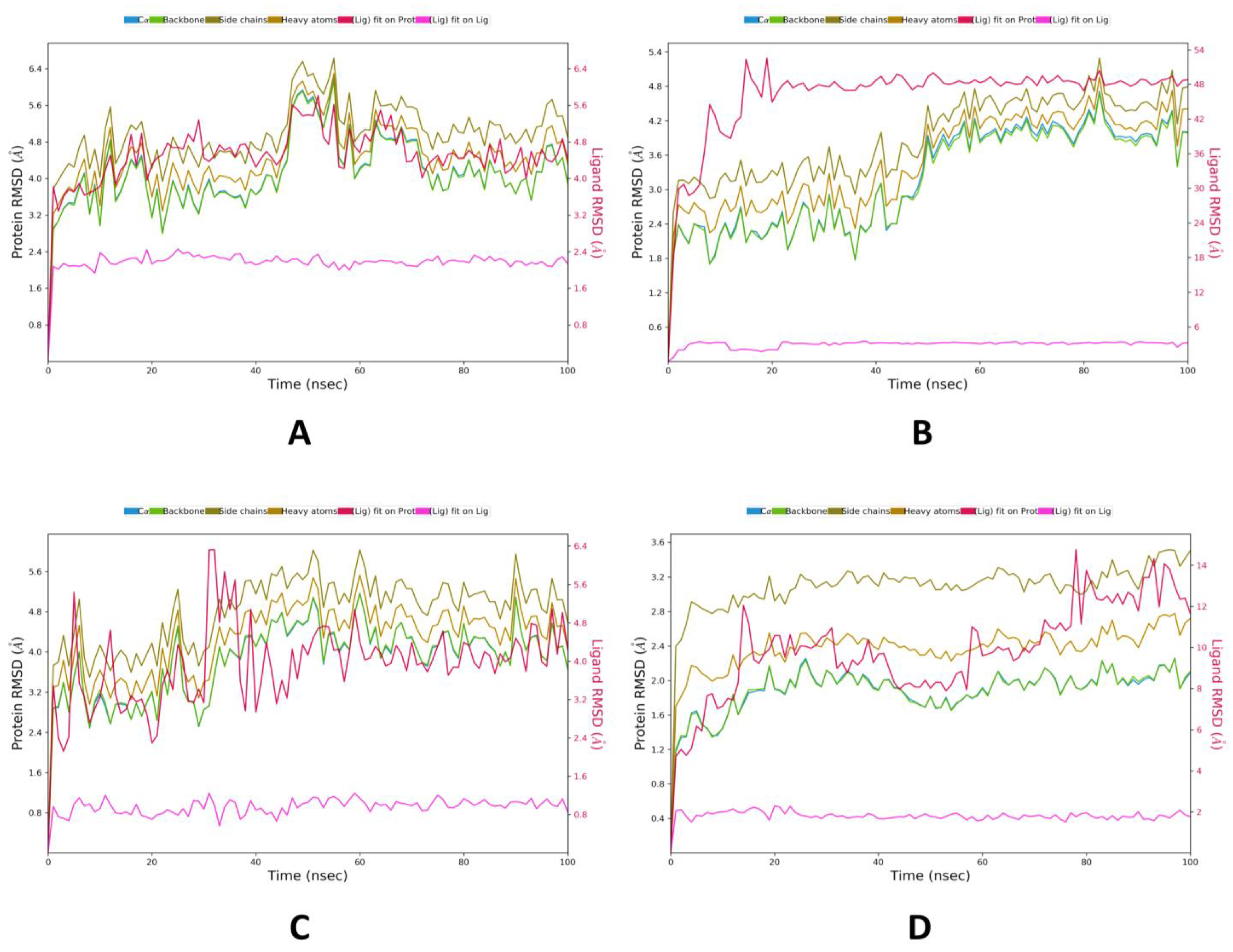
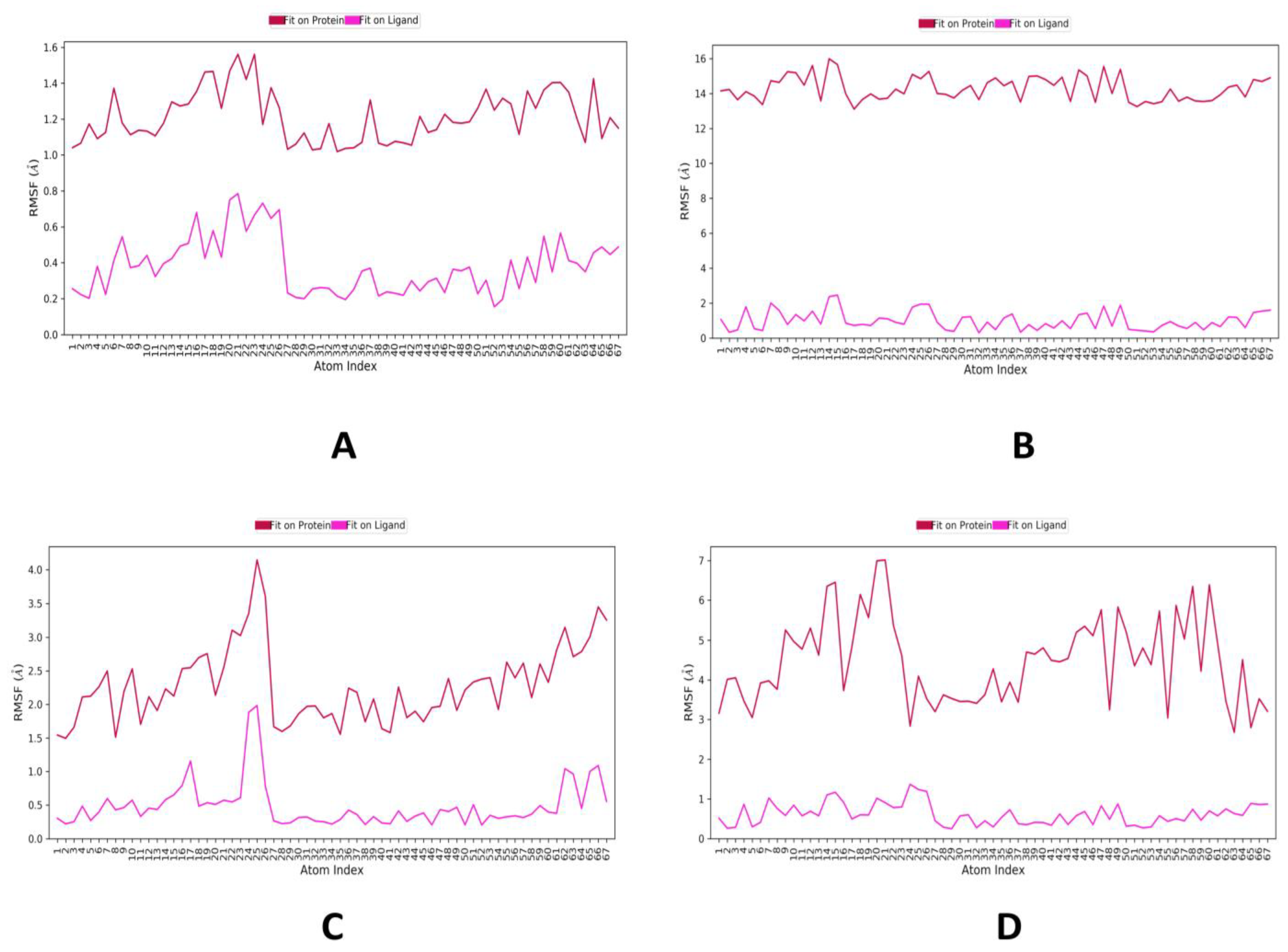
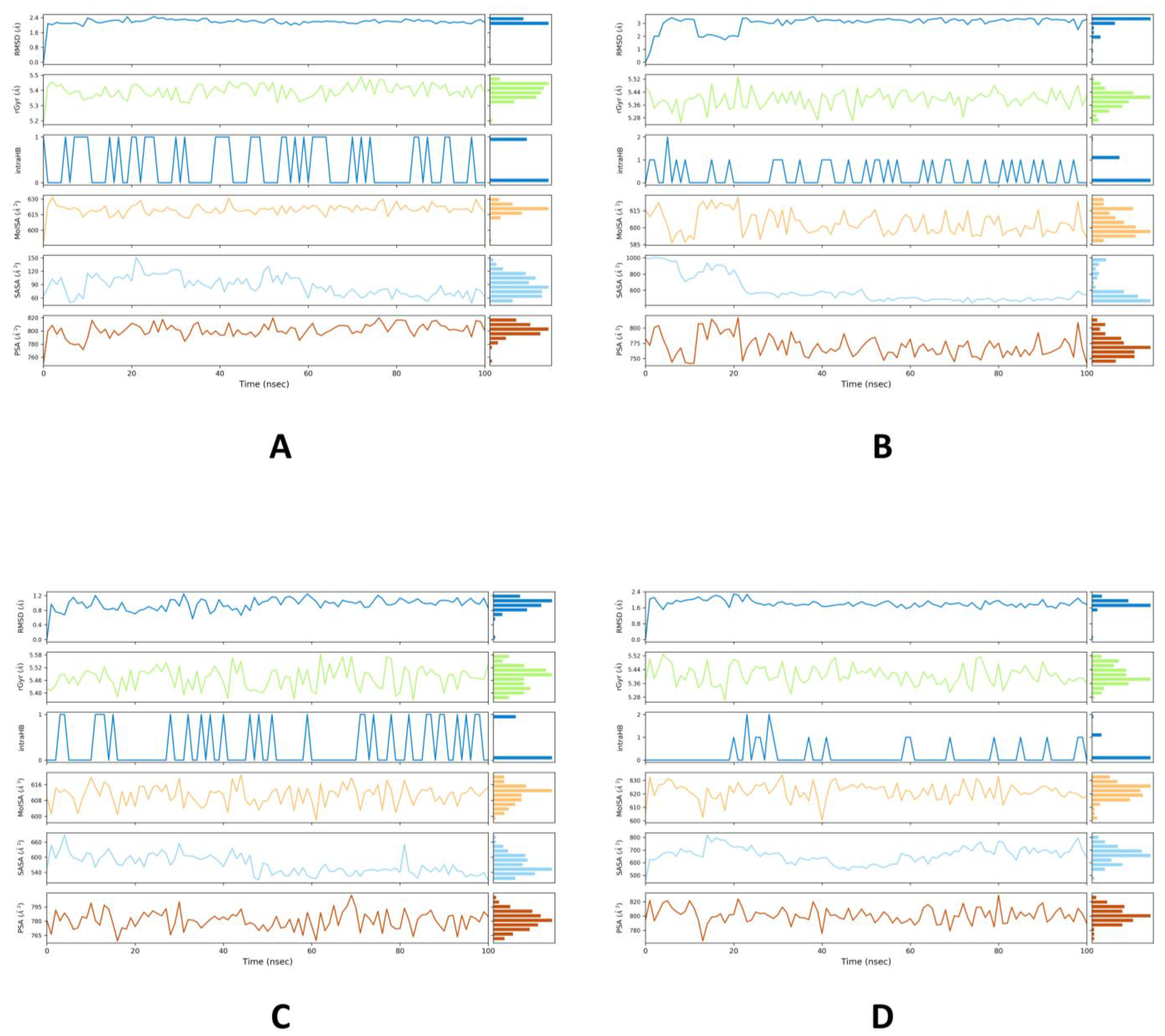


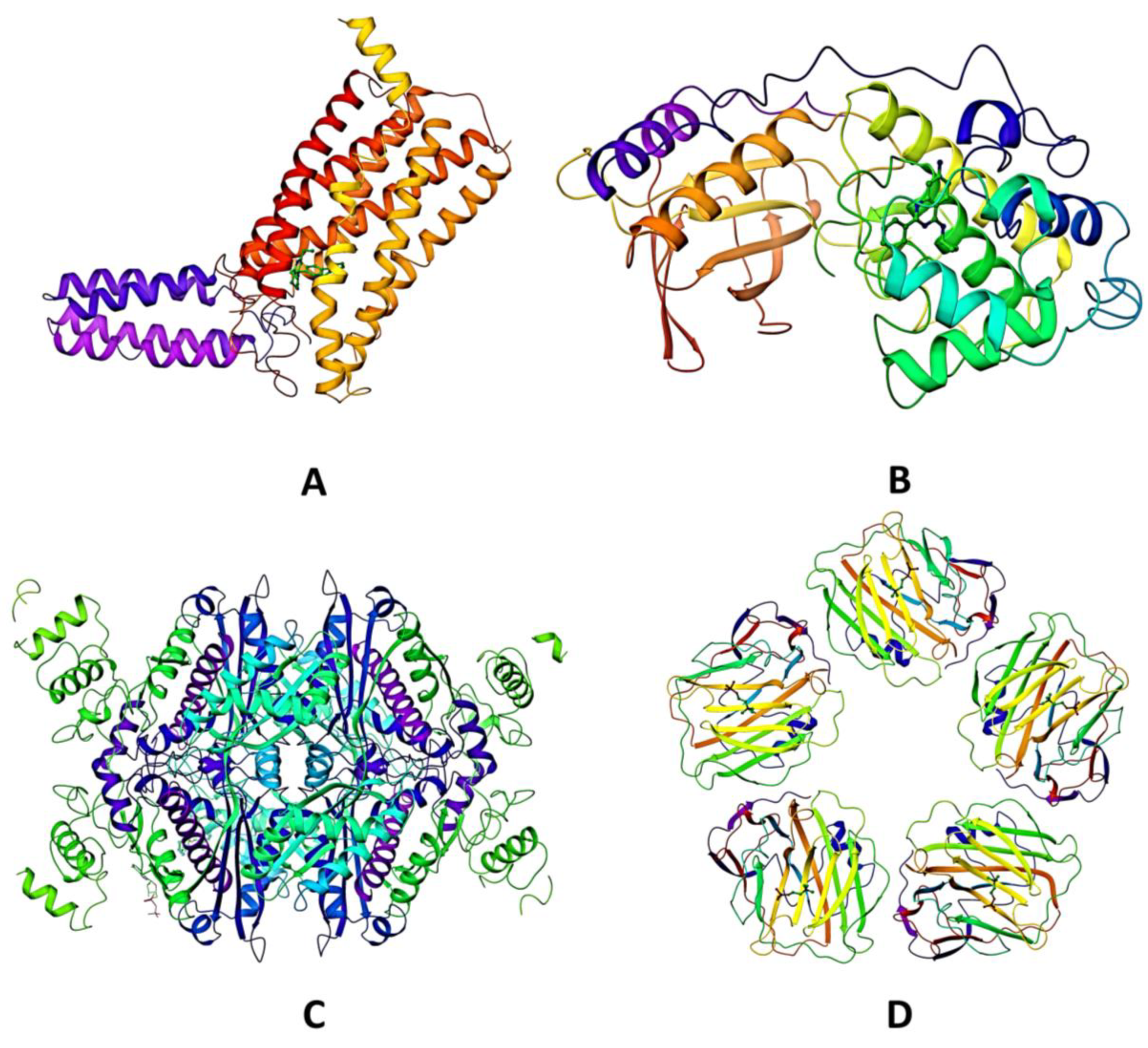
| Phytocompounds | 4YAY | 4DLI | 1HW9 | 1B09 |
|---|---|---|---|---|
| 3-O-Methylellagic-acid-3-rhamnoside | −7.668 | −4.082 | −4.411 | −2.601 |
| Arjunetin | −6.14 | −3.384 | −0.886 | −5.307 |
| Arjungenin | - | - | −2.534 | −1.71 |
| Arjunic acid | - | −2.774 | −2.587 | −1.315 |
| Arjunolic acid | - | - | −1.812 | −0.803 |
| Arjunone | −5.153 | −2.701 | −1.844 | −1.973 |
| beta-Sitosterol | −5.092 | −2.409 | 1.034 | −1.371 |
| Casuarinin | −18.276 | −9.216 | −10.685 | −9.678 |
| Catechin | −5.665 | −4.501 | −4.202 | −1.838 |
| Ellagic acid | −6.023 | −4.192 | −2.769 | - |
| Ethyl gallate | −6.776 | −4.652 | −3.616 | −3.072 |
| Gallic acid | −6.376 | −4.95 | −4.155 | −1.816 |
| Kaempferol | −5.498 | −5.151 | −3.026 | −2.473 |
| Leucocyanidin | −8.239 | −3.649 | −5.325 | −3.955 |
