Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction
Abstract
1. Introduction
- ✓
- ✓
- ✓
2. Results
2.1. SARS-CoV-2 Spike RBD/ACE2 Binding: Determination of KD
2.2. Validation of MDS Measurements
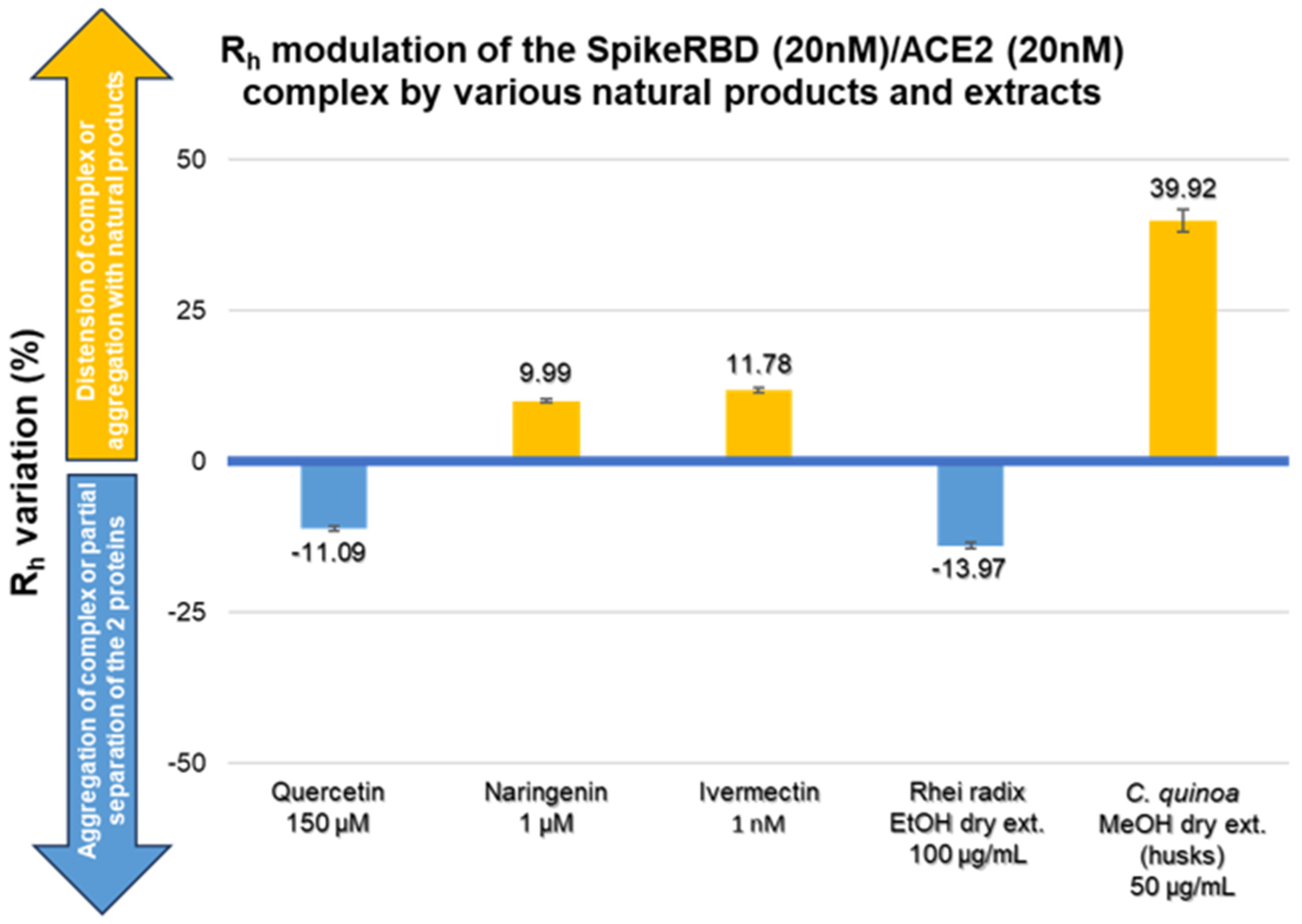
2.3. Modulation of SARS-CoV-2 Spike RBD/ACE2 Binding by Natural Compounds
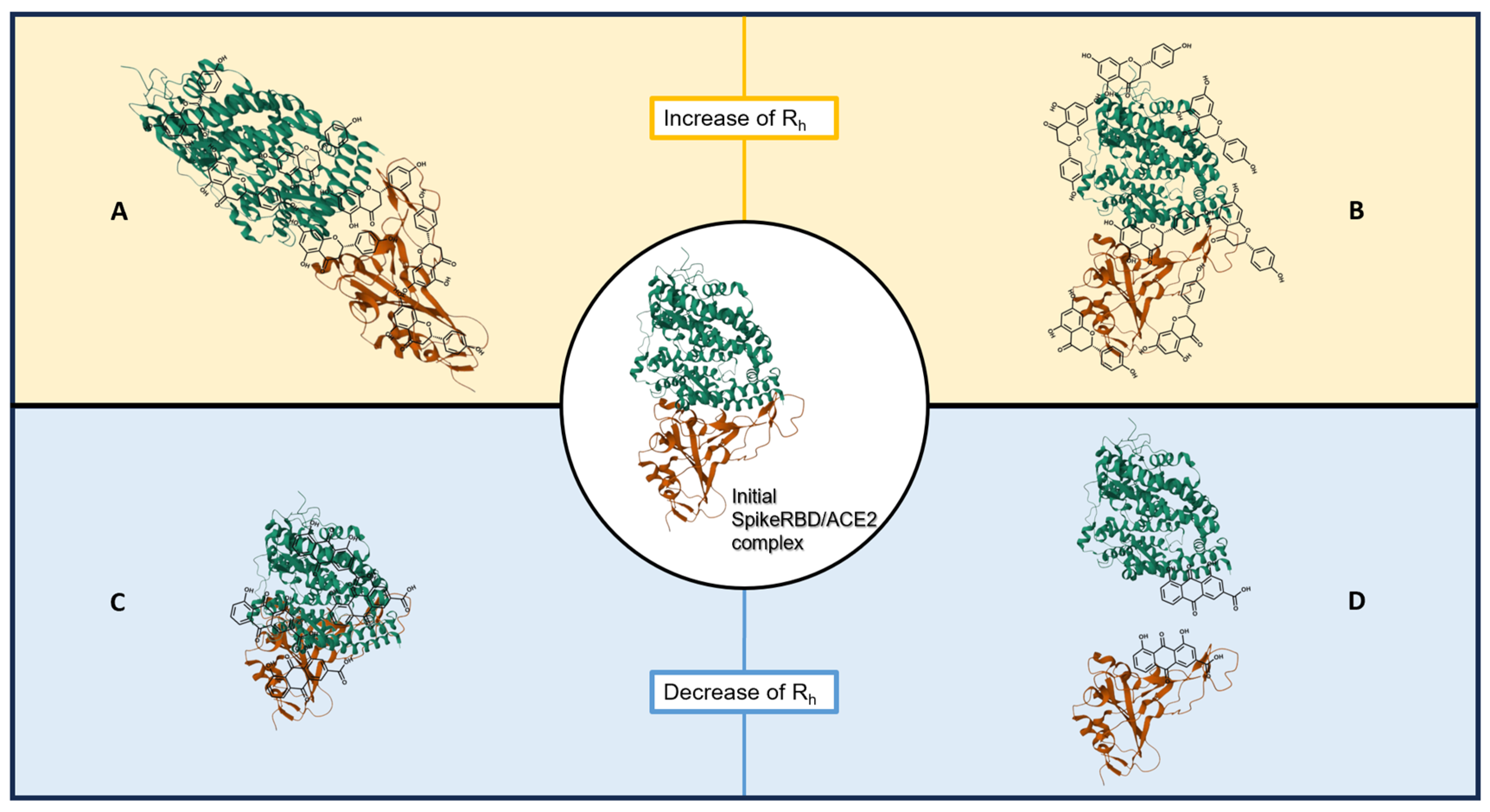
3. Discussion
- An increase in the hydrodynamic radius (higher Rh-complex) that could correspond either to a distension of the bound protein complex or to a clustering of natural compounds on the proteins; these should shift the affinity curve to the left and decrease KD.
