Abstract
Lipid membrane nanodomains or lipid rafts are 10–200 nm diameter size cholesterol- and sphingolipid-enriched domains of the plasma membrane, gathering many proteins with different roles. Isolation and characterization of plasma membrane proteins by differential centrifugation and proteomic studies have revealed a remarkable diversity of proteins in these domains. The limited size of the lipid membrane nanodomain challenges the simple possibility that all of them can coexist within the same lipid membrane domain. As caveolin-1, flotillin isoforms and gangliosides are currently used as neuronal lipid membrane nanodomain markers, we first analyzed the structural features of these components forming nanodomains at the plasma membrane since they are relevant for building supramolecular complexes constituted by these molecular signatures. Among the proteins associated with neuronal lipid membrane nanodomains, there are a large number of proteins that play major roles in calcium signaling, such as ionotropic and metabotropic receptors for neurotransmitters, calcium channels, and calcium pumps. This review highlights a large variation between the calcium signaling proteins that have been reported to be associated with isolated caveolin-1 and flotillin-lipid membrane nanodomains. Since these calcium signaling proteins are scattered in different locations of the neuronal plasma membrane, i.e., in presynapses, postsynapses, axonal or dendritic trees, or in the neuronal soma, our analysis suggests that different lipid membrane-domain subtypes should exist in neurons. Furthermore, we conclude that classification of lipid membrane domains by their content in calcium signaling proteins sheds light on the roles of these domains for neuronal activities that are dependent upon the intracellular calcium concentration. Some examples described in this review include the synaptic and metabolic activity, secretion of neurotransmitters and neuromodulators, neuronal excitability (long-term potentiation and long-term depression), axonal and dendritic growth but also neuronal cell survival and death.
Keywords:
flotillin; caveolin; ganglioside; lipid rafts; lipid membrane domains; calcium channel; NMDA; PMCA; P2XR; membrane domains; neuron; brain 1. Lipid Membrane Nanodomains Organization in the Neuronal Plasma Membrane
The classical model of the plasma membrane, named the fluid mosaic model, described by Jonathan Singer and Garth Nicolson in 1972, is excessively reductionist for properly accounting for the well-organized plasma membrane domains. Lipid rafts are plasma membrane large areas of 10 and 200 nm diameter in size enriched in cholesterol and sphingolipids [1]. The existence lipid rafts was initially a subject of debate between physical chemists and histologists due to difficulties in visualizing them and their ill-defined molecular composition [1,2]. In the last two decades, a number of new techniques such as single-molecule spectroscopy, super-resolution microscopy, fluorescence recovery after photobleaching, stimulated emission depletion, Förster resonance energy transfer (FRET), total internal reflection fluorescence, and fluorescence correlation spectroscopy techniques allowed to estimate the lower limit of lipid rafts in <20 nm [3,4,5,6]. Plasma membrane domains of 26 ± 13 nm radius have been observed in living cells diffusing as one entity for minutes [7]. Further work using stimulated emission depletion (STED) far-field fluorescence nanoscopy revealed spots sized 70-fold below the diffraction barrier transiently trapped between 10 and 20 ms, in cholesterol-mediated molecular complexes dwelling within <20-nm diameter areas [3]. The diffraction limit of visible light impedes domains smaller than 1 µm to be directly visualized and indeed large micrometer-sized lipid rafts domains are readily observed in artificial membranes [3]. Also, associated proteins can mask the direct observation of lipid rafts in living cells. A tentative attempt to determine analogous domains in living cells has been made based on homo-FRET efficiencies obtained through the rate of fluorescence anisotropy loss and using GFP labeled glycosyl-phosphatidylinositol-anchored proteins which allow an estimation of the upper size limit of lipid rafts at ~5 nm [8,9]. Yethiraj and Weisshaar have suggested that the spatial heterogeneity in cell membranes limits the transferability of the conclusion drawn from artificial membranes to live cells, as integral membrane proteins attached to the cytoskeleton act as obstacles that limit the size of lipid domains [8]. For all these reasons, we introduce the concept of lipid membrane domains in this review, arising from the fact that some membrane proteins form oligomers and clusters in the membranes, which formation is favored by cholesterol and other lipid species.
Regarding the protein components associated with lipid membrane domains, widely named in the bibliography as lipid rafts, a proteomic study identified up to 36 integral membrane proteins associated with lipid membrane domain and flotillin, as a marker of these membrane domains where identified in the human brain [10]. In another study, 175 membrane-associated proteins were identified by proteomics, including L-type calcium channels and the plasma membrane calcium ATPase (PMCA), using caveolin-1 (Cav-1) and flotillin-1 (Flot-1), as biomarkers of lipid membrane domains isolated from brain neonatal mice [11]. Similarly, a proteomic assessment of proteins present in isolated lipid membrane domains of adult mouse brains identified 133 proteins, using Flot-1 as a marker of plasma membrane domains [12]. This study also highlighted the colocalization of this protein with several calcium channel subunits [12]. In cultured hippocampal neurons, sphingolipid-cholesterol-enriched microdomains have been localized flotillin 1, Thy-1 cell surface antigen or CD90, as specific lipid membrane-domain markers, associated with the ganglioside named monosialotetrahexosylganglioside (GM1) [13]. It is worth to mention at this point that although GM1 is not a definite lipid membrane-domain marker, its distribution into lipid membrane domains depends on the concentration. At elevated concentration, GM1 can form its own domains organizing in the plasma membrane in non-lipid membrane-domain areas located predominantly in the Ld phase [14]. Very recent discoveries regarding the molecular architecture of lipid membrane nanodomains support their organization in planar tightly packed nanodisks of Cav-1, with a 140Å external diameter size [15]. It is also probable that a similar size supramolecular complex based on flotillin might exist, based on the observed structural conformations of stomatin, prohibitin, flotillin, and the modulator for HflB protease specific for phage lambda cII repressor (HflK/C) domains (SPFH domain) [16]. Also, some studies have reported the isolation of up to 4 types of domains in the plasma membrane at physiological conditions [17]. Given the existence of these nanostructures, a question arises regarding how many of the reported protein molecules in the aforementioned proteomic and non-proteomic studies [10,11,12,13] could fit within a single of these nanostructures on one neuronal lipid membrane domain. The quantity of proteins reported in the neuronal lipid membrane domain contrasts with the number of proteins that could fit within or surrounding a 140Å diameter size nanodisk, if this type of structure stands alone as the main component of neuronal planar-lipid membrane domains in the plasma membrane. Neuronal lipid membrane domains are different from those of the invaginated caveolae in a variety of cell types, which require the presence of the protein named cavin and higher-order interactions with other proteins [18,19,20]. Cav-1–cavin interaction seems required to form mature caveolae, which have a polygonal shape to induce curvature in non-neuronal cells [21,22,23,24]. Cavin is absent or released when conforming planar-non-invaginated lipid membrane domains [20,25,26,27], like those described in neuronal lipid membrane domains.
In addition to these studies, more efforts are required to ascertain whether Cav-1 nanodisks independently exist in neuronal cells, either as discrete entities supporting non-invaginated areas on the plasma membrane or as components of supramolecular structures analogous to those observed in invaginated caveolae [20]. Since supramolecular structures with a similar protein composition to that of caveolae do not exist in neurons, the presence of a high number of proteins located in lipid membrane domains raises questions regarding the number of proteins that one Cav-1 nanodisk can hold due to steric hindrance. Methods for lipid membrane-domain isolation based on differential gradient centrifugation cannot discern the existence of lipid membrane nanodomain subtypes. Particularly, cytochemical and histochemical studies combined with physicochemical techniques based on quantitative fluorescence energy transfer (FRET) techniques, as those conducted by the research group led by Prof. Gutierrez-Merino, have provided insights into this matter by identification of proteins in clusters complexing with protein markers of lipid membrane domains (caveolin and flotillin isoforms) at a distance <100 nm in studies performed in neurons and brain tissue using the appropriate secondary fluorescent antibodies against the primary antibody of the selected lipid membrane-domain marker (Box 1) [28,29,30,31,32,33,34,35,36]. As discussed in these articles, this is a particular case of FRET from one donor to multiple acceptors, a situation in which the maximum range of FRET distance is significantly expanded, as analyzed in detail in former studies with purified biological membranes [23,37,38,39]. These research findings might support the existence of clusters that could stand alone as individual entities, such as Cav-1 nanodisks, with a diverse variety of calcium transporter elements. The well-recognized and wide distribution of these transporters in neurons, functioning as partners of lipid membrane-domain markers, strongly suggests the potential existence of multiple lipid membrane-domain subtypes within neurons. A neuronal lipid membrane-domain subtype is defined in this work as a plasma membrane, synaptic or extrasynaptic structure characterized by the presence of a protein biomarker of lipid membrane nanodomain and a specific calcium transport systems. The existence of these subdomains might correlate with the function of calcium gradients associated with cytosolic calcium microcompartments, near the plasma membrane [33,40], and such patterns may arise under certain conditions [41,42,43,44].
In this context, it is intriguing and controversial whether different types of lipid membrane domains might exist within a single cell or across different cell types based on the complex lipid and protein composition of these domains. This issue might be particularly notorious in tissues such as the brain, where recent findings using single-cell sequencing and methods to map the spatial location of gene expression have unraveled the extraordinary cellular diversity existing within this tissue [45]. Strategies for isolating lipid membrane domains, named rafts in these studies, that utilized membrane tension generate large observable membrane domains or lipid rafts, that are converted into small ones when the tension was relieved [17]. This result lends support to the hypothesis that a myriad of not well-described plasma membrane nanodomains might exist.
Box 1. FRET from one donor to multiple acceptors.
Labeling of proteins with donor and acceptor secondary fluorescent antibodies forming a FRET pair is an approach that has been used to identify proteins clustered in lipid raft domains. This is a particular case of FRET from one donor to multiple acceptors because the density of labeling of commercial secondary fluorescent antibodies ranges between 2 and 10 dye molecules per antibody [46,47,48,49], and also because in theory, one primary IgG antibody can bind up to 2 secondary fluorescent IgG antibodies, one in each of the symmetrical domains of the primary antibody. Therefore, one dye molecule of the donor fluorescent antibody can form a FRET pair with 2–10 and 4–20 acceptor dyes bound to the acceptor secondary antibody for 1:1 and 1:2 stoichiometries of the primary/secondary antibody complex, respectively. The major advantage of a high density of labeling of the secondary fluorescent antibody is the amplification of the fluorescence intensity signal for fluorescence microscopy imaging of cells. In addition, it has another collateral advantage for FRET distance calculations, namely, that homotransfer between donors located in one fluorescent secondary antibody and time and space averaging of different orientations of donors and acceptors bound to different IgG molecules which should lead to a distribution close to a random orientation between donor and acceptors.
The number of acceptor dyes available to a donor dye bound to a fluorescent antibody for FRET will be larger when the target protein units form clusters within lipid membrane-domains. In this case, FRET will extend to acceptor dyes of secondary antibodies bound to the primary antibodies that stain all neighbor protein targets present in the cluster within the area accessible to the IgG complex of primary/secondary antibodies plus the effective FRET distance between the selected donor and acceptor dyes. Each 1:1 complex of primary/secondary IgG antibodies will reach proteins located up to ≈30 nm from the target protein, taking into account the size of IgG molecules and their rotational mobility. Therefore, this implies that donor dyes bound to a primary/secondary IgG/ protein-1 complex can make contacts with acceptor dyes bound to the primary/secondary IgG complex attached to protein-2 separated up to ≈ 60 nm in the same lipid membrane-domain. If there is more than one unit of the target protein-2 stained with the secondary fluorescent antibody labeled with the acceptor dye, the number of acceptors/donor available for FRET will be proportionally increased.
In addition, the overall rate (kT) of FRET can be written for these cases as the sum of the rate of FRET between each one of the possible donor/acceptor pairs that can be formed in the system under study, i.e., kT = Σ ki, see for example [50,51]. Therefore, the overall FRET efficiency is the sum of the efficiency of energy transfer between all the possible donor/acceptor pairs that can be formed in the system [50,51,52]. This further increases the effective FRET distance using donor and acceptor secondary fluorescent antibodies. A simple calculation can serve to illustrate this point. For FRET from 1 donor to 10 acceptor molecules located at an equidistant distance, the apparent distance for 50% efficiency of FRET will be ~10 × R0 from the target protein labeled with the donor secondary fluorescent antibody, where R0 is the value of this distance for a single donor/acceptor pair, which ranges between 5 and 6 for the most frequently used FRET pairs in fluorescence microscopy. Let us remind here that the useful donor/acceptor distance range for a single donor/acceptor pair is approximately up to twice the distance for 50% efficiency of FRET [51,53]. Note that 10 acceptors per donor can be reached in any of the following cases: (i) 1:2 stoichiometry of the primary/secondary antibody complex and an average density of labeling of the acceptor fluorescent antibody of 5 dye molecules per antibody, and only one unit of the target proteins in the lipid membrane-domain; and (ii) 1:1 stoichiometry of the primary/secondary antibody complex and an average density of labeling of the acceptor fluorescent antibody of 5 dye molecules per antibody, with two protein units labeled with the acceptor fluorescent antibody within 60 nm in the same lipid membrane-domain. In summary, the effective FRET distance range extends to 80–200 nm when donor and acceptor dyes are bound to secondary fluorescent IgG antibodies directed against different target proteins present in lipid membrane-domains.
Thus, FRET using donor and acceptor secondary fluorescent antibodies is a suitable approach to monitor the co-localization of proteins within lipid membrane-domains of 100–200 nm. Also, it follows from this analysis that when there is only one unit of one of the target proteins within each lipid membrane-domain, co-localization of proteins within smaller lipid membrane-domains of 40 or 20 nm can be studied with the use of fluorescent primary antibodies or antibody Fab fragments, respectively, instead of using fluorescent secondary antibodies.
For cells, application of membrane tension resulted in several types of large domains; one class of domains was identified as a lipid raft, defined here as lipid membrane domain. Furthermore, the distribution of protein components of lipid domains [54,55,56,57] in planar non-invaginated regions of the neuronal plasma membrane [20,25,26,27], may be considered a robust evidence for the existence of not-so-transient, underlying structures that support several membrane nanodomains in neurons. This structural arrangement may differ from that observed in other cell types, where membrane invaginated areas forming caveolae have been described involved in membrane trafficking, with a transient formation and elimination of the protein content of these domains.