| Luteolin | −7.705 | −5.137 | −4.134 | −2.103 |
| Oleanolic acid | - | −0.464 | −0.457 | −0.539 |
| Quercetin | −8.355 | −5.794 | −4.463 | −1.79 |
| Rutin | −10.668 | −6.619 | −7.787 | −7.684 |
| Terminic acid | - | - | −4.463 | −1.066 |
| Lisinopril | −7.722 | −5.654 | −3.573 | −4.715 |
| Phosphocholine | −5.401 | −2.933 | −4.565 | −4.609 |
| Losartan | −5.663 | −4.363 | −3.48 | −3.632 |
| Simvastatin | −5.286 | −1.434 | −1.732 | −2.18 |
| Losmapimod | −4.687 | −3.133 | −1.344 | −1.582 |
| Target Proteins | ∆GBind (kcal/mol) | ∆GvdW (kcal/mol) | ∆GCoulomb (kcal/mol) | ∆GH-bond (kcal/mol) | ∆GLipo (kcal/mol) | ∆GSolv GB (kcal/mol) |
|---|---|---|---|---|---|---|
| 4YAY | −79.43 | −68.58 | −89.27 | −6.26 | −24.07 | 103.86 |
| 4DLI | −14.72 | −12.45 | −44.26 | −1.35 | −4.83 | 48.91 |
| 1HW9 | −34.20 | −32.32 | −44.05 | −1.81 | −7.76 | 50.75 |
| 1B09 | −19.68 | −14.01 | −25.17 | −1.63 | −5.26 | 25.01 |