- A decrease in the hydrodynamic radius (lower Rh-complex), possibly indicating a collapse or folding of the complex or a partial separation of the two protein partners; this should shift the affinity curve to the right and increase KD.
3.1. Quercetin
3.2. Naringin and Naringenin
3.3. Ivermectin
3.4. Rhei Radix
3.5. Bitter Chenopodium Quinoa Husks
- In silico docking studies predict the following:
- Stigmastane-type steroidal saponins (vernonioside A2, vernonioside A4 and vernonioside D2) exhibit inhibitory potential against SARS-CoV-2 cysteine proteases [80].
- A series of saikosaponins favorably bind to the RDB region of the SARS-CoV-2 spike protein, with saikosaponin B4 as the best probable inhibitor [31], and saikosaponins bind to the NSP15 endoribonuclease and to the prefusion spike glycoprotein SARS-CoV-2, saikosaponins U and V, showing the highest affinity toward both proteins [33].
- In vitro studies indicate the following:
- Oleanane saikosaponin B2, at 6 µM, significantly inhibits viral attachment and penetration, impeding HCoV 229E infection in pre-, co-, and postinfection models [85].
- Derivatives of glycyrrhizic acid are 10 to 70 times more active than glycyrrhizin itself in inhibiting the replication of a SARS-CoV clinical isolate in Vero cells; however, some compounds lose advantages in terms of viral selectivity [88].
- Aescin (6 µM) and four glycyrrhizin and aescin derivatives (<100 μM) showed activities toward SARS-CoV (H.K. strain) in Vero cells [89].
3.6. Other Natural Products Tested
4. Materials and Methods
4.1. Tested Natural Products
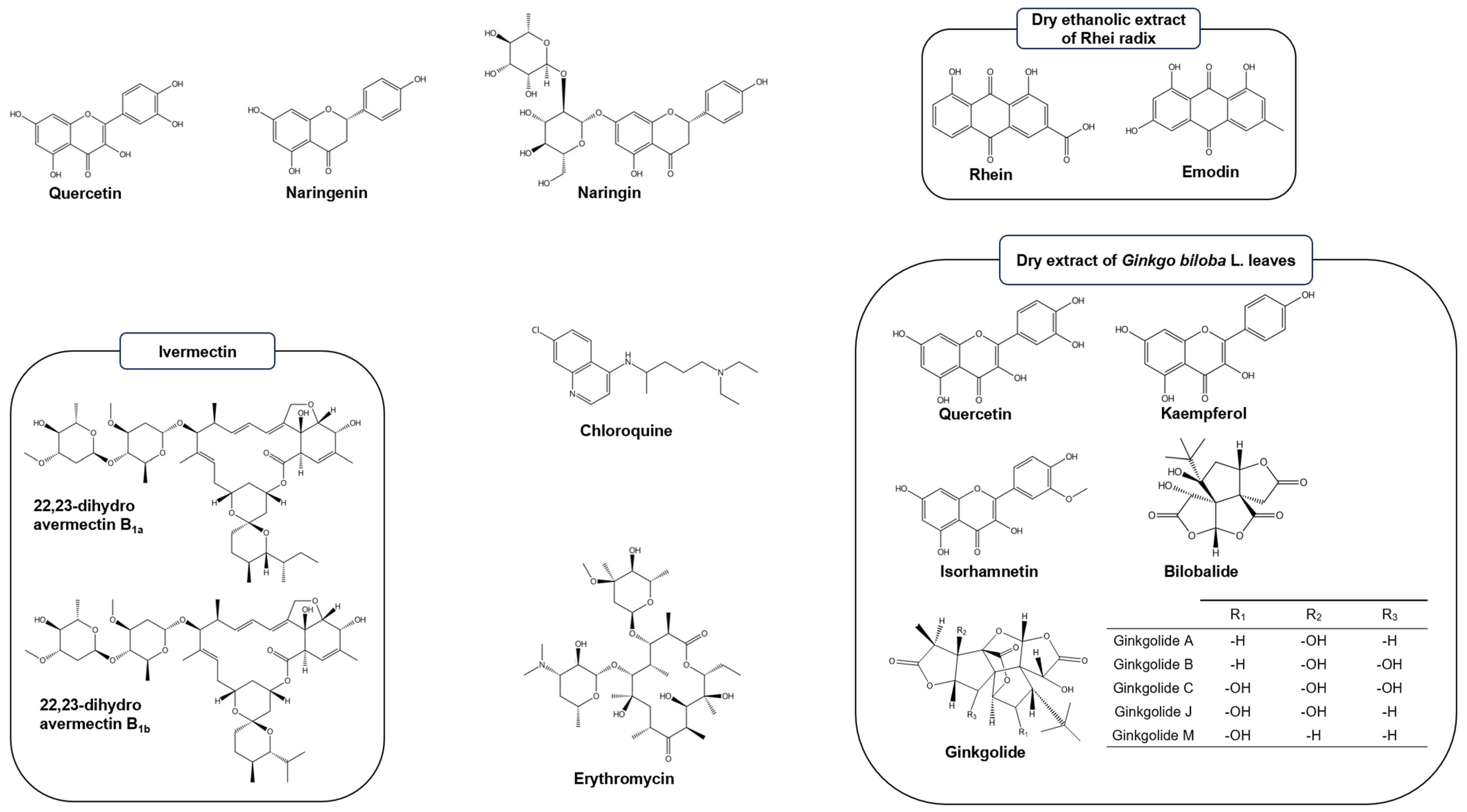
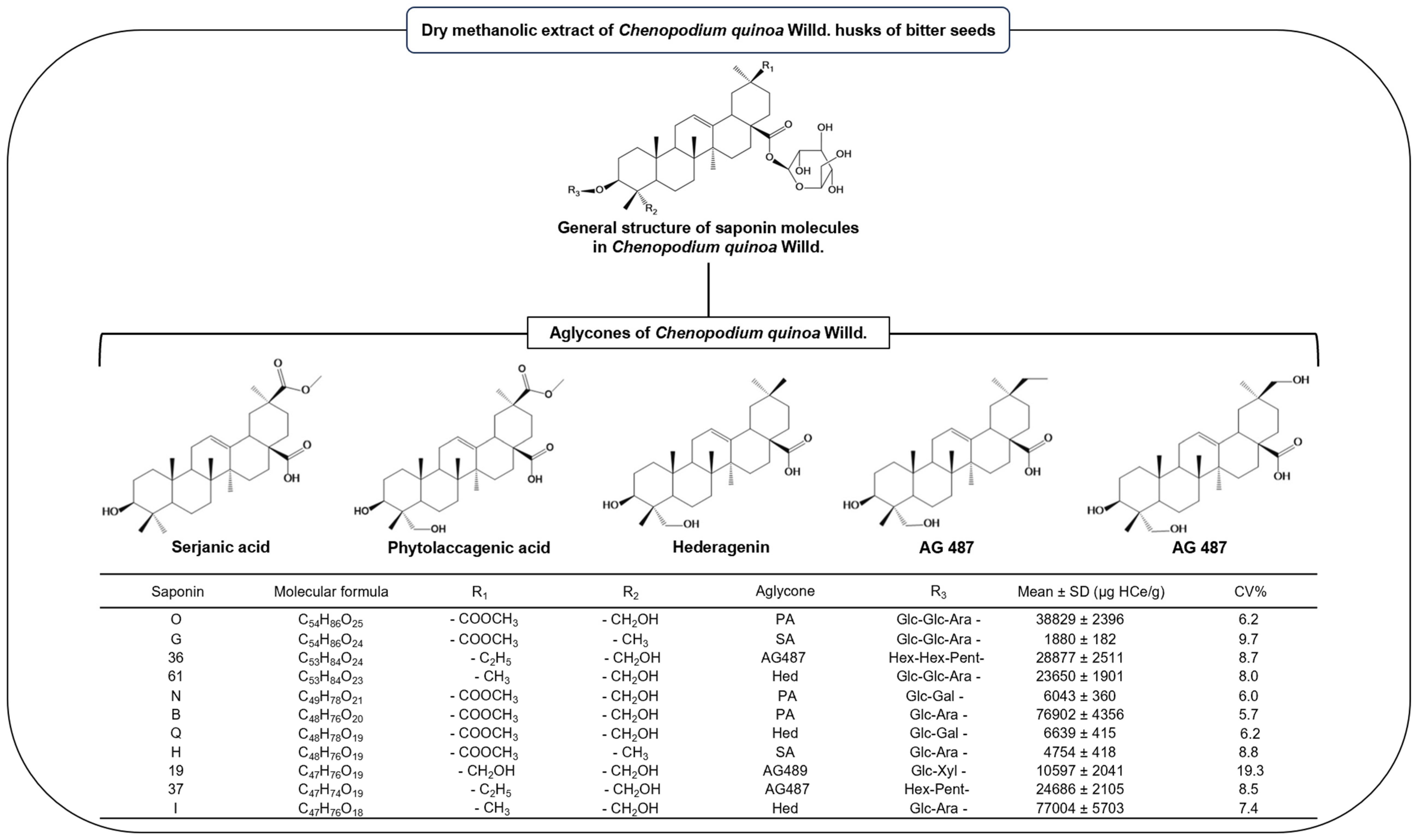
4.2. Proteins and Fluorescence Labeling
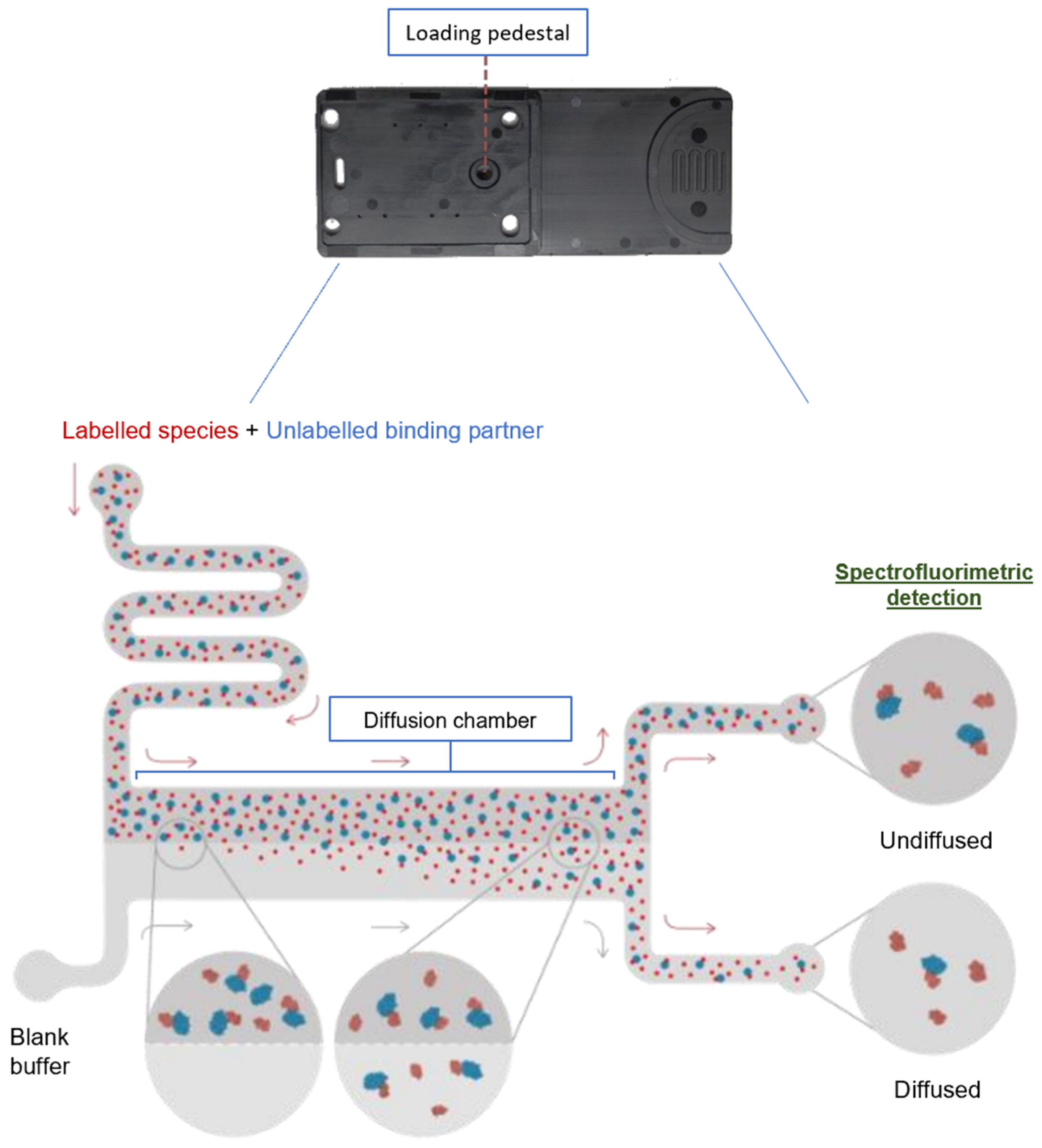
4.3. SARS-CoV-2 Spike RBD/ACE2 Affinity Measurement by Microfluidic Diffusional Sizing
4.4. Modulation of SARS-CoV-2 Spike RBD/ACE2 Affinity by Natural Compounds
4.5. Statistical Analysis of Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Health Orgarnization. Available online: https://covid19.who.int/table (accessed on 21 October 2023).