The objective of this review is to provide a comprehensive exploration and integrative analysis of information, suggesting the existence of lipid/protein-domain subtypes within neuronal cells. Several proteins that play major roles in neuronal calcium signaling have been described as components of lipid membrane domains [58], i.e., neurotransmitter receptors [59,60] and calcium transport systems [43,61], and they present a differential subcellular distribution within a single neuron and across different types of neurons, as shown in this review. The distribution pattern serves as a crucial tool for proposing the existence of diverse lipid membrane-domain subtypes in neurons.
2. Properties of Caveolin-, Flotillin- or Ganglioside-Containing Lipid Membrane Domains
Within neuronal lipid membrane domains, at least two classes of protein, named caveolin and flotillin, can scaffold cholesterol and have been used as biomarkers of these domains [62,63,64,65,66,67,68]. The differential spatial distribution of the caveolin-, flotillin- or some specific lipid-enriched domains of the neuronal plasma membrane suggests that various domains co-exist in one neuron. We will call them caveolin- and flotillin-enriched lipid membrane domains. Their differential association with plasma membrane receptors acting through calcium signaling, as well as with calcium channels and transport systems might be useful to classify lipid membrane nanodomains. Other lipids, such as gangliosides have been associated with both in certain contexts but not always [69,70,71]. This supports the idea that their presence might constitute a marker for additional lipid membrane nanodomain subtypes. The characterization and differentiation between these domains have been challenged by the limitations and insufficient resolution of the conventional methods for preparative isolation of lipid domains using a whole brain tissue or cells in culture (Figure 1). This is a major handicap for a proper classification of lipid membrane-domain subtypes. A potential dissection through immunohistochemical and immunocytochemical methods could offer insights of their precise intracellular and intercellular locations. Moreover, this dissection could contribute to a better comprehension of how key plasma membrane components in charge of calcium homeostasis are regulated in lipid membrane domains. The subsequent paragraphs of this review provide a brief account of the actual knowledge of these nanodomains in neurons.
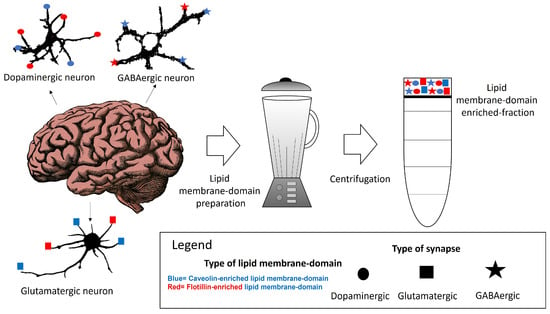
Figure 1.
Some of the potential lipid membrane domains that can be isolated from brain tissue are associated with different neuronal types.
2.1. Caveolin-Enriched Lipid Membrane Domains in Neurons
Cav-1 is the major component forming caveolae at the plasma membrane [27,72,73,74,75]. Several domains are recognized in the linear sequence of this protein related to its function and its interaction with lipids. Membrane binding, cholesterol recognition, and oligomerization functions have been attributed to the scaffolding domain (SD) of Cav-1 [76,77,78]. As part of the SD, a function for the intramembrane domain (IMD; residues 102−134) has been assigned, forming a unique α-helical hairpin that does not traverse the membrane [79,80,81]. Proteins associated with caveolin are characterized by the presence of an aromatic-rich caveolin binding motif (CBM) with the following compositions (ϕXϕXXXXϕ, ϕXXXXϕXXϕ or ϕXϕXXXXϕXXϕ, where ϕ is an aromatic and X an unspecified amino acid) [82,83,84]. Cav-1 also presents a cholesterol recognition/interaction amino acid consensus (CRAC) domain composed of the amino acid residues VTKYWFYR [85], which allows the interaction of this protein with cholesterol. It must be highlighted at this point that the presence of a CRAC domain in proteins is neither necessary nor sufficient for cholesterol binding [86,87]. In this sense, proteins including CRAC domains can be neutral with respect to cholesterol binding, and proteins lacking CRAC domain can bind cholesterol which is the case of transmembrane protein domains lacking a CRAC, a CARC or a tilted domain, as reviewed by Fantini and Barrantes [88]. For this reason, cholesterol interaction with caveolin might be beneficiated by additional interactions with the protein/membrane microenvironment.
Cav-1 (Figure 2, panel a), which is one of the units required for caveolae formation, can hetero- or homo-oligomerize in complexes composed of 14–16 monomers (200–400 kDa) [89,90]. Recently, the typical supramolecular structure of this protein has been described by cryo-electron microscopy [15]. Cav-1 overexpression in E. coli formed 8S-like complexes and oligomerize, forming heterologous caveolae (h-caveolae) and sculpting membranes, which are two of the essential functions of mammalian cells caveolae [91,92]. Cav-1 can assemble in protomers organized into a tightly packed disc with a planar membrane-embedded surface [15]. Several Cav-1 protomers (11 protomers) can oligomerize to form an 8S complex, a type of complex with a proposed biological role essential for caveolae biogenesis since 8S complexes are known to concentrate in endoplasmic reticulum (ER) exit sites [93]. Also, they accumulate at the Golgi, where they lose their diffusional mobility and associate with cholesterol [94,95] and eventually assemble into 70S complexes [93]. The cholesterol-rich membranes containing 70S Cav1 complexes are then transported to the cell surface. The formation of the 8S complex occurs in a cooperative process mediated by its oligomerization domain (OD), which is aided by its SD and signature motif (SM). The crystallography study revealed that the 11 Cav-1 protomers can organize into a disc-shaped complex with a diameter of ~140 Å and a height of ~34 Å to form the 8S complex [15]. The nanodisk contains an outer “rim”, a central β-barrel “hub”, and 11 curved α-helical “spokes” with Cav-1 C-terminal ends oriented towards the hub and N-terminal ends towards the rim (Figure 2, panel b). This study supports that caveolin complexes may stabilize flat membrane surfaces of polyhedral structures rather than imposing continuous membrane curvature [15]. Although this structure is formed in an almost cholesterol-depleted environment, since cholesterol synthesis in E. coli is present in only freshly isolated strains [96,97], this study provides evidence of the structural dependence that caveolae might have on other proteins but also cholesterol in the membranes [98]. Interestingly, the location of the cholesterol interacting domain on the Cav-1 nanodisk surrounding this structure (Figure 2, panel c and d) is compatible with the “lipid belt” model proposed to mediate the interaction between some lipids and proteins, including ion channels, some of them described as Cav-1 protein partners in lipid membrane domains (Figure 2, panel e). This observation suggests that Cav-1 nanodisks may be a part of a lipid belt or a “shell” constituting the immediate perimeter of the protein channel [38,99,100,101], in those channels where no cholesterol interacting domain has been described, complexing and conforming a lipid-protein membrane domain (Figure 2, panel e).
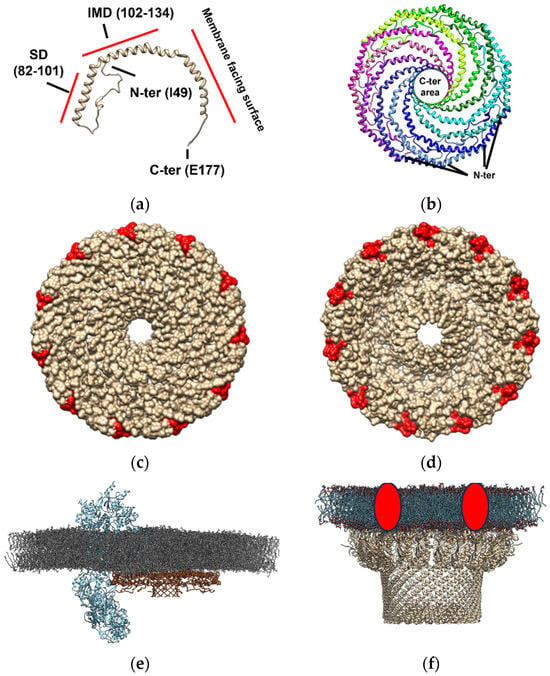
Figure 2.
Molecular architecture of caveolin- and flotillin-enriched domains based on Cav-1 forming nanodisks based on the PDB model 7SC0 and as reported in the bibliography [15] and similar proteins to flotillin constituting SPFH domains. Cav-1 constitutes the singular unit for the formation of these structures. The amino and carboxyl end of the protein are labeled as N- and C-end (panel (a)). The scaffolding domains (SD, residues 82–101) and the intermembrane domain (IMD, residues 102–134) formed are shown in respect to the Cav-1 region that faces the membrane as shown in this panel. Nanodisks as formed by 11 Cav-1 protomers (individually labeled with different colors) which are tightly packed disks locating in planar membrane-embedded surfaces (panel (b)). The location of the C-ends oriented to form a central β-barrel “hub” (~28 Å wide), and N-terminal sides forming an outer “rim” (~23 Å wide) for generation of the nanodisks with a 140 Å diameter in size is shown in this panel. Representation of the membrane-oriented Cav-1 nanodisk surface in respect to cholesterol binding site (labeled in red is shown in panel (c). Representation of the cytoplasmic Cav-1 nanodisk surface in respect to cholesterol binding site (labeled in red is shown in panel (d)). Location of the cholesterol binding site at the periphery of the nanodisk is compatible with the “lipid belt” proposed model for the interaction of some lipids with ion channels suggesting that cholesterol may be a part of a lipid belt or a “shell” constituting the immediate perimeter of the channel protein with could be mediated by complexation with Cav-1 nanodisks [38,99,100,101]. An artistic representation of a Cav-1 nanodisk (PDB: 7SC0, brown-colored backbone) interacting with voltage-dependent L-type calcium channel subunit α-1S (Cav1.1 subunit, PDB: 5GJW, blue-colored backbone) in a model membrane of dipalmitoyl phosphatidylcholine (colored in grey) is shown in panel (e). An artistic representation of the macromolecular structure of a flotillin-enriched domain based on that reported in the literature [102], (using 7VHP PDB model, light-brown-colored backbone) complexing with some proteases that might degrade misfolded/damaged membrane proteins or cytoplasmic proteins (red circles) at the membranes (panel (f)). Hydrophobic tails are represented in blue and polar heads in red, as described in bacterial membrane microdomains [102].
Regarding caveolin-enriched domains in neurons, certain studies have indicated that neuronal lipid membrane domains associated with caveolin are flat and do not have the invaginated appearance described for caveolae [103]. Caveolae curvature has been shown to be dependent upon cavin, and its release from lipid membrane domains has been associated with planar non-invaginated surfaces distinct from caveolae [20,25,26,27]. The crystallographic studies that provide evidence for the existence of macromolecular structures organized into Cav-1 nanodisks suggest that neuronal lipid membrane domains might at least be constituted by this structure, serving as fundamental units responsible for caveolin-enriched domains present in the plasma membrane of neurons.
Although the best-known endocytic route in cells is dependent upon clathrin and independent upon lipid membrane domains [104,105], alternative endocytic routes involving lipid membrane domains mediated by caveolae exist [106,107]. They rely on the protein named dynamin in some cases on Pacsin-2 and are dependent upon cholesterol, as shown by its sensitivity to cholesterol depletion [108,109,110]. They have been involved in the uptake of glycosylphosphatidylinositol (GPI)-anchored proteins (GPI-APs) and opportunistic ligands, including simian virus 40 and cholera toxin (CTx) [111]. Some authors have stated that distinct mechanisms of clathrin-independent endocytosis have unique sphingolipid requirements [112], but in many cases a role has been assigned to caveolin as an initiator of intracellular signaling via protein clustering, the segregation of proteins, and the protein trafficking to and from the membrane-associated with G proteins [113,114]. These processes can generally directly regulate channel permeability for calcium or modulate other components that regulate intracellular calcium concentration through the channel [82,115]. For example, secreted neurotrophins (including brain-derived neurotrophic factor (BDNF) and neurotrophic factors (NT): NT3, NT4, and NT5) can exert prolonged effects on presynaptic transmitter secretion or postsynaptic responses [116]. Neurotrophins binding to their receptors (tyrosine kinase (Trk)-A, Trk-B, Trk-C, etc.) occur in discontinuous regions of neuronal cell membranes associated with membrane lipid membrane domains [117].
Regarding the relevance of caveolin-enriched domains in brain neurons in in vivo studies, some of them have shown a correlation between Cav-1–knocking down (Cav-1–KD) and the disruption of Cav-1-enriched membrane domains found in neurodegenerative diseases, such Alzheimer’s disease where an alteration of signaling processes associated with lipid membrane domains has been also described [118]. Caveolin has also been implicated in synaptic vesicle exocytosis impairment ascribed to changes in synaptic vesicle dynamics driven by Cav-1 palmitoylation using a Cav-1- knock-out animals (Cav-1-KO) [119]. Oppositely, an increase in caveolin expression was found to improve and preserve motor and cognitive function after brain trauma using animal models [120]. These experiments support that Cav-1 levels might enhance cellular survival and growth. Also, some researchers support its role as a candidate for its level modulation to repair the injured and neurodegenerative brain [121,122]. The opposite effect has been observed in some animal models of Huntington’s disease, where a loss or reduction of Cav-1 expression rescues the phenotype in neurons and significantly delays the onset of motor decline and development of neurons. Therefore, aberrant interaction between Huntingtin and Cav-1 leading to altered cholesterol homeostasis in these diseases has been suggested [123].
2.2. Histological and Cytological Distribution of Caveolin-Enriched Lipid Membrane Domains in Neurons and Their Function in Calcium Signaling
Cav-1 has been identified as a component of lipid membrane domains localized within cell bodies and dendrites of primary culture of cerebellar granule neurons and Purkinje cells [33,119,124,125], soma and postsynapses of the anterior cingulate cortex neurons in tissue [126,127], cell body and puncta localized to areas of cellular outgrowth and synapses and dendritic spines of primary culture of hippocampal neurons [128,129,130].