| Compounds | Drug-Likeness | Toxicity Prediction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| cLogP (<5) | nrot (<5) | MW (<500 Da) | HBD (<5) | HBA (<10) | Lipinski Rule | Hepato-Toxicity | Carcino- Genicity | Cyto- Toxicity | LD50 (mg/kg) | |
| Casuarinin | −3.23 | 4 | 936.65 | 16 | 26 | No | No | No | No | 2170 (Class V) |
| Lisinopril | −1.46 | 13 | 405.49 | 4 | 7 | Yes | No | No | No | 8500 (Class VI) |
| Phosphocholine | −4.54 | 4 | 184.15 | 2 | 4 | Yes | No | No | No | 12,900 (Class VI) |
| Losartan | 3.36 | 8 | 422.91 | 2 | 5 | Yes | No | No | No | 300 (Class III) |
| Simvastatin | 3.77 | 7 | 418.57 | 1 | 5 | Yes | No | No | No | 1000 (Class IV) |
| Target Proteins | Amino Acids | Resolution | Chain Selected for Docking | Grid Box Coordinates |
|---|---|---|---|---|
| Human Angiotensin receptor (PDB ID: 4YAY) | 412 | 2.90 Å | Chain-A | x = −22.32; y = 6.81; z = 33.81 |
| P38 Mitogen-activated protein kinase (MAPK, PDB ID: 4DLI) | 360 | 1.91 Å | Chain-A | x = 24.22; y = −16.74; z = −10.1 |