- Bhakti, K. Insuffisance Respiratoire Hypoxémique Aigüe (Syndrome de Détresse Respiratoire Aiguë [SDRA], Ou [ARDS], Acute Respiratory Distress Syndrome). N. Engl. J. Med. 2006, 354, 2564–2575. [Google Scholar] [CrossRef]
- Patel, K.P.; Vunnam, S.R.; Patel, P.A.; Krill, K.L.; Korbitz, P.M.; Gallagher, J.P.; Suh, J.E.; Vunnam, R.R. Transmission of SARS-CoV-2: An update of current literature. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Goel, N.; Jain, A.; Kumari, A. The role of ACE2 receptor and its age related immunity in COVID-19. Int. J. Pharm. Sci. Rev. Res. 2020, 63, 190–194. [Google Scholar]
- Prashantha, C.N.; Gouthami, K.; Lavanya, L.; Bhavanam, S.; Jakhar, A.; Shakthiraju, R.G.; Suraj, V.; Sahana, K.V.; Sujana, H.S.; Guruprasad, N.M.; et al. Molecular screening of antimalarial, antiviral, anti-inflammatory and HIV protease inhibitors against spike glycoprotein of coronavirus. J. Mol. Graph. Model. 2021, 102, 107769. [Google Scholar] [CrossRef] [PubMed]
- Rajah, M.M.; Bernier, A.; Buchrieser, J.; Schwartz, O. The Mechanism and Consequences of SARS-CoV-2 Spike-Mediated Fusion and Syncytia Formation. J. Mol. Biol. 2022, 434, 167280. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Ye, F.; Guo, Y.; Xia, L.; Zhong, X.; Chi, X.; Zhou, Q. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res. 2021, 31, 717–719. [Google Scholar] [CrossRef]
- Camargo, S.M.R.; Vuille-Dit-Bille, R.N.; Meier, C.F.; Verrey, F. ACE2 and gut amino acid transport. Clin. Sci. 2020, 134, 2823–2833. [Google Scholar] [CrossRef]
- Wiese, O.; Zemlin, A.E.; Pillay, T.S. Molecules in pathogenesis: Angiotensin converting enzyme 2 (ACE2). J. Clin. Pathol. 2021, 74, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e610. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, S.; Piziorska, M.A.; Denninger, V.; Morgunov, A.S.; Ilsley, A.; Malik, A.Y.; Schneider, M.M.; Devenish, S.R.A.; Meisl, G.; Kosmoliaptsis, V.; et al. Antibody Affinity Governs the Inhibition of SARS-CoV-2 Spike/ACE2 Binding in Patient Serum. ACS Infect. Dis. 2021, 7, 2362–2369. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Müller, T.; Rajah, L.; Yates, E.V.; Aprile, F.A.; Zhang, Y.; Cohen, S.I.A.; White, D.A.; Herling, T.W.; De Genst, E.J.; et al. Microfluidic diffusion analysis of the sizes and interactions of proteins under native solution conditions. ACS Nano 2016, 10, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Alqathama, A.A.; Ahmad, R.; Alsaedi, R.B.; Alghamdi, R.A.; Abkar, E.H.; Alrehaly, R.H.; Abdalla, A.N. The vital role of animal, marine, and microbial natural products against COVID-19. Pharm. Biol. 2022, 60, 509–524. [Google Scholar] [CrossRef]
- Frediansyah, A.; Sofyantoro, F.; Alhumaid, S.; Al Mutair, A.; Albayat, H.; Altaweil, H.I.; Al-Afghani, H.M.; AlRamadhan, A.A.; AlGhazal, M.R.; Turkistani, S.A.; et al. Microbial natural products with antiviral activities, including anti-SARS-CoV-2: A review. Molecules 2022, 27, 4305. [Google Scholar] [CrossRef]
- Low, Z.; Lani, R.; Tiong, V.; Poh, C.; AbuBakar, S.; Hassandarvish, P. COVID-19 therapeutic potential of natural products. Int. J. Mol. Sci. 2023, 24, 9589. [Google Scholar] [CrossRef]
- Lingwan, M.; Shagun, S.; Pant, Y.; Nanda, R.; Masakapalli, S. Antiviral phytochemicals identified in Rhododendron arboreum petals exhibited strong binding to SARS-CoV-2 MPro and Human ACE2 receptor. Preprints 2020, 2020080530. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, W.; Cheng, L.; Li, M.; Huang, J.; Bao, S.; Xu, Q.; Ma, Z. Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Nat. Prod. Bioprospecting 2022, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Raghuvanshi, R.; Ceylan, F.D.; Bolling, B.W. Quercetin and Its Metabolites Inhibit Recombinant Human Angiotensin-Converting Enzyme 2 (ACE2) Activity. J. Agric. Food Chem. 2020, 68, 13982–13989. [Google Scholar] [CrossRef]
- Pan, B.; Fang, S.; Zhang, J.; Pan, Y.; Liu, H.; Wang, Y.; Li, M.; Liu, L. Chinese herbal compounds against SARS-CoV-2: Puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Comput. Struct. Biotechnol. J. 2020, 18, 3518–3527. [Google Scholar] [CrossRef]
- Priyandoko, D. Molecular Docking Study of the Potential Relevance of the Natural Compounds Isoflavone and Myricetin to COVID-19. Int. J. Bioautomation 2021, 25, 271–282. [Google Scholar] [CrossRef]
- Shakhsi-Niaei, M.; Soureshjani, E.H.; Babaheydari, A.K. In Silico Comparison of Separate or Combinatorial Effects of Potential Inhibitors of the SARS-CoV-2 Binding Site of ACE2. Iran J. Public Health 2021, 50, 1028–1036. [Google Scholar] [CrossRef]
- Tutunchi, H.; Naeini, F.; Ostadrahimi, A.; Hosseinzadeh-Attar, M.J. Naringenin, a flavanone with antiviral and anti-inflammatory effects: A promising treatment strategy against COVID-19. Phytother. Res. 2020, 30, 3137–3147. [Google Scholar] [CrossRef]
- Abdelli, I.; Hassani, F.; Bekkel Brikci, S.; Ghalem, S. In Silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J. Biomol. Struct. Dyn. 2020, 39, 3263–3276. [Google Scholar] [CrossRef]