A study has shown that Cav-1 partially colocalize with the N-methyl D-aspartate receptor subtype 2B (NR2B) subunit of the N-methyl-D-aspartic acid receptor (NMDAR), which is highly enriched in dendritic shafts and spines of rat cortical neurons at postsynaptic terminals [131,132]. NMDARs are glutamate-gated ion channels that mediate excitatory neurotransmission in the central nervous system (CNS) [133]. The presence of NMDARs at presynapses or postsynapses has a different function. In the case of presynapses, NMDA receptors have a function in neurotransmission and plasticity [134], and postsynaptic receptors are needed for spike-timing-dependent long-term depression (LTD) induction [135]. A study of Cav-1 overexpression in neurons showed that Cav-1 mediated expression of NMDAR subtypes promoting pathways dependent upon the membrane cholesterol associated with primary neuron arborization events [121]. Two regions on NR2B subunits (W635AFFAVIF642, and, F1042SFKSDRY1049) have been potentially suggested to interact with caveolin-binding motifs [84,132]. A disruption of the interaction between Cav-1 and NR2B has anti-nociceptive effects at the anterior cingulate cortex [126], which correlate with the observed effect of pain agonists promote a shift of the NR2B subunits of NMDA receptor subunit to non-lipid membrane-domain areas [132]. Also, an increased amount of caveolin promotes an enhanced surface level of NR2B in this brain area [126], which leads to an increase in cytosolic calcium concentration and activation of extracellular-signal-regulated kinase/cAMP response element (ERK/CREB) signaling pathways [136]. Thus, decreased caveolin expression in cells disrupts NMDAR signaling events, and reintroducing Cav-1 rescues proper NMDAR signaling. Since NR2B contains the binding site for glutamate [137], this suggests that caveolin is required for the signal transduction pathway activated by glutamate release from the presynaptic terminals [132]. It has been suggested that the regulation of the NR2B subunit by Cav-1 might be attributed to the modulation of proto-oncogene tyrosine-protein kinase (Src) activity since Cav-1 was observed to be essential for NMDA-mediated phosphorylation of Src and ERK1/2 activation [132], which is required for NMDA-mediated signaling (i.e.,: NMDA preconditioning stimuli) [121,132,138].
Src family tyrosine kinases (SFKs) serve as central regulators for the modulation of NMDAR signaling in normal and ischemic conditions and the induction of long-term potentiation (LTP) [139,140,141]. This modulation accounts for SFK-mediated tyrosine phosphorylation of NR2B, a subunit found highly phosphorylated in postsynaptic terminals [140,141]. Head and collaborators proposed that Cav-1, via its ability to scaffold key signaling components, mediates in the NMDAR localization to neuronal membrane domains, NMDAR/Src tyrosine kinase family/ERK signaling, and protection of neurons from ischemic injury and cell death [132]. Cav-1 promotes NR2B surface levels and has been shown to contribute to the modulation of chronic neuropathic pain in the anterior cingulate cortex [126].Cellular stress events (i.e., superoxide anion radical, osmotic stress, and UV exposure) can increase SFK-mediated phosphorylation of caveolin [142]. In addition, some studies early reported the existence of a negative regulatory feedback loop in non-neuronal cells in which Y14 phosphorylated Cav-1, would bind and activates C-terminal Src kinase (Csk) and subsequently phosphorylates and inactivates Src [143,144,145,146]. In neurons, the regulatory role of Cav-1 phosphorylation/dephosphorylation by Src/Csk has been shown to mediate axonal outgrowth of motor neurons in Xenopus neuromuscular development [147]. Regarding the regulation of the system by oxidative stress, it should be noted that indeed it can activate both Src-kinases and their negative regulator Csk and induces phosphorylation of Cav-1 as a targeting protein for Csk [145]. These results suggest that caveolin could mediate in events mediated by NMDAR, such as those associated with neuronal plasticity and injury that might be associated with oxidative stress [132,148], by regulation of its level of phosphorylation.
Presynaptic NMDA receptors play pivotal roles in excitatory neurotransmission, contributing to synaptic plasticity and facilitating presynaptic neurotransmitter release, functions that are crucial for synaptic maturation and plasticity during formative periods of brain development [134,149,150]. It has been reported that presynaptic NMDA receptors might modulate superoxide anion production by NADPH oxidases (NOXs) [151]. In turn, NMDA receptors may be modulated by superoxide anion by a similar mechanism in postsynapses [142], and locations where NR2B subunits have been found at presynapses as the cerebellum [152] and neocortex [153]. In this case, modulation by superoxide anion might be associated with superoxide anion producing enzymes of very specific sources, also clustering within lipid membrane domains [32]. Flavoproteins, such as the enzyme cytochrome b5 reductase (Cb5R), have been established to form complexes within plasma membrane lipid membrane domains of cerebellar granule neurons, as those described by our laboratory [28]. Cb5R is one of the major sources of superoxide anion in the plasma membrane lipid membrane domains of cerebellar granule neurons [29,31]. This protein holds the potential to facilitate certain superoxide anion-dependent adjustments of the NMDA receptor at the presynaptic terminals. The existence of these proteins associated with caveolin [33], might constitute an alternative form of caveolin-enriched lipid membrane-domain subtype in respect to those previously commented.
Both Cb5R and neuronal nitric oxide synthases (nNOS), as alternative redox flavoproteins located within the neuronal plasma membrane lipid membrane domains, have been proposed to form complexes associated with caveolin-enriched domains [32,33]. These complexes have been postulated to function as redox nanotransducers, in charge of controlling calcium transporters such as the L-type calcium channels and NMDA receptors. These microchip-like structure have been proposed to tightly orchestrate coupling between calcium and nitric oxide signaling in presynapses of glutamatergic cerebellar granule neurons (CGNs) [32]. The co-localization of these components agrees with the suggested effect of glutamate on the activation of NMDA receptors in neuronal terminals containing nNOS, leading to nitric oxide (NO•) formation and amplifying neurotransmitter release, a mechanism early hypothesized by Snyder and Dawson [154]. These specialized domains can promote a localized and transient increase in calcium concentration up to 1 µM within a nearby microcompartments of 100 nm with low calcium buffering capacity [32]. nNOS is inactive at low calcium concentrations, but it active when calcium concentration is high enough to afford a significant saturation of calmodulin (EC50 ≈ 0.2–0.4 μM). The mechanism by which nNOS is regulated by caveolin remains unknown. The modulation of nNOS activity by Cav-1 seems to be distinct from the one observed to regulate endothelial NOS [155].
In hippocampal and cortical neuron cultures, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) has been associated with caveolin-enriched lipid membrane domains [156]. The AMPAR present in lipid membrane domains is regulated by the activity of NMDAR and NO•-mediated pathways [129,156]. This regulation might be potentially interconnected with redox nanotransducers described above in adjacent domains observed in cerebellar granule neurons, particularly in presynaptic membranes [32]. NO• has a mimicking similar effect to that of NMDA, leading to the recruitment of AMPARs to the surface since lipid membrane domains are required for receptor insertion into the membrane [156]. Cholesterol depletion leads to instability of surface AMPAR, a gradual loss of synapses (both inhibitory and excitatory), and loss of dendritic spines [129].
Metabotropic glutamate receptors (mGluRs) are responsible for so-called slow synaptic transmission associated with the effects of peptide neurotransmitters and non-peptide neuromodulators [157,158]. Metabotropic receptors are G-protein-associated receptors enriched at excitatory synapses [159,160]. There are eight subtypes of mGluRs classified in presynapses as group I mGluRs (formed by mGluR1/mGluR5 subtypes) selectively activated by 3,5-dihydroxyphenylglycine and coupled to inositol phospholipid hydrolysis, group II mGluRs (formed by GluR2/mGluR3 subtypes) and group III mGluRs (formed by mGluR4/mGluR6/mGluR7/mGluR8 subtypes) [161,162,163]. Association of metabotropic mGluR with caveolin has been shown for group I and II [159,164,165], which might have a wide different location depending on the receptor type [166]. These group I of metabotropic glutamate receptors are modulated by Cav-1 [128] through the caveolin binding motif of the mGlu1 receptor (FVTLIFVLY-ϕXXXXϕXXϕ). Cav-1 interacts with mGluR1 through a motif contained within the last segment of the first transmembrane (TM) domain and the first intracellular loop of the receptor [159]. A second putative Cav-1 binding motif contained within i3 and the first segment of TM6 is also present in mGluR1/5 [159]. Localization of mGluR1/5 in lipid-protein membrane domains is promoted by Cav-1, which controls the rate of constitutive mGluR1 internalization and, therefore, regulates the expression of the receptor at the cell surface [128,159]. Indeed, the control of constitutive mGluR internalization rate and the surface level of mGlu1 has been shown to be dependent upon caveolar/lipid membrane-domain-dependent endocytosis associated with Cav-1 [159]. In addition, activation of other mGluR induced through complex to estrogen receptor subunits has been associated with different caveolin isoforms, including Cav-3 expression, in different brain areas: striatum, the estrogen receptor (ER) alpha (Erα)/Cav-1/mGLUR5/Gq GTPases (Gq) complex and the ERα or ERβ/Cav-3/mGLUR3/Gi/o proteins complex; hippocampus, ERα/Cav-1/mGLUR1a/Gq complex and the and the ERα or ERβ/Cav3/mGLUR2/Gi/o complex; the arcuate nucleus, ERα/Cav-1/mGLUR1a/Gq; astrocytes (hypothalamus) ERα/Cav-1/mGLUR1a/Gq; dorsal root ganglion neurons, ERα/Cav3/mGLUR2/Gi/o [167].
A G-protein-dependent intracellular calcium release by activation of phospholipase C (PLC), inositol-3-phosphate (IP3) pathway, and the transient receptor potential canonical channel (TRPC) are components associated with the group I of metabotropic receptors. These proteins are all present in lipid membrane domains [168,169,170,171]. Using dihydroxyphenylglycerol, an agonist of the group I mGluR, an increase in the mGluR1α clustering level to phosphorylated caveolin was found [172]. Other studies have shown that, the interaction between Cav-1 and group I mGluRs regulates mGluR-dependent phosphorylation/activation of MAPKs [159]. Lipid membrane-domain disruption with methyl-β-cyclodextrin induced a block in the agonist-dependent mGluR1α internalization, being the implication of caveolin suggested in synaptic plasticity in the cerebellum [173].
L-type calcium channels are known to regulate synaptic activity, contributing to the initiation of endosome recycling, which regulates the abundance of synaptic molecules such as AMPA-type glutamate receptors in neuronal dendrites [174]. This function might support the existence of L-type calcium channels associated with caveolin-enriched domain as a lipid membrane nanodomain subtype located at postsynaptic membranes [174]. Some subunits of the L-type calcium channel, such as A2δ-2 subunits, colocalize with proteins binding to gangliosides in alternative lipid membrane-domain structures to those described associated with caveolin [175]. Although non-invaginated caveolar structures have been suggested to exist in neurons, internalization of neurotrophins activated tyrosine kinases receptors (TrkA) [176] and TrkB [118], at growth cones might be dependent upon caveolin-associated endocytosis [177,178]. L-type calcium channels are also very sensitive to oxidative stress, as reported by the NMDA receptor, but in this case, by direct effect since these complexes present an allosteric thiol-containing “redox switch” that controls the activity of the L-type calcium channel [179].
Regulation of N-type calcium channel by Cav-1 has been observed in caveolin-enriched lipid membrane domains of neuroblastoma NG108-15 cell lines [180]. Downregulation of Cav-1 production in these cells induced a 79% reduction in the N-type current density without significant changes in the channel’s activation and inactivation time course. The regulation of the channel by membrane cholesterol associated with caveolin was observed to be responsive to this effect rather than induced by direct modulation by caveolin [180]. A similar modulatory effect was observed for R-type voltage calcium channels and neurokinin receptors using kidney cell lines, where cholesterol was responsible for its modulation since intracellular diffusion of Cav-1 scaffolding peptide or overexpression of Cav-1 unaffected the channel function [180].
Localization of the PMCA has also been found at caveolin-enriched lipid membrane domains [33]. The cerebellar synaptosome isoform 4 of the PMCA was specifically localized in this domain with respect to other isoforms locating at non-lipid membrane domains [181]. Some studies show the stimulation of PMCA by acidic phospholipids such as phosphatidylserine [182]. This lipid is normally located at the inner leaflet of plasma membranes and enriched in caveolin-enriched domains in non-neuronal cells [183]. Phosphatidylserine externalization is typical of cell death processes associated with apoptosis [184], and this event might modulate PMCA activity and the interaction this lipid [185].
Purinergic receptors (P2X) have been associated with Cav-1-enriched lipid membrane domains [186,187,188]. Cooperatively, CaMKIIα and Cav-1 drive ATP-induced membrane delivery of the P2X3 receptor as reported in dorsal root ganglion neurons [187]. The NH2-terminus of the P2X3 receptor was identified to interact with caveolin through the ‘T12KSVVVKSWTI22’ motif and the extended motif ‘F6FTYETTKSVVVKSWTI22’ was engaged to CaMKIIα binding [187]. P2X3 receptors are associated with calcium influxes, which further activate the calcium/calmodulin-dependent protein kinase IIa (CaMKIIa), and are primarily expressed in primary sensory neurons located in dorsal root ganglion (DRG) responsible for pain [189,190]. Upon receptor phosphorylation, an increase in P2X3 interaction with Cav-1 has been observed, providing a mechanism for P2X3 receptor sensitization in pain development [187]. It is particularly noteworthy that immunoreactivity of P2X3 in the plasma membrane was not decreased by the cholesterol depletion with methyl-β-cyclodextrin and cholesterol sequestering had no effect on P2X3- or P2X2/3-mediated inward currents [191]. This result support that the P2X3 receptor may be diffusely distributed in lipid membrane domains and in non-lipid membrane domains in primary sensory neurons [191].