| HMG-Co A reductase (PDB ID: 1HW9) | 467 | 2.33 Å | Chain-A | x = −5.94; y = −0.9; z = −19.46 |
| Human C-reactive Protein (PDB ID: 1B09) | 206 | 2.50 Å | Chain-A | x = −5.94; y = −0.9; z = −19.46 |
| Sr. No | Compounds/Drugs | Formula | Compound ID |
|---|---|---|---|
| 1 | Arjunetin | C36H58O10 | 21152828 |
| 2 | Arjunic acid | C30H48O5 | 15385516 |
| 3 | Arjunolic acid | C30H48O5 | 73641 |
| 4 | Arjunone | C19H20O6 | 14034821 |
| 5 | Arjungenin | C30H48O6 | 12444386 |
| 6 | β-sitosterol | C29H50O | 222284 |
| 7 | Casuarinin | C41H28O26 | 13834145 |
| 8 | Ellagic acid | C14H6O8 | 5281855 |
| 9 | Ethyl gallate | C9H10O5 | 13250 |
| 10 | Gallic acid | C7H6O5 | 370 |
| 11 | Luteolin | C15H10O6 | 5280445 |
| 12 | Quercetin | C15H10O7 | 5280343 |
| 13 | Terminic acid | C30H48O4 | 132568257 |
| 14 | (+)-Catechin | C15H14O6 | 9064 |
| 15 | Rutin | C27H30O16 | 5280805 |
| 16 | Kaempferol | C15H10O6 | 5280863 |
| 17 | Leucocyanidin | C15H14O7 | 71629 |
| 18 | 3-O-Methylellagic acid 3’-rhamnoside | C21H18O12 | 5319609 |