- Badraoui, R.; Saoudi, M.; Hamadou, W.S.; Elkahoui, S.; Siddiqui, A.J.; Alam, J.M.; Jamal, A.; Adnan, M.; Suliemen, A.M.E.; Alreshidi, M.M.; et al. Antiviral effects of artemisinin and Its derivatives against SARS-CoV-2 main protease: Computational evidences and interactions with ACE2 allelic variants. Pharmaceuticals 2022, 15, 129. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, L.; Gao, C.; Chen, W.; Vong, C.T.; Yao, P.; Yang, Y.; Li, X.; Tang, X.; Wang, S.; et al. Astragali Radix (Huangqi): A promising edible immunomodulatory herbal medicine. J. Ethnopharmacol. 2020, 258, 112895. [Google Scholar] [CrossRef] [PubMed]
- Goswami, T.; Bagchi, B. Molecular Docking study of Receptor Binding Domain of SARS-CoV-2 Spike Glycoprotein with Saikosaponin, a Triterpenoid Natural Product. ChemRxiv. Camb. Camb. Open Engag. 2020. [Google Scholar] [CrossRef]
- Rolta, R.; Salaria, D.; Sharma, P.; Sharma, B.; Kumar, V.; Rathi, B.; Verma, M.; Sourirajan, A.; Baumler, D.J.; Dev, K. Phytocompounds of Rheum emodi, Thymus serpyllum, and Artemisia annua Inhibit spike protein of SARS-CoV-2 binding to ACE2 receptor: In silico approach. Curr. Pharmacol. Rep. 2021, 7, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Shakya, A.; Prasad, S.K.; Singh, S.; Gurav, N.S.; Prasad, R.S.; Gurav, S.S. An in-silico evaluation of different saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2021, 39, 3244–3255. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.-S.; Yu, W.-D.; Wang, H.-X.; Liu, J.-Y.; Xu, P.-P. A potential antiviral activity of esculentoside A against binding interactions of SARS-COV-2 spike protein and angiotensin converting enzyme 2 (ACE2). Int. J. Biol. Macromol. 2021, 183, 2248–2261. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Altayb, H.N.; Al-Abbasi, F.A.; Kumar, V.; Kamal, M.A. The computational intervention of macrolide antibiotics in the treatment of COVID-19. Curr. Pharm. Des. 2021, 27, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Braz, H.L.B.; Silveira, J.A.d.M.; Marinho, A.D.; de Moraes, M.E.A.; Moraes Filho, M.O.d.; Monteiro, H.S.A.; Jorge, R.J.B. In Silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 56, 106119. [Google Scholar] [CrossRef]
- Kalhor, H.; Sadeghi, S.; Abolhasani, H.; Kalhor, R.; Rahimi, H. Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches. J. Biomol. Struct. Dyn. 2022, 40, 1299–1315. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.; Rheinstein, P.H. Ivermectin docks to the SARS-CoV-2 spike receptor-binding domain attached to ACE2. In Vivo 2020, 34, 3023–3026. [Google Scholar] [CrossRef]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef]
- Du, H.-X.; Zhu, J.-Q.; Chen, J.; Zhou, H.-F.; Yang, J.-H.; Wan, H.-T. Revealing the therapeutic targets and molecular mechanisms of emodin-treated coronavirus disease 2019 via a systematic study of network pharmacology. Aging 2021, 13, 14571–14589. [Google Scholar] [CrossRef]
- Zhan, Y.; Ta, W.; Tang, W.; Hua, R.; Wang, J.; Wang, C.; Lu, W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021, 82, 1124–1130. [Google Scholar] [CrossRef]
- Gangadevi, S.; Badavath, V.N.; Thakur, A.; Yin, N.; De Jonghe, S.; Acevedo, O.; Jochmans, D.; Leyssen, P.; Wang, K.; Neyts, J.; et al. Kobophenol A Inhibits Binding of Host ACE2 Receptor with Spike RBD Domain of SARS-CoV-2, a Lead Compound for Blocking COVID-19. J. Phys. Chem. Lett. 2021, 12, 1793–1802. [Google Scholar] [CrossRef]
- Perrella, F.; Coppola, F.; Petrone, A.; Platella, C.; Montesarchio, D.; Stringaro, A.; Ravagnan, G.; Fuggetta, M.P.; Rega, N.; Musumeci, D. Interference of Polydatin/Resveratrol in the ACE2:Spike Recognition during COVID-19 Infection. A Focus on Their Potential Mechanism of Action through Computational and Biochemical Assays. Biomolecules 2021, 11, 1048. [Google Scholar] [CrossRef]
- Chen, R.H.; Yang, L.J.; Hamdoun, S.; Chung, S.K.; Lam, C.W.-K.; Zhang, K.X.; Guo, X.; Xia, C.; Law, B.Y.K.; Wong, V.K.W. 1,2,3,4,6-Pentagalloyl Glucose, a RBD-ACE2 Binding Inhibitor to Prevent SARS-CoV-2 Infection. Front. Pharmacol. 2021, 12, 634176. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.J.; Chen, R.H.; Hamdoun, S.; Coghi, P.; Ng, J.P.L.; Zhang, D.W.; Guo, X.; Xia, C.; Law, B.Y.K.; Wong, V.K.W. Corilagin prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding. Phytomedicine 2021, 87, 153591. [Google Scholar] [CrossRef] [PubMed]
- Balkrishna, A.; Pokhrel, S.; Singh, H.; Joshi, M.; Mulay, V.P.; Haldar, S.; Varshney, A. Withanone from Withania somnifera attenuates SARS-CoV-2 RBD and host ACE2 interactions to rescue spike protein induced pathologies in humanized zebrafish model. Drug Des. Devel. Ther. 2021, 15, 1111–1133. [Google Scholar] [CrossRef] [PubMed]
- Caohuy, H.; Eidelman, O.; Chen, T.; Liu, S.; Yang, Q.; Bera, A.; Walton, N.I.; Wang, T.T.; Pollard, H.B. Common cardiac medications potently inhibit ACE2 binding to the SARS-CoV-2 Spike, and block virus penetration and infectivity in human lung cells. Sci. Rep. 2021, 11, 22195. [Google Scholar] [CrossRef]