2.3. Flotillin and Neuronal Lipid Membrane Domains
Domains formed by flotillin in the plasma membrane differ from those in which Cav-1 is present. Furthermore, they are dynamic and bud into the cell [192]. The main protein components of these domains are the flotillin isoforms, Flot-1 and Flot-2, which share 50% sequence identity [193]. They are in charge of membrane curvature induction in non-neuronal cells, the formation of plasma-membrane invaginations morphologically similar to caveolae, and the accumulation of intracellular vesicles [192]. Early studies suggested flotillin proteins organization into stable tetramers in membrane microdomains [194]. Some studies suggested the possible role of flotillin as a new marker of caveolae [194], and subsequent studies have shown that flotillin and caveolin do not always co-localize [56]. Nevertheless, it cannot be discarded that a certain amount of flotillin could be enriched at caveolae [195]. An estimation of the size dimension of flotillin-enriched lipid membrane domains by immunolabelling suggests the formation of patches ranging 40–200 nm in neurons [196]. These studies correlate with a description of flotillin protein complexes as part of a family of proteins named SPFH (stomatin, prohibitin, flotillin, and HflK/C) forming an operon with NfeD proteins [197]. The ancient origin of SPFH-domain proteins and the Nodulation efficiency protein D (NfeD) protein and the stomatin operon partner protein (STOPP) can be traced back to the ancient living cells that diverged and evolved to Archaea and Bacteria to constitute the main binding region of apolar polyisoprenoids as well as cholesterol, contributing to lipid membrane-domain formation [197].
SPFH are proteins enriched in the plasma membranes and also in other subcellular membranes, of prokaryotic and eukaryotic cells [111,198]. Electron microscopy studies have shown a wide distribution of Flot-1 in cells localizing at the cytoplasmic side of the plasma membrane, the cytoplasmic side of primary and secondary lysosome membrane, lipofuscin, multivesicular bodies, Golgi saccules, the cytoplasmic leaflet of the vesicles associated with Golgi apparatus and the lumen side of ER of neuronal cells of rat brain [196]. They have an SPFH domain in common in their structure formed by an N-terminal hydrophobic region that associates proteins to the membrane [111,198]. Flotillin isoforms contain a conserved domain C-terminal to the SPFH domain, called the ‘flotillin domain’, although is not present in the other SPFH domain-containing proteins [193]. SPFHs can form high ordered structures complexes organized as circular structures comprising homo- or hetero-oligomers [102,199,200]. Several structural membrane microdomain organizations by SPFH family proteins have been proposed [102] (Figure 2, panel f). In flotillin structure, two domains with unclear functions have been shown to be present. The first SPFH domain contains sites for acylation [201,202]. In contrast, the C-terminal domain mediates the oligomerization and contains Ala-Glu repeats and phosphorable Tyr residues [203,204,205], which are important for flotillin function.
In brain, anatomical and physiological studies have shown that Flot-1 enhances the formation of glutamatergic synapses but not GABAergic synapses, and it has been suggested that this protein might have a role in neurodevelopmental disorders and axon regeneration and growth [206]. Flotillin is recognized as essential for growth cone elongation and regeneration in retinal ganglion cells and mouse hippocampal neurons [207,208]. Notably, when flotillin isoforms are downregulated, and the signaling pathways that govern actin dynamics are disrupted, axon formation fails to occur [209].
Some studies have demonstrated that flotillin directly regulates the formation of cadherin complexes [210,211]. Flotillin-enriched domains have been observed to be required for the dynamic association, stabilization of cadherins at cell–cell junctions [212], transducing extracellular signals into intracellular signaling events, and modulating alterations in the cytoskeleton in response to various external stimuli [213], signal transduction of Trk receptors, and participates in cellular trafficking pathways [214]. However, the molecular mechanism of action of this protein in these processes is not well understood [215].
It is known that Flot-1 acylation determines this protein traffic from the endoplasmic reticulum toward the plasma membrane [210]. Palmitoylated Flot-1 efflux from the endoplasmic reticulum also mediates Cav-1 traffic to the plasma membrane, avoiding the endoplasmic reticulum stress by inhibiting the synthesis of Cav-1 [210]. Once Flot-1 reaches the plasma membrane, it hetero-oligomerizes with Flot-2 and undergoes depalmitoylation/repalmitoylation, which evokes prolonged insulin-like growth factor-1 (IGF-1) signaling [210]. Recently, a role of Flot-1 in mediating the membrane expression and cellular responses of the transient receptor potential vanilloid type 2 (TRPV2) has been described in primary neuronal culture of dorsal root ganglion [216]. This suggests a crosstalk between TRPV2 and lipid membrane-domain components may influence the cellular morphology and play critical roles in nociception and pain [216]. Also, flotillin depalmitoylation has been linked to receptor cycling between the plasma membrane and endosomes alone or with Flot-2 [210].
Although palmitoylation/palmitoylation of flotillins regulate this protein location into lipid membrane domains, the regulatory role of palmitoylation is not exclusive for this protein. Cav-1 can be palmitoylated on multiple cysteine residues although palmitoylation is not necessary for localization of caveolin to caveolae [217]. Palmitoylated Cav-1 has been involved in signaling molecules assembly in plasma membrane caveolae and in intracellular cholesterol transport [218]. Also, cav-1 palmiltoylation for example, can regulate synaptic vesicle dynamics events [119], which are processes associated with SNARE machinery [219] linked with different plasma membrane domains [220]. Some of the proteins constituting the SNARE complexes might eventually be associated with lipid membrane domains [221,222]. Therefore, this process should not be directly associated with lipid membrane domains [223].
Glebov and collaborators have suggested flotillin participation in a third endocytosis pathway different from those described for clathrin and caveolin [224]. Flot-1 can colocalize in endosomes with the fluid-phase marker dextran, the glycosylphosphatidylinositol-anchored CD59 (GPI-AP CD59), and CTx and is required for a dynamin-independent endocytic pathway that mediates receptor-independent fluid-phase endocytosis and these markers [224]. This supports that gangliosides colocalization might be used to track endocytosis processes, as also suggested for caveolin-enriched lipid membrane domains. In neurons, flotillin was initially discovered in caveolin-independent cholesterol- and glycosphingolipid-enriched membrane microdomains expressed during axon regeneration [212].
2.4. Histological Cytological Distribution of Flotillin-Enriched Lipid Membrane Domains in Neurons and Function Calcium Signaling
Flotillin isoforms have been widely used as a lipid membrane-domain biomarker. Flotillin isoforms have been observed to colocalize with calcium channel α1 subunit CaV2.1, which are subunits of P/Q type calcium channels located presynaptic areas of the brain [175,225], GPI-enriched areas [226] and small uniform puncta of pre and postsynapse of hippocampal neurons [206,227], soma and postsynapses of rat cerebral cortex [127]. Flotillin-enriched lipid membrane domains are abundant in the axonal plasma membrane and are found in less amount in somatodendritic membranes [228]. This correlated with electrophysiological results using whole-cell patch clamp, showing that Flot-1 increases in the frequency of miniature excitatory postsynaptic currents but not miniature inhibitory postsynaptic currents. In contrast, amplitude and decay kinetics of either type of synaptic current were unaffected, linking these domains with calcium homeostasis [206].
One-third of the NMDAR clusters with flotillin in cultured hippocampal neurons [227]. In hippocampal neurons, both NR2A and NR2B subunits of NMDARs interact with Flot-1 [227]. Flot-1 has been associated with the NR1 subunit preferentially at synaptic areas rather than non-synaptic NR1-enriched areas of hippocampal neurons [206]. It has been suggested that NMDAR interaction with flotillin is involved in recruiting NMDARs into lipid membrane domains to initiate second messenger signaling cascades linked with receptor depletion for neuronal protection during NMDAR-induced excitotoxicity [229]. Indeed, some lipoprotein receptor involved in cholesterol traffic from astrocytes to neurons, such as low-density lipoprotein receptor-related protein 1 (LRP1) [230,231,232], has been suggested to influence the composition of postsynaptic protein complexes through NMDA-induced degradation of the postsynaptic density protein 95 (PSD-95) [233], which might link this process with cholesterol homeostasis and regulation of lipid membrane domains enriched on PSD-95. NMDARs can associate with scaffold protein PSD-95 and form signaling complexes that differ in composition depending on whether they are found in the postsynaptic density or the presynaptic lipid membrane domains. Recently, enhancement in the formation of glutamatergic synapses but not gamma-aminobutyric acid-dependent (GABAergic) synapses has been observed by modulation of Flot-1 level, which suggests further exploration of Flot-1 effect in neurodevelopmental disorders [206]. The authors have postulated that flotillin might have a role in the endocytic internalization of the NMDA receptors after high neuronal stimulation, thereby implicating a subtype of flotillin-enriched domain in the modulation of this process [227]. Flot-1 acylation determines this protein traffic from the endoplasmic reticulum toward the plasma membrane and supports the idea that these domains might be involved with the trafficking of these receptors toward the membrane [168].
Flot-1 and Flot-2 are associated with Ras-binding family of small GTPase 11A (Rab11A) and sorting nexin 4 (SNX4) binding proteins that participates in the recycling and co-transportation of PSD-95, N-cadherin, the glutamate receptors GluA1 and GluN1 to be delivered to the postsynaptic membrane in spines of hippocampal neurons [234]. The mechanism of action remains to be determined [234].
The Cav 2.1 subunit (also known as α (1A) subunit) is a component of the P- and Q-type calcium channels [235], which have different locations and properties than the L-type calcium channels associated with caveolin domains. The α2δ-2 subunit of P- and Q-type calcium channels [236,237,238], partitions with Cav2.1 subunit into flotillin-enriched lipid domains isolated from the cerebellum [175].
PMCA has also been found in isolated flotillin-enriched lipid membrane domains from dissociated cortical and hippocampal primary neurons in culture, and its activity has been affected by cholesterol depletion [181,239]. The PMCA activity in these domains has been described to be higher than the PMCA activity excluded from these microdomains [240]. The activity decreased when cholesterol was depleted from these domains [240].
2.5. Gangliosides as a Lipid Membrane-Domain Biomarkers for Some Caveolin- and Flotillin-Enriched Lipid Membrane Domains
The presence of gangliosides has been observed in both caveolin- and flotillin-enriched lipid membrane domains [241,242,243], although they are not specifically localized at the plasma membrane and their properties are not exclusively dependent on their polar head group [244]. This type of lipid is strongly abundant in the brain, i.e., in cerebellar granule neurons, they are 5% of total amphipathic lipids [245]. The resulting ganglioside-driven membrane organization are reliant on its production pattern, which is tightly regulated [244]. Not all gangliosides colocalize at the same type of plasma membrane domains [246]. Some authors have concluded that proteins binding to plasma membrane gangliosides can be divided into host plasma membrane proteins and extracellular proteins [247]. Some gangliosides such as GM1 are known to be particularly enriched in the outer leaflet of neuronal lipid membrane domains and exhibit a nearly exclusive presence within these domains compared to non-lipid membrane domains regions. The lipid membrane domain/non-lipid membrane domain ratio values range from 10 to 1000 [248]. Recent molecular dynamics simulation data have shown that three different subpopulations of gangliosides such as GM1 can be characterized in the same lipid membrane domain [14,249], distributed into the central, peripheric and edge areas, which defines their mobility from less to high [247]. Gangliosides at the edge adopt the typical chalice or butterfly-like (open wings) dimeric conformation [250], although conformational possibilities might be further extended by the biochemical diversity of gangliosides. Ganglioside concentration in the same lipid membrane domain creates a large negative electrostatic surface potential, which is one of the essential properties of lipid membrane domains for protein, toxin, or pathogenic agents easily binding due to the electropositive potential [247].
Two types of gangliosides binding domains (GBD) have been described in proteins present in lipid membrane domains:
- -
- Type 1 GBD, or GBD-1, comprises any membrane protein ganglioside-binding domain able to form a stoichiometric (1:1, mol:mol) complex with a single ganglioside molecule [247]. GBD-1 is generally present at the flexible juxta membrane region interacting with transmembrane glycoproteins [113]. The serotonin 5-HT1A receptor, the tumor stem cell marker CD133 are candidates the EGF and PDGF receptors and ion transporters [247]. These membrane proteins are expected to reside at the edge of a lipid raft.
- -
- Type 2 GBD, or GBD-2 are represented by protein dimeric structures resembling a flower chalice or the open wings of a butterfly [250,251]. The typical protein insertion processes have been associated with these domains in which proteins with a hairpin loop interact with the ganglioside, leading to a conformational change that implicates a deep interaction with the ganglioside [251]. This type of ganglioside-dependent insertion process accounts at the edge of a lipid raft or at the periphery since they need to have sufficient conformational flexibility to accommodate the loop [251]. Chalice-shaped ganglioside dimers are required for HIV fusion with host cell membranes [247,252] and the formation of oligomeric calcium permeable amyloid pores [247,253].
In this organization, it is unclear which proteins present in flotillin and caveolin-enriched domains, and more specifically in the brain, might contain GBDs. Cav-1 and Flot 1 have been shown to colocalize with 5-hydroxytryptamine receptor (5-HT1A) [254], and CD133 colocalize with Cav-1-enriched lipid membrane domains [255], which present GBD-1. Caveolin, but no flotillin [256], has been associated with HIV infection and latency [257,258], and this might correlate with the presence of HIV proteins associated with GBD-2 domain. Increased GM1 concentrations have been found in cerebrospinal fluid ganglioside, indicating neuronal involvement in all stages of HIV-1 infection [259].
2.6. Histological Cytological Distribution of Gangliosides-Enriched Lipid Membrane Domains in Neurons and Function Calcium Signaling
Regarding calcium transport systems, gangliosides are well-known modulators of calcium homeostasis [260]. PMCA2 and 3 are known to be regulated by endogenous ganglioside content, such as the asialoGM1 that promotes a decrease in pump activity [261,262]. This correlates with the identification of PMCA location in caveolin-enriched lipid membrane domains in cerebellar granule neurons by Marques-da-Silva and Gutiérrez-Merino [33]. The highest PMCA activity is present in the lipid membrane domains enriched in cholesterol and gangliosides [263], which correlates with a report showing that neuraminidase treatment and D-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (d-PDMP), a used inhibitor of glycosphingolipid biosynthesis, induce a decrease in PMCA activity [261]. However, the mechanism of PMCA inhibition by GM1 is still under discussion. Some researchers have suggested that GM1 affects the PMCA interaction via calmodulin modulation of calcium pump affinity and the Vmax [262]. This contrasts with the suggestion of modulation based on the interaction with the calmodulin-binding domain stimulating the phosphatase activity of PMCA by stabilizing E(2) conformer [264,265]. Total lipid membrane domains associated PMCA activity is higher than the PMCA activity excluded from lipid membrane-microdomains [240]. Depletion of cellular cholesterol dramatically inhibited the activity of the lipid membrane-domain-associated PMCA with no effect on the activity of the non-lipid membrane-domain pool [240]. This modulatory function of gangliosides contrasts with that inducing activation of L-type calcium channels, as shown in N18 neuroblastoma cells by the same gangliosides [266].