| 19 | Oleanolic acid | C30H48O3 | 10494 |
| 20 | Phosphocholine (PC) | C5H15NO4P+ | 1014 |
| 21 | Simvastatin | C25H38O5 | 54454 |
| 24 | Lisinopril | C21H31N3O5 | 5362119 |
| 25 | Losartan | C22H23ClN6O | 3961 |
| 26 | Losmapimod | C22H26FN3O2 | 11552706 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, V.; Sharma, N.; Orfali, R.; Patel, C.N.; Alnajjar, R.; Saini, R.; Sourirajan, A.; Khosla, P.K.; Dev, K.; Perveen, S. Multitarget Potential of Phytochemicals from Traditional Medicinal Tree, Terminalia arjuna (Roxb. ex DC.) Wight & Arnot as Potential Medicaments for Cardiovascular Disease: An In-Silico Approach. Molecules 2023, 28, 1046. https://doi.org/10.3390/molecules28031046
Kumar V, Sharma N, Orfali R, Patel CN, Alnajjar R, Saini R, Sourirajan A, Khosla PK, Dev K, Perveen S. Multitarget Potential of Phytochemicals from Traditional Medicinal Tree, Terminalia arjuna (Roxb. ex DC.) Wight & Arnot as Potential Medicaments for Cardiovascular Disease: An In-Silico Approach. Molecules. 2023; 28(3):1046. https://doi.org/10.3390/molecules28031046
Chicago/Turabian StyleKumar, Vikas, Nitin Sharma, Raha Orfali, Chirag N. Patel, Radwan Alnajjar, Rakshandha Saini, Anuradha Sourirajan, Prem Kumar Khosla, Kamal Dev, and Shagufta Perveen. 2023. "Multitarget Potential of Phytochemicals from Traditional Medicinal Tree, Terminalia arjuna (Roxb. ex DC.) Wight & Arnot as Potential Medicaments for Cardiovascular Disease: An In-Silico Approach" Molecules 28, no. 3: 1046. https://doi.org/10.3390/molecules28031046
APA StyleKumar, V., Sharma, N., Orfali, R., Patel, C. N., Alnajjar, R., Saini, R., Sourirajan, A., Khosla, P. K., Dev, K., & Perveen, S. (2023). Multitarget Potential of Phytochemicals from Traditional Medicinal Tree, Terminalia arjuna (Roxb. ex DC.) Wight & Arnot as Potential Medicaments for Cardiovascular Disease: An In-Silico Approach. Molecules, 28(3), 1046. https://doi.org/10.3390/molecules28031046