- Chitsike, L.; Krstenansky, J.; Duerksen-Hughes, P.J. ACE2: S1 RBD interaction-targeted peptides and small molecules as potential COVID-19 therapeutics. Adv. Pharmacol. Pharm. Sci. 2021, 2021, e1828792. [Google Scholar] [CrossRef] [PubMed]
- Clementi, N.; Scagnolari, C.; D’Amore, A.; Palombi, F.; Criscuolo, E.; Frasca, F.; Pierangeli, A.; Mancini, N.; Antonelli, G.; Clementi, M.; et al. Naringenin is a powerful inhibitor of SARS-CoV-2 infection in vitro. Pharmacol. Res. 2021, 163, 105255. [Google Scholar] [CrossRef] [PubMed]
- van Breemen, R.B.; Muchiri, R.N.; Bates, T.A.; Weinstein, J.B.; Leier, H.C.; Farley, S.; Tafesse, F.G. Cannabinoids block cellular entry of SARS-CoV-2 and the emerging variants. J. Nat. Prod. 2022, 85, 176–184. [Google Scholar] [CrossRef]
- Kim, T.; Jeon, S.; Jang, Y.; Gotina, L.; Won, J.; Ju, Y.; Kim, S.; Jang, M.; Won, W.; Park, M.; et al. Platycodin D, a natural component of Platycodon grandiflorum, prevents both lysosome- and TMPRSS2-driven SARS-CoV-2 infection by hindering membrane fusion. Exp. Mol. Med. 2021, 53, 956–972. [Google Scholar] [CrossRef]
- Senthil Kumar, K.J.; Gokila Vani, M.; Wang, C.-S.; Chen, C.-C.; Chen, Y.-C.; Lu, L.-P.; Huang, C.-H.; Lai, C.-S.; Wang, S.-Y. Geranium and lemon essential oils and their active compounds downregulate angiotensin-converting enzyme 2 (ACE2), a SARS-CoV-2 spike receptor-binding domain, in epithelial cells. Plants 2020, 9, 770. [Google Scholar] [CrossRef]
- Xu, H.; Liu, B.; Xiao, Z.; Zhou, M.; Ge, L.; Jia, F.; Liu, Y.; Jin, H.; Zhu, X.; Gao, J.; et al. Computational and experimental studies reveal that thymoquinone blocks the entry of coronaviruses into in vitro cells. Infect. Dis. Ther. 2021, 10, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Große, M.; Ruetalo, N.; Layer, M.; Hu, D.; Businger, R.; Rheber, S.; Setz, C.; Rauch, P.; Auth, J.; Fröba, M.; et al. Quinine Inhibits Infection of Human Cell Lines with SARS-CoV-2. Viruses 2021, 13, 647. [Google Scholar] [CrossRef]
- Wang, N.; Han, S.; Liu, R.; Meng, L.; He, H.; Zhang, Y.; Wang, C.; Lv, Y.; Wang, J.; Li, X.; et al. Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus. Phytomedicine 2020, 79, 153333. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Li, C.-C.; Hsiang, C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007, 74, 92–101. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; van der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife 2021, 10, e70658. [Google Scholar] [CrossRef] [PubMed]
- Laffeber, C.; de Koning, K.; Kanaar, R.; Lebbink, J.H.G. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. J. Mol. Biol. 2021, 433, 167058. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Jiang, C.; Liang, B.; Reynoso, L.; et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021, 31, 720–722. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Nutalai, R.; Tuekprakhon, A.; et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211.e7. [Google Scholar] [CrossRef] [PubMed]
- Prévost, J.; Richard, J.; Gasser, R.; Ding, S.; Fage, C.; Anand, S.P.; Adam, D.; Vergara, N.G.; Tauzin, A.; Benlarbi, M.; et al. Impact of temperature on the affinity of SARS-CoV-2 Spike for ACE2. bioRxiv 2021, 451812. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- ReactionBiology. Reaction Biology. Available online: https://www.reactionbiology.com/sites/default/files/Images/Content/Biophysical_Assay/SPR_S%20protein%20ACE2_ReactionBiology_V2.pdf (accessed on 25 October 2023).
- Allen, J.D.; Watanabe, Y.; Chawla, H.; Newby, M.L.; Crispin, M. Subtle Influence of ACE2 Glycan Processing on SARS-CoV-2 Recognition. J. Mol. Biol. 2021, 433, 166762. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A.; Arukuusk, P.; Härk, H.; Juronen, E.; Langel, Ü.; Ustav, M.; Järv, J. ACE2 peptide fragment interacts with several sites on the SARS-CoV-2 spike protein S1. bioRxiv 2020, 424682. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Smith, M.D.; Smith, J.C. Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface. ChemRxiv, 2020; preprint. [Google Scholar]
- Charles, C.; Nachtergael, A.; Ouedraogo, M.; Belayew, A.; Duez, P. Effects of chemopreventive natural products on non-homologous end-joining DNA double-strand break repair. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2014, 768, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Rebello, C.J.; Beyl, R.A.; Lertora, J.J.L.; Greenway, F.L.; Ravussin, E.; Ribnicky, D.M.; Poulev, A.; Kennedy, B.J.; Castro, H.F.; Campagna, S.R.; et al. Safety and pharmacokinetics of naringenin: A randomized, controlled, single-ascending-dose clinical trial. Diabetes Obes. Metab. 2020, 22, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Biber, A.; Harmelin, G.; Lev, D.; Ram, L.; Shaham, A.; Nemet, I.; Kliker, L.; Erster, O.; Mandelboim, M.; Schwartz, E. The effect of ivermectin on the viral load and culture viability in early treatment of nonhospitalized patients with mild COVID-19—A double-blind, randomized placebo-controlled trial. Int. J. Infect. Dis. 2022, 122, 733–740. [Google Scholar] [CrossRef] [PubMed]