An almost complete colocalization of NMDARs with the lipid membrane-domain marker ganglioside GM1 has been found in postsynaptic densities close to GM1 [267]. GM1 has been shown to reduce the neurotoxicity of NMDAR, which suggests that receptors located at this location might differentially response to glutamate in this location. However, GM1 does not suppress the function of the NMDAR channel directly [268,269,270]. This protection might be associated with endocytic internalization of the NMDA receptors associated with flotillin-enriched lipid membrane domains, as indicate above [227].
By electron microscopy, a subpopulation of synaptic membrane fractions has been found to be enriched in GM1, and 46 percent of the labeled vesicles are also labeled the GluR2 subunit of the AMPAR [271]. SFKs has been associated with gangliosides and caveolin-enriched lipid membrane domains [272]. They are important since they also mediate the phosphorylation of the AMPARs [273], and they can mediate GluA2-binding protein exchange through endocytosis of GluA2-containing synaptic AMPARs [60]. This might constitute an additional subtype of lipid membrane domains enriched in gangliosides and implicated in endocytic processes or the same associated with Src and NMDA receptors at excitatory synapses. Location studies suggest that AMPAR within PSD are segregated from NMDA receptor clusters [274,275]. In addition, a study has shown that GM1-bound to GluR2-containing AMPARs are functionally segregated from the AMPAR-trafficking complexes (ATCs) containing Thorase, n-ethylmaleimide-sensitive factor attachment protein gamma (γ-SNAP), N-ethylmaleimide sensitive fusion protein (NSF), and nicalin bind selectively to trisialoganglioside gt1b (GT1b) [276], which could define alternative AMPAR domains at the plasma membrane.
GM1 modulation of calcium channels was first described in neurons using N18 neuroblastoma cells [266,277,278] and primary neurons [279,280]. Studies with N18 cells showed that GM1 blocked the intracellular calcium increase sensitive to dihydropyridine blockers at a concentration of 5 mM [266], proposing GM1 function as a constitutive inhibitor of L-type calcium channels [260]. GM1 functions as neuritogenic molecules in neuronal differentiation phases [278]. Upregulation of this lipid has been found in the plasma and nuclear membranes during axonogenesis [278]. In the presence of neuraminidase (N’ase), an enzyme that increases the cell surface content of GM1, a prolific outgrowth of neurites has been found in Neuro-2a and NG108-15 cells [278]. This effect can be blocked by the cholera toxin B, a biochemical tool extensively used for labeling lipid membrane domains using fluorescent conjugates, which potentiated the effect of N’ase in NG108-15 cells [278].
Although cholera toxin binding to ganglioside GM1 supports that this regulation is mediated by lowering free GM1 concentration in the plasma membrane, it remains to be known whether cholera toxin can be sequestered the GM1 localizing in lipid membrane domains, which might modulate the L-type calcium channels associated with these domains. Neurite outgrowth correlated with the influx of extracellular calcium, which correlates with the reported modulation of calcium channels by gangliosides [260].
Using synaptosomes, the N-type calcium channels has also been found to be activated by GM1 ganglioside, followed by the P-type, and very weakly influencing other channels in cerebrocortical synapses [281]. Based on previous indications showing gangliosides with association with caveolin- and flotillin-enriched lipid membrane domains, it is not clear if calcium transporter elements modulated by this lipid might constitute a population implicated in the endocytic process or just be simply subjected to endocytosis.
3. The Summary of the Distribution Map
A wide range of possible complexes enriched in lipid membrane nanodomain subtypes in the same or different glutamatergic neurons has been described. The organization of NMDAR, L-P/Q calcium channels, some metabotropic receptors, and PMCA located in the synapses of glutamatergic neurons are shown in Figure 3.
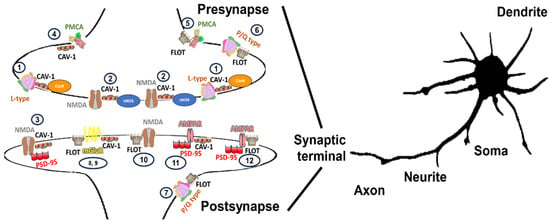
Figure 3.
Illustration of a variety of caveolin- and flotillin-enriched lipid membrane domains location complexing with calcium transporter elements (NMDAR, L-P/Q calcium channels, some metabotropic receptors, and PMCA) in synaptic terminals described to exist in glutamatergic neuronal cells. Calcium transporter elements have been differentially described to be present in many neuronal locations, including somas, neurites, axons, dendrites, spines, and synaptic terminals. In synaptic terminals a variability of subunits may yield specific calcium transporters for that location (i.e.,: presynaptic and postsynaptic NMDAR might be differentiated by the type of subunits that configure them in hippocampal neurons [282]) that might differ in configuration from those distributed in other neuronal locations and vary in respect to the neuronal cell type [135]. In this figure, we are focusing on calcium-transporting elements associated with caveolin- and flotillin-enriched lipid membrane domains, that should be added to those elements that are not located in lipid membrane-domain areas (not shown in this figure) and omitted in synaptic terminals that comprise areas of 0.5 to 2 μm size [283]. Lipid membrane domains associated with gangliosides are suggested to be involved in endocytic processes in some membranes and have been omitted from this figure for the sake of clarity. NMDA (1) and L-type calcium channels (2) located in caveolin-enriched domains might function as redox nanotransducers in charge of the control of these calcium transporters working as a microchip-like structure for a tighter functional coupling between calcium, nitric oxide and superoxide anion signaling in presynapses [32,154,155] and postsynapses (3) also sensitive to superoxide anion [132,142], in glutamatergic CGNs. Also, in associated caveolin-enriched domains at presynapses, we can allocate PMCA (4), which are susceptible of inhibition by GM1 contained in these subdomains [33,261,262]. PMCA has also been described to be present in flotillin-enriched lipid membrane domains (5), which are very sensitive to cholesterol content. The activity in these domains is higher than the one not present in lipid membrane domains [240]. The differential response to endogenous cholesterol and gangliosides seems to support that caveolin and flotillin-enriches domains constitute different lipid membrane nanodomain subtypes in presynaptic terminals. We can also find P/Q-type calcium channels at presynaptic terminals (6) associated with flotillin-enriched domains and GPI-enriched areas [175,225,226]. This type of cluster can also be found in postsynaptic terminals (7) [206], and the physiological behavior has been characterized by the presence of Flot-1 and has been related to and increases in the frequency of miniature excitatory postsynaptic currents. Several subunits of metabotropic receptors have been described to colocalize in caveolin- and flotillin-enriched domains (8) and (9). Subunit interaction with caveolin has been better described than for flotillin. Several motifs of mGlu subunits have been described to interact with cav-1 [128,159], which controls the rate of receptor internalization and location at the surface [128,159]. A function of recruitment of NMDARs into lipid membrane domains at postsynapses to initiate second messenger signaling cascades linked with receptor depletion for neuronal protection in NMDAR-induced excitotoxicity has been suggested for NMDAR located at flotillin-enriched domains (10) [212]. As previously indicated, NMDARs can associate with scaffold protein PSD-95 and form signaling complexes that differ in their composition. Some subunits of the AMPAR have also been located in caveolin- (11) and flotillin-enriched domains (12) at post synaptic terminals associated with PSD-95. NO• has a similar effect mimicking that of NMDA, recruiting AMPARs to lipid membrane-domain surface which suggest a counterplay with lipid membrane domains associated with postsynaptic domains (3) or presynaptic (1) and (2) domains since NO• can reach this location by diffusion from presynaptic sources.
A summary of the components implicated in calcium signaling in neurons and their association and function with each lipid membrane-domain subtype can be found in the Table 1.

Table 1.
Calcium signaling components and distribution map in lipid raft-domain subtypes.
4. Conclusions
Future work should further elucidate the relationship between caveolin- and flotillin-enriched domains and the proteins and lipid partners present in each type of platform that, as shown in this review, may form different lipid membrane-domain subtypes. This includes the effect of cholesterol in calcium signaling and the potential modulation of elements in charge, including calcium channels, that might differentially interact with this lipid in neurons, concerning the same population of protein that might present in non- lipid membrane-domain areas. The cumulative experimental evidence analyzed in this review suggest that lipid membrane-domain subtypes are likely to exist in neurons, largely based on the well-known location and distribution of calcium transporter elements differentially interacting with caveolin- and flotillin-enriched domains.
There is a need for a better characterization of the molecular components of different lipid membrane-domain subtypes in different types of neurons, and of the role of protein-protein and protein-lipid interactions in the functional modulation of the components of these domains. One of the open questions to be answered is associated with the role of cholesterol and its effects, induced by direct interaction with proteins or by changes in the physical-chemical properties of the membranes. Cholesterol enantiomers are potential tools that might help to answer this question since they have identical physical properties to cholesterol but opposite three-dimensional configurations compared to cholesterol [300]. An additional question that needs to be addressed in the future concerns the presence of proteins such as cavin that are present in caveolae of non-neuronal cells and seem to be required for the plasma membrane curvature. Neuronal plasma membranes are non-invaginated, suggesting that cavin is not present in these structures, but it should not be discarded the presence of other Cav-1 homologous partners at the neuronal lipid membrane domains that have been determined to be present in non-neuronal cells such as caveolin-2 (Cav-2). Cav-2 is a protein that has also been located at neuronal plasma membrane lipid membrane domains. Indeed, antibodies against this protein have been shown to be helpful in inhibiting some of the protein activities associated with plasma membrane lipid membrane nanodomains of synaptosomes, such as Cb5R activity [29,31]. Although an antagonist role has been described for Cav-2 with respect to Cav-1 due to the ability of Cav-2 to bind cholesterol [301], it cannot be discarded the presence of Cav-2 in the same domains or its role as a major component of some lipid membrane nanodomain subtype. Besides calcium transport channels, the majority of the proteins associated with lipid membrane domains are lipid-anchored proteins [302,303]. Cholesterol might also modulate the dynamics of bulk phases in membranes, altering membrane proteins’ folding and stability, and impacting energetics for protein oligomerization [304]. The hypothetical role of recently discovered molecular architectures enriched in caveolin-forming nanodisks [15], in buffering, distributing, or controlling cholesterol availability for neuronal plasma membrane proteins deserves to be studied in future studies.
Author Contributions
Conceptualization, A.K.S.-A. and C.G.-M.; writing—original draft preparation, A.K.S.-A.; writing—review and editing, A.K.S.-A., J.P., D.M.-d.-S., O.H.M.-C. and C.G.-M.; visualization, A.K.S.-A.; supervision, C.G.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We would like to thank Molecules and MDPI for the support and organization of the Special Issue dedicated to Carlos Gutiérrez-Merino: “Themed Issue in Honor of Carlos Gutiérrez Merino: Forty Years of Research Excellence in the Field of Membrane Proteins and Bioenergetics.”.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
5-hydroxytryptamine receptor (5-HT1A); NSF attachment protein (γ-SNAP); α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR); AMPAR-trafficking complexes (ATCs); brain-derived neurotrophic factor (BDNF); calcium/calmodulin-dependent protein kinase IIa (CaMKIIa); caveolin-1 (Cav-1); Cav-1–knocking down (Cav-1–KD); caveolin-2 (Cav-2); cytochrome b5 reductase (Cb5R); cerebellar granule neurons (CGN); caveolin binding motif (CMB); central nervous system (CNS); Cholesterol Recognition/Interaction Amino Acid Consensus (CRAC); C-terminal Src kinase (CsK); cholera toxin (CTx); dorsal root ganglion (DRG); estrogen receptor (ER); fluorescence energy transfer (FRET); ganglioside binding domains (GBD); monosialotetrahexosylganglioside (GM1); glycosylphosphatidylinositol (GPI); trisialoganglioside gt1b (GT1b); insulin-like growth factor-1 (IGF-1); intermembrane domain (IMD); inositol-3-phosphate (IP3); long-term depression (LTD); low-density lipoprotein receptor-related protein 1 (LRP1); metabotropic glutamate receptors (mGluRs); neuraminidase (N’ase); modulation efficiency protein D (NfeD); N-methyl-D-aspartate receptor (NMDAr); neuronal nitric oxide synthase (nNOS); NADPH oxidases (NOXs); N-ethylmaleimide sensitive fusion protein (NSF); NMDAr subtype 2B subunit (NR2B); oligomerization domain (OD); purinergic P2X receptor (P2XR); protein data bank (PBD); phospholipase C (PLC); plasma membrane calcium ATPase (PMCA); postsynaptic density protein 95 (PSD-95); scaffolding domain (SD); Src tyrosine kinase family (SFK); signature motif (SM); stomatin, prohibitin, flotillin, and HflK/C domains (SPFH); stomatin operon partner protein (STOPP); transmembrane (TM); tyrosine kinase receptors (Trk); transient receptor potential canonical channel (TRPC); transient receptor potential vanilloid type 2 (TRPV2).
References
- Pike, L.J. Rafts Defined: A Report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006, 47, 1597–1598. [Google Scholar] [CrossRef] [PubMed]
- Goñi, F.M. “Rafts”: A Nickname for Putative Transient Nanodomains. Chem. Phys. Lipids 2019, 218, 34–39. [Google Scholar] [CrossRef]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schönle, A.; et al. Direct Observation of the Nanoscale Dynamics of Membrane Lipids in a Living Cell. Nature 2009, 457, 1159–1162. [Google Scholar] [CrossRef] [PubMed]
- Klotzsch, E.; Schütz, G.J. A Critical Survey of Methods to Detect Plasma Membrane Rafts. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120033. [Google Scholar] [CrossRef]
- Kusumi, A.; Fujiwara, T.K.; Tsunoyama, T.A.; Kasai, R.S.; Liu, A.-A.; Hirosawa, K.M.; Kinoshita, M.; Matsumori, N.; Komura, N.; Ando, H.; et al. Defining Raft Domains in the Plasma Membrane. Traffic 2020, 21, 106–137. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Adame, P.L.; Meza, U.; Rodríguez-Menchaca, A.A.; Sánchez-Armass, S.; Ruiz-García, J.; Gomez, E. Determination of the Size of Lipid Rafts Studied through Single-Molecule FRET Simulations. Biophys. J. 2021, 120, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Pralle, A.; Keller, P.; Florin, E.L.; Simons, K.; Hörber, J.K. Sphingolipid-Cholesterol Rafts Diffuse as Small Entities in the Plasma Membrane of Mammalian Cells. J. Cell Biol. 2000, 148, 997–1008. [Google Scholar] [CrossRef]
- Yethiraj, A.; Weisshaar, J.C. Why Are Lipid Rafts Not Observed In Vivo? Biophys. J. 2007, 93, 3113–3119. [Google Scholar] [CrossRef]
- Sharma, P.; Varma, R.; Sarasij, R.C.; Ira; Gousset, K.; Krishnamoorthy, G.; Rao, M.; Mayor, S. Nanoscale Organization of Multiple GPI-Anchored Proteins in Living Cell Membranes. Cell 2004, 116, 577–589. [Google Scholar] [CrossRef]
- Martosella, J.; Zolotarjova, N.; Liu, H.; Moyer, S.C.; Perkins, P.D.; Boyes, B.E. High Recovery HPLC Separation of Lipid Rafts for Membrane Proteome Analysis. J. Proteome Res. 2006, 5, 1301–1312. [Google Scholar] [CrossRef]
- Yu, H.; Wakim, B.; Li, M.; Halligan, B.; Tint, G.S.; Patel, S.B. Quantifying Raft Proteins in Neonatal Mouse Brain by “tube-Gel” Protein Digestion Label-Free Shotgun Proteomics. Proteome Sci. 2007, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska, M.; Castillo, C.; Francesconi, A. Quantitative Profiling of Brain Lipid Raft Proteome in a Mouse Model of Fragile X Syndrome. PLoS ONE 2015, 10, e0121464. [Google Scholar] [CrossRef]
- Ledesma, M.D.; Da Silva, J.S.; Schevchenko, A.; Wilm, M.; Dotti, C.G. Proteomic Characterisation of Neuronal Sphingolipid-Cholesterol Microdomains: Role in Plasminogen Activation. Brain Res. 2003, 987, 107–116. [Google Scholar] [CrossRef]
- Galimzyanov, T.R.; Lyushnyak, A.S.; Aleksandrova, V.V.; Shilova, L.A.; Mikhalyov, I.I.; Molotkovskaya, I.M.; Akimov, S.A.; Batishchev, O.V. Line Activity of Ganglioside GM1 Regulates the Raft Size Distribution in a Cholesterol-Dependent Manner. Langmuir 2017, 33, 3517–3524. [Google Scholar] [CrossRef]
- Porta, J.C.; Han, B.; Gulsevin, A.; Chung, J.M.; Peskova, Y.; Connolly, S.; Mchaourab, H.S.; Meiler, J.; Karakas, E.; Kenworthy, A.K.; et al. Molecular Architecture of the Human Caveolin-1 Complex. Sci. Adv. 2022, 8, eabn7232. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, H.; Matsui, I. Higher-Order Structure Formation Using Refined Monomer Structures of Lipid Raft Markers, Stomatin, Prohibitin, Flotillin, and HflK/C-Related Proteins. FEBS Open Bio 2023, 13, 926–937. [Google Scholar] [CrossRef]
- Ayuyan, A.G.; Cohen, F.S. Raft Composition at Physiological Temperature and pH in the Absence of Detergents. Biophys. J. 2008, 94, 2654–2666. [Google Scholar] [CrossRef]
- Lamaze, C.; Tardif, N.; Dewulf, M.; Vassilopoulos, S.; Blouin, C.M. The Caveolae Dress Code: Structure and Signaling. Curr. Opin. Cell Biol. 2017, 47, 117–125. [Google Scholar] [CrossRef]
- Stoeber, M.; Schellenberger, P.; Siebert, C.A.; Leyrat, C.; Helenius, A.; Grünewald, K. Model for the Architecture of Caveolae Based on a Flexible, Net-like Assembly of Cavin1 and Caveolin Discs. Proc. Natl. Acad. Sci. USA 2016, 113, E8069–E8078. [Google Scholar] [CrossRef]
- Matthaeus, C.; Sochacki, K.A.; Dickey, A.M.; Puchkov, D.; Haucke, V.; Lehmann, M.; Taraska, J.W. The Molecular Organization of Differentially Curved Caveolae Indicates Bendable Structural Units at the Plasma Membrane. Nat. Commun. 2022, 13, 7234. [Google Scholar] [CrossRef]
- Lee, J.; Glover, K.J. The Transmembrane Domain of Caveolin-1 Exhibits a Helix-Break-Helix Structure. Biochim. Biophys. Acta 2012, 1818, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; Tillu, V.; McMahon, K.-A.; Collins, B.M. Key Phases in the Formation of Caveolae. Curr. Opin. Cell Biol. 2021, 71, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Jarsch, I.K.; Daste, F.; Gallop, J.L. Membrane Curvature in Cell Biology: An Integration of Molecular Mechanisms. J. Cell Biol. 2016, 214, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Has, C.; Das, S.L. Recent Developments in Membrane Curvature Sensing and Induction by Proteins. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129971. [Google Scholar] [CrossRef] [PubMed]
- Gambin, Y.; Ariotti, N.; McMahon, K.-A.; Bastiani, M.; Sierecki, E.; Kovtun, O.; Polinkovsky, M.E.; Magenau, A.; Jung, W.; Okano, S.; et al. Single-Molecule Analysis Reveals Self Assembly and Nanoscale Segregation of Two Distinct Cavin Subcomplexes on Caveolae. Elife 2013, 3, e01434. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.; Köster, D.; Ruez, R.; Gonnord, P.; Bastiani, M.; Abankwa, D.; Stan, R.V.; Butler-Browne, G.; Vedie, B.; Johannes, L.; et al. Cells Respond to Mechanical Stress by Rapid Disassembly of Caveolae. Cell 2011, 144, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; McMahon, K.-A.; Wu, Y. Caveolae: Formation, Dynamics, and Function. Curr. Opin. Cell Biol. 2020, 65, 8–16. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; García-Bereguiaín, M.A.; Martín-Romero, F.J.; Gutiérrez-Merino, C. Regionalization of Plasma Membrane-Bound Flavoproteins of Cerebellar Granule Neurons in Culture by Fluorescence Energy Transfer Imaging. J. Fluoresc. 2006, 16, 393–401. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; Garcia-Bereguiain, M.A.; Martin-Romero, F.J.; Gutierrez-Merino, C. Clustering of Plasma Membrane-Bound Cytochrome B5 Reductase within “lipid Raft” Microdomains of the Neuronal Plasma Membrane. Mol. Cell Neurosci. 2009, 40, 14–26. [Google Scholar] [CrossRef]
- Marques-da-Silva, D.; Samhan-Arias, A.K.; Tiago, T.; Gutierrez-Merino, C. L-Type Calcium Channels and Cytochrome B5 Reductase Are Components of Protein Complexes Tightly Associated with Lipid Rafts Microdomains of the Neuronal Plasma Membrane. J. Proteom. 2010, 73, 1502–1510. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; Marques-da-Silva, D.; Yanamala, N.; Gutierrez-Merino, C. Stimulation and Clustering of Cytochrome B5 Reductase in Caveolin-Rich Lipid Microdomains Is an Early Event in Oxidative Stress-Mediated Apoptosis of Cerebellar Granule Neurons. J. Proteom. 2012, 75, 2934–2949. [Google Scholar] [CrossRef]
- Marques-da-Silva, D.; Gutierrez-Merino, C. L-Type Voltage-Operated Calcium Channels, N-Methyl-d-Aspartate Receptors and Neuronal Nitric-Oxide Synthase Form a Calcium/Redox Nano-Transducer within Lipid Rafts. Biochem. Biophys. Res. Commun. 2012, 420, 257–262. [Google Scholar] [CrossRef]
- Marques-da-Silva, D.; Gutierrez-Merino, C. Caveolin-Rich Lipid Rafts of the Plasma Membrane of Mature Cerebellar Granule Neurons Are Microcompartments for Calcium/Reactive Oxygen and Nitrogen Species Cross-Talk Signaling. Cell Calcium 2014, 56, 108–123. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; López-Sánchez, C.; Marques-da-Silva, D.; Lagoa, R.; Garcia-Lopez, V.; García-Martínez, V.; Gutierrez-Merino, C. High Expression of Cytochrome B5 Reductase Isoform 3/Cytochrome B5 System in the Cerebellum and Pyramidal Neurons of Adult Rat Brain. Brain Struct. Funct. 2016, 221, 2147–2162. [Google Scholar] [CrossRef]
- Poejo, J.; Salazar, J.; Mata, A.M.; Gutierrez-Merino, C. Binding of Amyloid β(1-42)-Calmodulin Complexes to Plasma Membrane Lipid Rafts in Cerebellar Granule Neurons Alters Resting Cytosolic Calcium Homeostasis. Int. J. Mol. Sci. 2021, 22, 1984. [Google Scholar] [CrossRef]
- Poejo, J.; Orantos-Aguilera, Y.; Martin-Romero, F.J.; Mata, A.M.; Gutierrez-Merino, C. Internalized Amyloid-β (1–42) Peptide Inhibits the Store-Operated Calcium Entry in HT-22 Cells. Int. J. Mol. Sci. 2022, 23, 12678. [Google Scholar] [CrossRef]
- Bonini, I.C.; Antollini, S.S.; Gutiérrez-Merino, C.; Barrantes, F.J. Sphingomyelin Composition and Physical Asymmetries in Native Acetylcholine Receptor-Rich Membranes. Eur. Biophys. J. 2002, 31, 417–427. [Google Scholar] [CrossRef]
- Antollini, S.S.; Soto, M.A.; Bonini de Romanelli, I.; Gutiérrez-Merino, C.; Sotomayor, P.; Barrantes, F.J. Physical State of Bulk and Protein-Associated Lipid in Nicotinic Acetylcholine Receptor-Rich Membrane Studied by Laurdan Generalized Polarization and Fluorescence Energy Transfer. Biophys. J. 1996, 70, 1275–1284. [Google Scholar] [CrossRef]
- Gutiérrez-Merino, C.; Bonini de Romanelli, I.C.; Pietrasanta, L.I.; Barrantes, F.J. Preferential Distribution of the Fluorescent Phospholipid Probes NBD-Phosphatidylcholine and Rhodamine-Phosphatidylethanolamine in the Exofacial Leaflet of Acetylcholine Receptor-Rich Membranes from Torpedo Marmorata. Biochemistry 1995, 34, 4846–4855. [Google Scholar] [CrossRef]
- Parekh, A.B. Ca2+ Microdomains near Plasma Membrane Ca2+ Channels: Impact on Cell Function. J. Physiol. 2008, 586, 3043–3054. [Google Scholar] [CrossRef]
- Mironov, S.L. Rethinking Calcium Profiles around Single Channels: The Exponential and Periodic Calcium Nanodomains. Sci. Rep. 2019, 9, 17196. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Tschanz, A.; Krupnik, L.; Ries, J. Quantitative Data Analysis in Single-Molecule Localization Microscopy. Trends Cell Biol. 2020, 30, 837–851. [Google Scholar] [CrossRef]
- Wang, L.-Y.; Augustine, G.J. Presynaptic Nanodomains: A Tale of Two Synapses. Front. Cell. Neurosci. 2015, 8, 455. [Google Scholar] [CrossRef]
- Chen, Y.; Matveev, V. Stationary Ca2+ Nanodomains in the Presence of Buffers with Two Binding Sites. Biophys. J. 2021, 120, 1942–1956. [Google Scholar] [CrossRef]
- Kwon, D. The Quest to Map the Mouse Brain. Nature 2023, 620, 685–687. [Google Scholar] [CrossRef]
- Holmes, K.L.; Lantz, L.M.; Russ, W. Conjugation of Fluorochromes to Monoclonal Antibodies. In Current Protocols in Cytometry; Chapter 4, Unit 4.2; Wiley Periodicals LLC.: Hoboken, NJ, USA, 2001. [Google Scholar] [CrossRef]
- Haugland, R.P. Antibody Conjugates for Cell Biology. In Current Protocols in Molecular Biology; Chapter 16, Unit 16.5; Wiley Periodicals LLC.: Hoboken, NJ, USA, 2001. [Google Scholar] [CrossRef]
- Haugland, R.P. Coupling of Monoclonal Antibodies with Fluorophores. Methods Mol. Biol. 1995, 45, 205–221. [Google Scholar] [CrossRef]
- Vira, S.; Mekhedov, E.; Humphrey, G.; Blank, P.S. Fluorescent-Labeled Antibodies: Balancing Functionality and Degree of Labeling. Anal. Biochem. 2010, 402, 146–150. [Google Scholar] [CrossRef]
- Gutierrez-Merino, C. Quantitation of the Förster Energy Transfer for Two-Dimensional Systems. II. Protein Distribution and Aggregation State in Biological Membranes. Biophys. Chem. 1981, 14, 259–266. [Google Scholar] [CrossRef]
- Gutierrez-Merino, C.; Centeno, F.; Garcia-Martin, E.; Merino, J.M. Fluorescence Energy Transfer as a Tool to Locate Functional Sites in Membrane Proteins. Biochem. Soc. Trans. 1994, 22, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Dewey, T.G.; Hammes, G.G. Calculation on Fluorescence Resonance Energy Transfer on Surfaces. Biophys. J. 1980, 32, 1023–1035. [Google Scholar] [CrossRef]
- Stryer, L. Fluorescence Energy Transfer as a Spectroscopic Ruler. Annu. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef]
- Pol, A.; Morales-Paytuví, F.; Bosch, M.; Parton, R.G. Non-Caveolar Caveolins—Duties Outside the Caves. J. Cell Sci. 2020, 133, jcs241562. [Google Scholar] [CrossRef]
- Simons, K.; Toomre, D. Lipid Rafts and Signal Transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Lang, D.M.; Lommel, S.; Jung, M.; Ankerhold, R.; Petrausch, B.; Laessing, U.; Wiechers, M.F.; Plattner, H.; Stuermer, C.A. Identification of Reggie-1 and Reggie-2 as Plasmamembrane-Associated Proteins Which Cocluster with Activated GPI-Anchored Cell Adhesion Molecules in Non-Caveolar Micropatches in Neurons. J. Neurobiol. 1998, 37, 502–523. [Google Scholar] [CrossRef]
- Lipid Raft Microdomains and Neurotransmitter Signalling|Nature Reviews Neuroscience. Available online: https://www.nature.com/articles/nrn2059 (accessed on 26 August 2023).