- González Canga, A.; Sahagún Prieto, A.M.; Diez Liébana, M.J.; Fernández Martínez, N.; Sierra Vega, M.; García Vieitez, J.J. The pharmacokinetics and interactions of ivermectin in humans—A mini-review. AAPS J. 2008, 10, 42–46. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, J.M.; Kim, C. Pharmacokinetic analysis of rhein in Rheum undulatum L. J. Ethnopharmacol. 2003, 84, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, X.-M.; Zhang, L.; Li, X.-Y.; Wang, B.-X. Pharmacokinetic of rhein in healthy male volunteers following oral and retention enema administration of rhubarb extract: A single dose study. Am. J. Chin. Med. 2005, 33, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Mieres-Castro, D.; Mora-Poblete, F. Saponins: Research Progress and Their Potential Role in the Post-COVID-19 Pandemic Era. Pharmaceutics 2023, 15, 348. [Google Scholar] [CrossRef]
- Ogunyemi, O.M.; Gyebi, G.A.; Ibrahim, I.M.; Olaiya, C.O.; Ocheje, J.O.; Fabusiwa, M.M.; Adebayo, J.O. Dietary stigmastane-type saponins as promising dual-target directed inhibitors of SARS-CoV-2 proteases: A structure-based screening. RSC Adv. 2021, 11, 33380–33398. [Google Scholar] [CrossRef]
- Rehan, M.; Shafiullah. Medicinal plant-based saponins targeting COVID-19 Mpro in silico. Tradit Med. Res. 2021, 6, 21–30. [Google Scholar] [CrossRef]
- Falade, V.A.; Adelusi, T.I.; Adedotun, I.O.; Abdul-Hammed, M.; Lawal, T.A.; Agboluaje, S.A. In Silico investigation of saponins and tannins as potential inhibitors of SARS-CoV-2 main protease (Mpro). Silico Pharmacol. 2021, 9, 9. [Google Scholar] [CrossRef]
- Chen, H.; Du, Q. Potential Natural Compounds for Preventing SARS-CoV-2 (2019-nCoV) Infection. Preprints 2020. [Google Scholar] [CrossRef]
- Yan, Y.; Shen, X.; Cao, Y.; Zhang, J.; Wang, Y.; Cheng, Y. Discovery of Anti-2019-nCoV Agents from Chinese Patent Drugs via Docking Screening. Preprints 2020, 2020020254. [Google Scholar] [CrossRef][Green Version]
- Cheng, P.W.; Ng, L.T.; Chiang, L.C.; Lin, C.C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006, 33, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.; Lu, H.T.; Fan, K.W.; Cheng, V.C.; Tsui, W.H.; Hung, I.F.; Lee, T.S.; et al. In Vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hoever, G.; Baltina, L.; Michaelis, M.; Kondratenko, R.; Baltina, L.; Tolstikov, G.A.; Doerr, H.W.; Cinatl, J., Jr. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005, 48, 1256–1259. [Google Scholar] [CrossRef]
- Wu, C.Y.; Jan, J.T.; Ma, S.H.; Kuo, C.J.; Juan, H.F.; Cheng, Y.S.; Hsu, H.H.; Huang, H.C.; Wu, D.; Brik, A.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012–10017. [Google Scholar] [CrossRef]
- Yu, K.; Chen, F.; Li, C. Absorption, disposition, and pharmacokinetics of saponins from chinese medicinal herbs: What do we know and what do we need to know more? Curr. Drug Metab. 2012, 13, 577–598. [Google Scholar] [CrossRef] [PubMed]
- Taco, V.; Savarino, P.; Benali, S.; Villacrés, E.; Raquez, J.-M.; Gerbaux, P.; Duez, P.; Nachtergael, A. Deep eutectic solvents for the extraction and stabilization of Ecuadorian quinoa (Chenopodium quinoa Willd.) saponins. J. Clean. Prod. 2022, 363, 132609. [Google Scholar] [CrossRef]
- Sha, A.; Liu, Y.; Hao, H. Current state-of-the-art and potential future therapeutic drugs against COVID-19. Front. Cell Dev. Biol. 2023, 11, 1238027. [Google Scholar] [CrossRef]
- Wu, A.; Shi, K.; Wang, J.; Zhang, R.; Wang, Y. Targeting SARS-CoV-2 entry processes: The promising potential and future of host-targeted small-molecule inhibitors. Eur. J. Med. Chem. 2024, 263, 115923. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Wu, C.-F.; Huang, Y.-T. Phenols from the roots of Rheum palmatum attenuate chemotaxis in rat hepatic stellate cells. Planta Med. 2008, 74, 1246–1252. [Google Scholar] [CrossRef] [PubMed]
- Sigma-Aldrich. Available online: https://www.sigmaaldrich.com/certificates/Graphics/COfAInfo/fluka/pdf/rtc/PHR1380_LRAC6468.pdf (accessed on 18 November 2023).
- van Beek, T.A.; Montoro, P. Chemical analysis and quality control of Ginkgo biloba leaves, extracts, and phytopharmaceuticals. J. Chromatogr. A 2009, 1216, 2002–2032. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Huang, Y.-J.; Chen, L.-G.; Lee, L.-T.; Yang, L.-L. Inducible nitric oxide synthase inhibitors of chinese herbs III. Rheum palmatum. Planta Med. 2002, 68, 869–874. [Google Scholar] [CrossRef] [PubMed]
- EMA. Assessment Report on Rheum palmatum L. and Rheum Officinale Baillon, Radix; HMPC—European Medicines Agency: Amsterdam The Netherlands, 2020.
- Fluidic Analytics. Available online: https://www.fluidic.com/ (accessed on 21 October 2023).
- Fluidic Analytics. User Guide for Fluidity™ One-M IFU-0011 v4; Fluidic Analytics: Cambridge, UK, 2022; p. 12. [Google Scholar]