- Muallem, S.; Chung, W.Y.; Jha, A.; Ahuja, M. Lipids at Membrane Contact Sites: Cell Signaling and Ion Transport. EMBO Rep. 2017, 18, 1893–1904. [Google Scholar] [CrossRef]
- Grassi, S.; Giussani, P.; Mauri, L.; Prioni, S.; Sonnino, S.; Prinetti, A. Lipid Rafts and Neurodegeneration: Structural and Functional Roles in Physiologic Aging and Neurodegenerative Diseases. J. Lipid Res. 2020, 61, 636–654. [Google Scholar] [CrossRef]
- Hayashi, T. Membrane Lipid Rafts Are Required for AMPA Receptor Tyrosine Phosphorylation. Front. Synaptic Neurosci. 2022, 14, 921772. [Google Scholar] [CrossRef]
- Chen, J.; Sitsel, A.; Benoy, V.; Sepúlveda, M.R.; Vangheluwe, P. Primary Active Ca2+ Transport Systems in Health and Disease. Cold Spring Harb. Perspect. Biol. 2020, 12, a035113. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Qi, W.; Wang, L.-J.; Miao, H.-H.; Qu, Y.-X.; Li, B.-L.; Song, B.-L. Flotillins Play an Essential Role in Niemann-Pick C1-like 1-Mediated Cholesterol Uptake. Proc. Natl. Acad. Sci. USA 2011, 108, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Roitbak, T.; Surviladze, Z.; Tikkanen, R.; Wandinger-Ness, A. A Polycystin Multiprotein Complex Constitutes a Cholesterol-Containing Signalling Microdomain in Human Kidney Epithelia. Biochem. J. 2005, 392, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Volonte, D.; Galbiati, F.; Li, S.; Nishiyama, K.; Okamoto, T.; Lisanti, M.P. Flotillins/Cavatellins Are Differentially Expressed in Cells and Tissues and Form a Hetero-Oligomeric Complex with Caveolins in Vivo. Characterization and Epitope-Mapping of a Novel Flotillin-1 Monoclonal Antibody Probe. J. Biol. Chem. 1999, 274, 12702–12709. [Google Scholar] [CrossRef]
- Yang, G.; Xu, H.; Li, Z.; Li, F. Interactions of Caveolin-1 Scaffolding and Intramembrane Regions Containing a CRAC Motif with Cholesterol in Lipid Bilayers. Biochim. Biophys. Acta 2014, 1838, 2588–2599. [Google Scholar] [CrossRef]
- Hanafusa, K.; Hayashi, N. The Flot2 Component of the Lipid Raft Changes Localization during Neural Differentiation of P19C6 Cells. BMC Mol. Cell Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Wåhlén, E.; Olsson, F.; Söderberg, O.; Lennartsson, J.; Heldin, J. Differential Impact of Lipid Raft Depletion on Platelet-Derived Growth Factor (PDGF)-Induced ERK1/2 MAP-Kinase, SRC and AKT Signaling. Cell. Signal. 2022, 96, 110356. [Google Scholar] [CrossRef]
- Ouweneel, A.B.; Thomas, M.J.; Sorci-Thomas, M.G. The Ins and Outs of Lipid Rafts: Functions in Intracellular Cholesterol Homeostasis, Microparticles, and Cell Membranes. J. Lipid Res. 2020, 61, 676–686. [Google Scholar] [CrossRef]
- Davidović, D.; Kukulka, M.; Sarmento, M.J.; Mikhalyov, I.; Gretskaya, N.; Chmelová, B.; Ricardo, J.C.; Hof, M.; Cwiklik, L.; Šachl, R. Which Moiety Drives Gangliosides to Form Nanodomains? J. Phys. Chem. Lett. 2023, 14, 5791–5797. [Google Scholar] [CrossRef]
- Matsubara, T.; IIjima, K.; Kojima, T.; Hirai, M.; Miyamoto, E.; Sato, T. Heterogeneous Ganglioside-Enriched Nanoclusters with Different Densities in Membrane Rafts Detected by a Peptidyl Molecular Probe. Langmuir 2021, 37, 646–654. [Google Scholar] [CrossRef]
- Sipione, S.; Monyror, J.; Galleguillos, D.; Steinberg, N.; Kadam, V. Gangliosides in the Brain: Physiology, Pathophysiology and Therapeutic Applications. Front. Neurosci. 2020, 14, 572965. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhong, W.; Hu, Z.; Tang, X. A Review of the Role of Cav-1 in Neuropathology and Neural Recovery after Ischemic Stroke. J. Neuroinflammation 2018, 15, 348. [Google Scholar] [CrossRef] [PubMed]
- Simón, L.; Campos, A.; Leyton, L.; Quest, A.F.G. Caveolin-1 Function at the Plasma Membrane and in Intracellular Compartments in Cancer. Cancer Metastasis Rev. 2020, 39, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Yang, M.; Zhao, H.; Han, Y.; Jiang, N.; Yang, J.; Chen, W.; Li, C.; Liu, Y.; Zhao, C.; et al. Caveolin-1 Regulates Cellular Metabolism: A Potential Therapeutic Target in Kidney Disease. Front. Pharmacol. 2021, 12, 768100. [Google Scholar] [CrossRef]
- Razani, B.; Engelman, J.A.; Wang, X.B.; Schubert, W.; Zhang, X.L.; Marks, C.B.; Macaluso, F.; Russell, R.G.; Li, M.; Pestell, R.G.; et al. Caveolin-1 Null Mice Are Viable but Show Evidence of Hyperproliferative and Vascular Abnormalities. J. Biol. Chem. 2001, 276, 38121–38138. [Google Scholar] [CrossRef]
- Schlegel, A.; Schwab, R.B.; Scherer, P.E.; Lisanti, M.P. A Role for the Caveolin Scaffolding Domain in Mediating the Membrane Attachment of Caveolin-1. The Caveolin Scaffolding Domain Is Both Necessary and Sufficient for Membrane Binding in Vitro. J. Biol. Chem. 1999, 274, 22660–22667. [Google Scholar] [CrossRef]
- Wong, T.H.; Khater, I.M.; Joshi, B.; Shahsavari, M.; Hamarneh, G.; Nabi, I.R. Single Molecule Network Analysis Identifies Structural Changes to Caveolae and Scaffolds Due to Mutation of the Caveolin-1 Scaffolding Domain. Sci. Rep. 2021, 11, 7810. [Google Scholar] [CrossRef]
- Reese, C.F.; Chinnakkannu, P.; Tourkina, E.; Hoffman, S.; Kuppuswamy, D. Multiple Subregions within the Caveolin-1 Scaffolding Domain Inhibit Fibrosis, Microvascular Leakage, and Monocyte Migration. PLoS ONE 2022, 17, e0264413. [Google Scholar] [CrossRef]
- Aoki, S.; Thomas, A.; Decaffmeyer, M.; Brasseur, R.; Epand, R.M. The Role of Proline in the Membrane Re-Entrant Helix of Caveolin-1. J. Biol. Chem. 2010, 285, 33371–33380. [Google Scholar] [CrossRef]
- Root, K.T.; Julien, J.A.; Glover, K.J. Secondary Structure of Caveolins: A Mini Review. Biochem. Soc. Trans. 2019, 47, 1489–1498. [Google Scholar] [CrossRef]
- Yang, G.; Dong, Z.; Xu, H.; Wang, C.; Li, H.; Li, Z.; Li, F. Structural Study of Caveolin-1 Intramembrane Domain by Circular Dichroism and Nuclear Magnetic Resonance. Pept. Sci. 2015, 104, 11–20. [Google Scholar] [CrossRef]
- Fielding, C.J.; Fielding, P.E. Role of Cholesterol in Signal Transduction from Caveolae. In Lipid Rafts and Caveolae; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; pp. 91–113. ISBN 978-3-527-60807-2. [Google Scholar]
- Kenworthy, A.K. The Building Blocks of Caveolae Revealed: Caveolins Finally Take Center Stage. Biochem. Soc. Trans. 2023, 51, 855–869. [Google Scholar] [CrossRef]
- Couet, J.; Li, S.; Okamoto, T.; Ikezu, T.; Lisanti, M.P. Identification of Peptide and Protein Ligands for the Caveolin-Scaffolding Domain. Implications for the Interaction of Caveolin with Caveolae-Associated Proteins. J. Biol. Chem. 1997, 272, 6525–6533. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yao, Z.; Degenhardt, B.; Teper, G.; Papadopoulos, V. Cholesterol Binding at the Cholesterol Recognition/ Interaction Amino Acid Consensus (CRAC) of the Peripheral-Type Benzodiazepine Receptor and Inhibition of Steroidogenesis by an HIV TAT-CRAC Peptide. Proc. Natl. Acad. Sci. USA 2001, 98, 1267–1272. [Google Scholar] [CrossRef]
- Sheng, R.; Chen, Y.; Yung Gee, H.; Stec, E.; Melowic, H.R.; Blatner, N.R.; Tun, M.P.; Kim, Y.; Källberg, M.; Fujiwara, T.K.; et al. Cholesterol Modulates Cell Signaling and Protein Networking by Specifically Interacting with PDZ Domain-Containing Scaffold Proteins. Nat. Commun. 2012, 3, 1249. [Google Scholar] [CrossRef]
- Epand, R.M. Proteins and Cholesterol-Rich Domains. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1576–1582. [Google Scholar] [CrossRef]
- Fantini, J.; Barrantes, F.J. How Cholesterol Interacts with Membrane Proteins: An Exploration of Cholesterol-Binding Sites Including CRAC, CARC, and Tilted Domains. Front. Physiol. 2013, 4, 31. [Google Scholar] [CrossRef]
- Monier, S.; Parton, R.G.; Vogel, F.; Behlke, J.; Henske, A.; Kurzchalia, T.V. VIP21-Caveolin, a Membrane Protein Constituent of the Caveolar Coat, Oligomerizes in Vivo and in Vitro. Mol. Biol. Cell 1995, 6, 911–927. [Google Scholar] [CrossRef]
- Sargiacomo, M.; Scherer, P.E.; Tang, Z.; Kübler, E.; Song, K.S.; Sanders, M.C.; Lisanti, M.P. Oligomeric Structure of Caveolin: Implications for Caveolae Membrane Organization. Proc. Natl. Acad. Sci. USA 1995, 92, 9407–9411. [Google Scholar] [CrossRef]
- Ariotti, N.; Rae, J.; Leneva, N.; Ferguson, C.; Loo, D.; Okano, S.; Hill, M.M.; Walser, P.; Collins, B.M.; Parton, R.G. Molecular Characterization of Caveolin-Induced Membrane Curvature. J. Biol. Chem. 2015, 290, 24875–24890. [Google Scholar] [CrossRef] [PubMed]
- Walser, P.J.; Ariotti, N.; Howes, M.; Ferguson, C.; Webb, R.; Schwudke, D.; Leneva, N.; Cho, K.-J.; Cooper, L.; Rae, J.; et al. Constitutive Formation of Caveolae in a Bacterium. Cell 2012, 150, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Hayer, A.; Stoeber, M.; Bissig, C.; Helenius, A. Biogenesis of Caveolae: Stepwise Assembly of Large Caveolin and Cavin Complexes. Traffic 2010, 11, 361–382. [Google Scholar] [CrossRef] [PubMed]
- Hayer, A.; Stoeber, M.; Ritz, D.; Engel, S.; Meyer, H.H.; Helenius, A. Caveolin-1 Is Ubiquitinated and Targeted to Intralumenal Vesicles in Endolysosomes for Degradation. J. Cell Biol. 2010, 191, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Copeland, C.A.; Tiwari, A.; Kenworthy, A.K. Assembly and Turnover of Caveolae: What Do We Really Know? Front. Cell Dev. Biol. 2016, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Panchishina, M.V. [Cholesterol synthesis by several strains of Escherichia]. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1979, 9, 65–68. [Google Scholar]
- Santoscoy, M.C.; Jarboe, L.R. Production of Cholesterol-like Molecules Impacts Escherichia Coli Robustness, Production Capacity, and Vesicle Trafficking. Metab. Eng. 2022, 73, 134–143. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; ISBN 978-0-8153-3218-3. [Google Scholar]
- Barrantes, F.J. Structural Basis for Lipid Modulation of Nicotinic Acetylcholine Receptor Function. Brain Res. Brain Res. Rev. 2004, 47, 71–95. [Google Scholar] [CrossRef]
- Criado, M.; Eibl, H.; Barrantes, F.J. Effects of Lipids on Acetylcholine Receptor. Essential Need of Cholesterol for Maintenance of Agonist-Induced State Transitions in Lipid Vesicles. Biochemistry 1982, 21, 3622–3629. [Google Scholar] [CrossRef]
- Marsh, D.; Barrantes, F.J. Immobilized Lipid in Acetylcholine Receptor-Rich Membranes from Torpedo Marmorata. Proc. Natl. Acad. Sci. USA 1978, 75, 4329–4333. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, C.; Luo, D.; Yan, L.; Yang, W.; Li, N.; Gao, N. Structural Insights into the Membrane Microdomain Organization by SPFH Family Proteins. Cell Res. 2022, 32, 176–189. [Google Scholar] [CrossRef]
- Head, B.P.; Insel, P.A. Do Caveolins Regulate Cells by Actions Outside of Caveolae? Trends Cell Biol. 2007, 17, 51–57. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of Clathrin-Mediated Endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Smith, S.M.; Smith, C.J. Capturing the Mechanics of Clathrin-Mediated Endocytosis. Curr. Opin. Struct. Biol. 2022, 75, 102427. [Google Scholar] [CrossRef]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-Independent Pathways of Endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, a016758. [Google Scholar] [CrossRef] [PubMed]