- Fluidic Analytics. User Manual for Fluidity One-W and Fluidity One-W Serum IFU-0007v8; Fluidic Analytics: Cambridge, UK, 2021; p. 9. [Google Scholar]
- Overduin, M.; Esmaili, M. Memtein: The fundamental unit of membrane-protein structure and function. Chem. Phys. Lipids 2019, 218, 73–84. [Google Scholar] [CrossRef]
- Fluidic Analytics. Available online: https://www.fluidic.com/resources/hydrodynamicradius-and-protein-weight/ (accessed on 9 April 2022).
- Fluidic Analytics. Available online: https://www.fluidic.com/calculators-page/ (accessed on 23 October 2023).
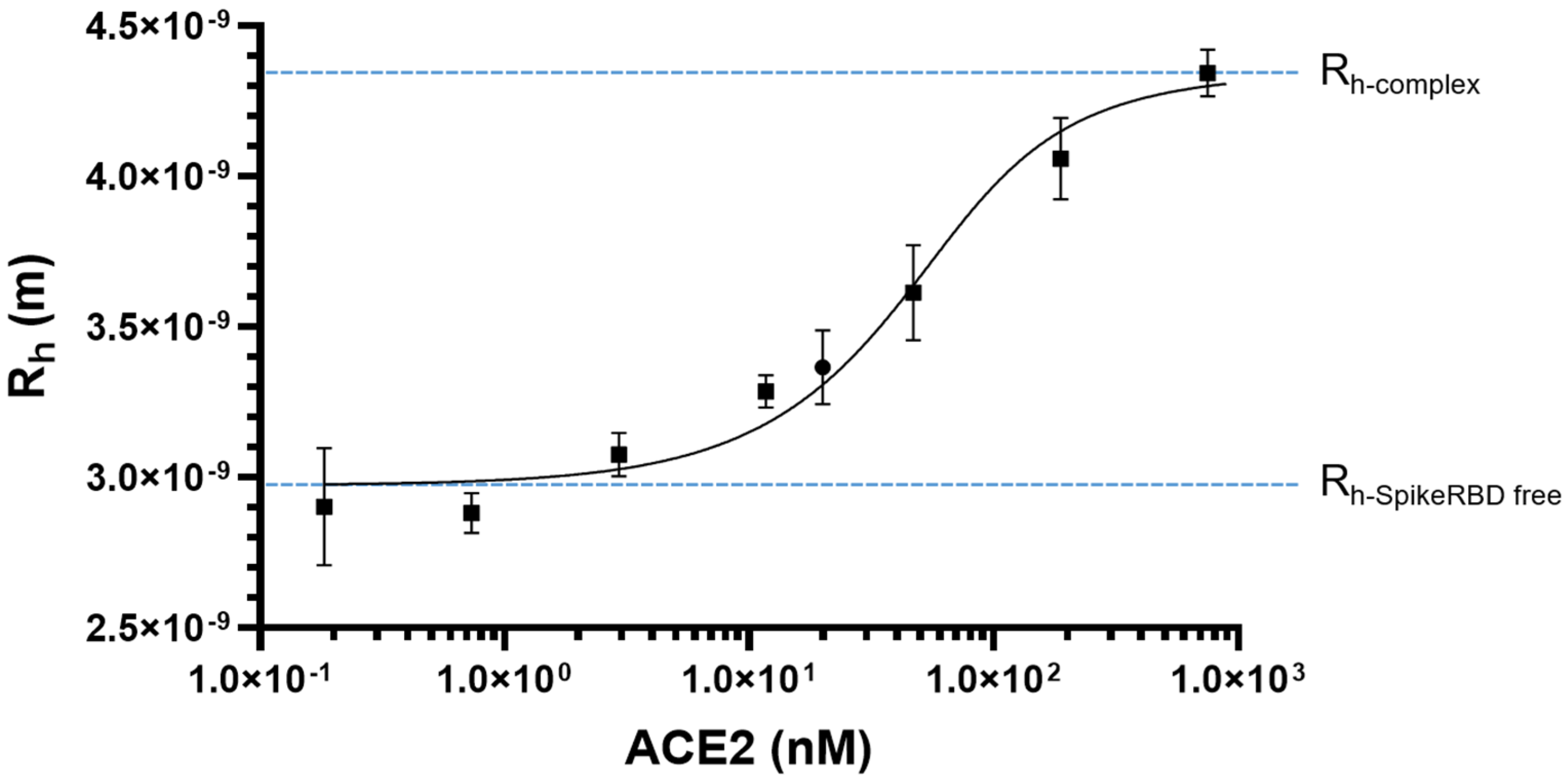
| Ligand-Receptor | KD Determination Method | KD (nM) | Reference |
|---|---|---|---|
| SARS-CoV-2 RBD/ACE2 | SPR | 4.7 | [58] |
| SPR | 63.0 | [59] | |
| SPR | 24.1 | [24] | |
| SPR | 17.0 | [60] | |
| SPR | 5.8 | [61] | |
| SPR | 44.2 | [62] | |
| BLI | 75.1 | [63] | |
| BLI | 161 | [64] | |
| BLI | 172 | [44] | |
| SARS-CoV-2 Spike/ACE2 | SPR | 14.7 | [65] |
| SPR | 29.1 | [66] | |
| SPR | 76 | [67] | |
| BLI | 12.8 | [68] | |
| BLI | 1.2 | [69] | |
| BLI | 133.0 | [70] |
| Criterion | What Is Assessed? | Result | Full Data (Supplementary Materials) |
|---|---|---|---|
| Selectivity | Eventual fluorescence interferences | No | Figure S1 |
| Between-chips reproducibility | 1 chip for 1 Rh measurement (n = 6) | CV = 3.99% | Table S1 |
| Within-chip reproducibility | 1 chip for Rh measurement X times at the same concentration (n = 8) | CV = 31.4% | Table S2 |
| Determination of KD over a single chip | 1 chip for 1 KD determination (7 points) (n = 3) | Erroneous values | Figure S2 |
| Quality of adjustment | Coefficient of determination (R2) | 0.854–0.963 | Table S7 |
| Accuracy of Rh for the Spike RBDlabelled | Comparison of experimental Rh with a value predicted from a range of protein standards with globular conformation | 105.4 ± 5.7% (n = 13) | Point 3 |
| Precision | Rh—Intraday precision (n = 4) | CV = 2.90% | Table S5 |
| Rh—Total precision (n = 6) | CV = 6.27% | Table S4 | |
| KD—Total precision (n = 5) | CV = 20.8% | Table S6 |
| Natural Product (NP) | Quercetin | Naringenin | Ivermectin | Rhei Radix EtOH Dry Extract | Chenopodium quinoa Willd. MeOH Dry Extract (Husks) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Range Tested | 1st Effective Concentration | Range Tested | 1st Effective Concentration | Range Tested | 1st Effective Concentration | Range Tested | 1st Effective Concentration | Range Tested | 1st Effective Concentration | |
| 0.1–150 µM | 150 µM | 0.1–50 µM | 1 µM | 1–100 nM | 1 nM | 1–100 µg/mL | 100 µg/mL | 1–200 µg/mL | 50 µg/mL | |
| Rh of daily control (nm) (mean ± SD) | 3.28 ± 0.12 | 2.91 ± 0.05 | 3.19 ± 0.07 | 3.03 ± 0.08 | 2.14 ± 0.02 | |||||
| Rh in the presence of NP (nm) (mean ± SD) | 3.35 ± 0.07 to 2.92 ± 0.03 | 2.92 ± 0.03 | 3.09 ± 0.05 to 3.21 ± 0.07 | 3.21 ± 0.07 | 3.57 ± 0.07 to 4.09 ± 0.08 | 3.57 ± 0.07 | 3.03 ± 0.07 to 2.61 ± 0.05 | 2.61 ± 0.05 | 2.34 ± 0.22 to 5.33 ± 0.31 | 3.0 ± 0.1 |
| Direction of Rh variation | Decrease | Increase | Increase | Decrease | Increase | |||||
| Rh variation (%) (mean ± SD) | 2.12 ± 0.09 to −11.09 ± 0.43 | −11.09 ± 0.43 | 6.08 ± 0.15 to 19.01 ± 0.85 | 9.99 ± 0.29 | 11.78 ± 0.40 to 27.97 ± 0.82 | 11.78 ± 0.40 | −0.15 ± 0.01 to −13.97 ± 0.46 | −13.97 ± 0.46 | 9.19 ± 0.87 to 148.98 ± 8.76 | 39.92 ± 1.91 |
| Natural Product (NP) | Naringin | Chloroquine | Erythromycin | Gingko biloba L. Dry Extract (Leaves) |
|---|---|---|---|---|
| Range tested | 0.1–50 µM | 1–1000 µM | 0.1–50 µM | 1–200 µg/mL |
| Rh of daily control (nm) (mean ± SD) | 3.31 ± 0.06 | 2.87 ± 0.04 | 3.14 ± 0.05 | 2.39 ± 0.11 |
| Rh of NP (nm) (mean ± SD) | 3.26 ± 3.38–3.46 ± 0.12 | 2.76 ± 0.13–2.94 ± 0.03 | 3.06 ± 0.07–3.11 ± 0.08 | 2.35 ± 0.03–2.34 ± 0.16 |
| Rh variation (%) (mean ± SD) | −1.64 ± 0.03–4.5 ± 0.2 | −3.92 ± 0.20–2.11 ± 0.03 | −2.29 ± 0.06–−0.73 ± 0.02 | −1.75 ± 0.08–−2.10 ± 0.17 |
| p Value | >0.9999–0.1256 | 0.4134–>0.9999 | 0.8427–>0.9999 | >0.9999–>0.9999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fauquet, J.; Carette, J.; Duez, P.; Zhang, J.; Nachtergael, A. Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction. Molecules 2023, 28, 8072. https://doi.org/10.3390/molecules28248072
Fauquet J, Carette J, Duez P, Zhang J, Nachtergael A. Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction. Molecules. 2023; 28(24):8072. https://doi.org/10.3390/molecules28248072
Chicago/Turabian StyleFauquet, Jason, Julie Carette, Pierre Duez, Jiuliang Zhang, and Amandine Nachtergael. 2023. "Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction" Molecules 28, no. 24: 8072. https://doi.org/10.3390/molecules28248072
APA StyleFauquet, J., Carette, J., Duez, P., Zhang, J., & Nachtergael, A. (2023). Microfluidic Diffusion Sizing Applied to the Study of Natural Products and Extracts That Modulate the SARS-CoV-2 Spike RBD/ACE2 Interaction. Molecules, 28(24), 8072. https://doi.org/10.3390/molecules28248072