- Ripa, I.; Andreu, S.; López-Guerrero, J.A.; Bello-Morales, R. Membrane Rafts: Portals for Viral Entry. Front. Microbiol. 2021, 12, 631274. [Google Scholar] [CrossRef] [PubMed]
- Gusmira, A.; Takemura, K.; Lee, S.Y.; Inaba, T.; Hanawa-Suetsugu, K.; Oono-Yakura, K.; Yasuhara, K.; Kitao, A.; Suetsugu, S. Regulation of Caveolae through Cholesterol-Depletion-Dependent Tubulation Mediated by PACSIN2. J. Cell Sci. 2020, 133, jcs246785. [Google Scholar] [CrossRef] [PubMed]
- Nabi, I.R.; Le, P.U. Caveolae/Raft-Dependent Endocytosis. J. Cell Biol. 2003, 161, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key Principles and Methods for Studying the Endocytosis of Biological and Nanoparticle Therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Browman, D.T.; Hoegg, M.B.; Robbins, S.M. The SPFH Domain-Containing Proteins: More than Lipid Raft Markers. Trends Cell Biol. 2007, 17, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.-J.; Singh, R.D.; Sharma, D.K.; Holicky, E.L.; Hanada, K.; Marks, D.L.; Pagano, R.E. Distinct Mechanisms of Clathrin-Independent Endocytosis Have Unique Sphingolipid Requirements. Mol. Biol. Cell 2006, 17, 3197–3210. [Google Scholar] [CrossRef]
- Stern, C.M.; Mermelstein, P.G. Caveolin Regulation of Neuronal Intracellular Signaling. Cell Mol. Life Sci. 2010, 67, 3785–3795. [Google Scholar] [CrossRef]
- Monier, S.; Dietzen, D.J.; Hastings, W.R.; Lublin, D.M.; Kurzchalia, T.V. Oligomerization of VIP21-Caveolin in Vitro Is Stabilized by Long Chain Fatty Acylation or Cholesterol. FEBS Lett. 1996, 388, 143–149. [Google Scholar] [CrossRef]
- Boulware, M.I.; Kordasiewicz, H.; Mermelstein, P.G. Caveolin Proteins Are Essential for Distinct Effects of Membrane Estrogen Receptors in Neurons. J. Neurosci. 2007, 27, 9941–9950. [Google Scholar] [CrossRef]
- Poo, M.M. Neurotrophins as Synaptic Modulators. Nat. Rev. Neurosci. 2001, 2, 24–32. [Google Scholar] [CrossRef]
- Zhong, W.; Huang, Q.; Zeng, L.; Hu, Z.; Tang, X. Caveolin-1 and MLRs: A Potential Target for Neuronal Growth and Neuroplasticity after Ischemic Stroke. Int. J. Med. Sci. 2019, 16, 1492–1503. [Google Scholar] [CrossRef]
- Head, B.P.; Peart, J.N.; Panneerselvam, M.; Yokoyama, T.; Pearn, M.L.; Niesman, I.R.; Bonds, J.A.; Schilling, J.M.; Miyanohara, A.; Headrick, J.; et al. Loss of Caveolin-1 Accelerates Neurodegeneration and Aging. PLoS ONE 2010, 5, e15697. [Google Scholar] [CrossRef]
- Koh, S.; Lee, W.; Park, S.M.; Kim, S.H. Caveolin-1 Deficiency Impairs Synaptic Transmission in Hippocampal Neurons. Mol. Brain 2021, 14, 53. [Google Scholar] [CrossRef]
- Egawa, J.; Schilling, J.M.; Cui, W.; Posadas, E.; Sawada, A.; Alas, B.; Zemljic-Harpf, A.E.; Fannon-Pavlich, M.J.; Mandyam, C.D.; Roth, D.M.; et al. Neuron-Specific Caveolin-1 Overexpression Improves Motor Function and Preserves Memory in Mice Subjected to Brain Trauma. FASEB J. 2017, 31, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Head, B.P.; Hu, Y.; Finley, J.C.; Saldana, M.D.; Bonds, J.A.; Miyanohara, A.; Niesman, I.R.; Ali, S.S.; Murray, F.; Insel, P.A.; et al. Neuron-Targeted Caveolin-1 Protein Enhances Signaling and Promotes Arborization of Primary Neurons. J. Biol. Chem. 2011, 286, 33310–33321. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Head, B.P. Caveolin-1 in Stroke Neuropathology and Neuroprotection: A Novel Molecular Therapeutic Target for Ischemic-Related Injury. Curr. Vasc. Pharmacol. 2019, 17, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Trushina, E.; Canaria, C.A.; Lee, D.Y.; McMurray, C.T. Loss of Caveolin-1 Expression in Knock-in Mouse Model of Huntington’s Disease Suppresses Pathophysiology In Vivo. Hum. Mol. Genet. 2014, 23, 129–144. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3857950/ (accessed on 11 May 2023). [CrossRef]
- Fortalezas, S.; Marques-da-Silva, D.; Gutierrez-Merino, C. Methyl-β-Cyclodextrin Impairs the Phosphorylation of the Β₂ Subunit of L-Type Calcium Channels and Cytosolic Calcium Homeostasis in Mature Cerebellar Granule Neurons. Int. J. Mol. Sci. 2018, 19, 3667. [Google Scholar] [CrossRef] [PubMed]
- Atlas, D. Voltage-Gated Calcium Channels Function as Ca2+-Activated Signaling Receptors. Trends Biochem. Sci. 2014, 39, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-X.; Hua, L.; Li, Y.-Q.; Jiang, Y.-Y.; Han, D.; Liu, H.; Tang, Q.-Q.; Yang, X.-N.; Yin, C.; Hao, L.-Y.; et al. Caveolin-1 in the Anterior Cingulate Cortex Modulates Chronic Neuropathic Pain via Regulation of NMDA Receptor 2B Subunit. J. Neurosci. 2015, 35, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Bigford, G.E.; Alonso, O.F.; Dietrich, W.D.; Keane, R.W. A Novel Protein Complex in Membrane Rafts Linking the NR2B Glutamate Receptor and Autophagy Is Disrupted Following Traumatic Brain Injury. J. Neurotrauma 2009, 26, 703–720. [Google Scholar] [CrossRef] [PubMed]
- Roh, S.-E.; Hong, Y.H.; Jang, D.C.; Kim, J.; Kim, S.J. Lipid Rafts Serve as Signaling Platforms for mGlu1 Receptor-Mediated Calcium Signaling in Association with Caveolin. Mol. Brain 2014, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Hering, H.; Lin, C.-C.; Sheng, M. Lipid Rafts in the Maintenance of Synapses, Dendritic Spines, and Surface AMPA Receptor Stability. J. Neurosci. 2003, 23, 3262–3271. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, S.B.; Chabot, C.; Gratton, J.-P.; Poirier, J. The Caveolin Scaffolding Domain Modifies 2-Amino-3-Hydroxy-5-Methyl-4-Isoxazole Propionate Receptor Binding Properties by Inhibiting Phospholipase A2 Activity. J. Biol. Chem. 2004, 279, 356–362. [Google Scholar] [CrossRef]
- Li, X.-H.; Miao, H.-H.; Zhuo, M. NMDA Receptor Dependent Long-Term Potentiation in Chronic Pain. Neurochem. Res. 2019, 44, 531–538. [Google Scholar] [CrossRef]
- Head, B.P.; Patel, H.H.; Tsutsumi, Y.M.; Hu, Y.; Mejia, T.; Mora, R.C.; Insel, P.A.; Roth, D.M.; Drummond, J.C.; Patel, P.M. Caveolin-1 Expression Is Essential for N-Methyl-D-Aspartate Receptor-Mediated Src and Extracellular Signal-Regulated Kinase 1/2 Activation and Protection of Primary Neurons from Ischemic Cell Death. FASEB J. 2008, 22, 828–840. [Google Scholar] [CrossRef]
- Hansen, K.B.; Yi, F.; Perszyk, R.E.; Furukawa, H.; Wollmuth, L.P.; Gibb, A.J.; Traynelis, S.F. Structure, Function, and Allosteric Modulation of NMDA Receptors. J. Gen. Physiol. 2018, 150, 1081–1105. [Google Scholar] [CrossRef]
- Banerjee, A.; Larsen, R.S.; Philpot, B.D.; Paulsen, O. Roles of Presynaptic NMDA Receptors in Neurotransmission and Plasticity. Trends Neurosci. 2016, 39, 26–39. [Google Scholar] [CrossRef]
- Carter, B.C.; Jahr, C.E. Postsynaptic, Not Presynaptic NMDA Receptors Are Required for Spike-Timing-Dependent LTD Induction. Nat. Neurosci. 2016, 19, 1218–1224. [Google Scholar] [CrossRef]
- Paul, S.; Connor, J.A. NR2B-NMDA Receptor Mediated Increases in Intracellular Ca2+ Concentration Regulate the Tyrosine Phosphatase, STEP, and ERK MAP Kinase Signaling. J. Neurochem. 2010, 114, 1107–1118. [Google Scholar] [CrossRef]
- Laube, B.; Hirai, H.; Sturgess, M.; Betz, H.; Kuhse, J. Molecular Determinants of Agonist Discrimination by NMDA Receptor Subunits: Analysis of the Glutamate Binding Site on the NR2B Subunit. Neuron 1997, 18, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Stary, C.; Tsutsumi, Y.; Patel, P.; Head, B.; Patel, H.; Roth, D. Caveolins: Targeting pro-Survival Signaling in the Heart and Brain. Front. Physiol. 2012, 3, 393. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.M.; Roder, J.C.; Davidow, J.; Salter, M.W. Src Activation in the Induction of Long-Term Potentiation in CA1 Hippocampal Neurons. Science 1998, 279, 1363–1367. [Google Scholar] [CrossRef]
- Rostas, J.A.; Brent, V.A.; Voss, K.; Errington, M.L.; Bliss, T.V.; Gurd, J.W. Enhanced Tyrosine Phosphorylation of the 2B Subunit of the N-Methyl-D-Aspartate Receptor in Long-Term Potentiation. Proc. Natl. Acad. Sci. USA 1996, 93, 10452–10456. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Komai, S.; Tezuka, T.; Hisatsune, C.; Umemori, H.; Semba, K.; Mishina, M.; Manabe, T.; Yamamoto, T. Characterization of Fyn-Mediated Tyrosine Phosphorylation Sites on GluR Epsilon 2 (NR2B) Subunit of the N-Methyl-D-Aspartate Receptor. J. Biol. Chem. 2001, 276, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Volonté, D.; Galbiati, F.; Pestell, R.G.; Lisanti, M.P. Cellular Stress Induces the Tyrosine Phosphorylation of Caveolin-1 (Tyr(14)) via Activation of P38 Mitogen-Activated Protein Kinase and c-Src Kinase. Evidence for Caveolae, the Actin Cytoskeleton, and Focal Adhesions as Mechanical Sensors of Osmotic Stress. J. Biol. Chem. 2001, 276, 8094–8103. [Google Scholar] [CrossRef]
- Grande-García, A.; Echarri, A.; de Rooij, J.; Alderson, N.B.; Waterman-Storer, C.M.; Valdivielso, J.M.; del Pozo, M.A. Caveolin-1 Regulates Cell Polarization and Directional Migration through Src Kinase and Rho GTPases. J. Cell Biol. 2007, 177, 683–694. [Google Scholar] [CrossRef]
- Radel, C.; Rizzo, V. Integrin Mechanotransduction Stimulates Caveolin-1 Phosphorylation and Recruitment of Csk to Mediate Actin Reorganization. Am. J. Physiol.-Heart Circ. Physiol. 2005, 288, H936–H945. [Google Scholar] [CrossRef][Green Version]
- Cao, H.; Sanguinetti, A.R.; Mastick, C.C. Oxidative Stress Activates Both Src-Kinases and Their Negative Regulator Csk and Induces Phosphorylation of Two Targeting Proteins for Csk: Caveolin-1 and Paxillin. Exp. Cell Res. 2004, 294, 159–171. [Google Scholar] [CrossRef]
- Okada, M. Regulation of the Src Family Kinases by Csk. Int. J. Biol. Sci. 2012, 8, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Breuer, M.; Berger, H.; Borchers, A. Caveolin 1 Is Required for Axonal Outgrowth of Motor Neurons and Affects Xenopus Neuromuscular Development. Sci. Rep. 2020, 10, 16446. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, S.B.; Blain, J.-F.; Gratton, J.-P.; Poirier, J. A Role for Caveolin-1 in Post-Injury Reactive Neuronal Plasticity. J. Neurochem. 2005, 92, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Lituma, P.J.; Kwon, H.-B.; Alviña, K.; Luján, R.; Castillo, P.E. Presynaptic NMDA Receptors Facilitate Short-Term Plasticity and BDNF Release at Hippocampal Mossy Fiber Synapses. eLife 2021, 10, e66612. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, F.B.; Min, R.; Nevian, T. Presynaptic NMDA Receptors Influence Ca2+ Dynamics by Interacting with Voltage-Dependent Calcium Channels during the Induction of Long-Term Depression. Neural Plast. 2022, 2022, e2900875. [Google Scholar] [CrossRef] [PubMed]
- Brennan, A.M.; Suh, S.W.; Won, S.J.; Narasimhan, P.; Kauppinen, T.M.; Lee, H.; Edling, Y.; Chan, P.H.; Swanson, R.A. NADPH Oxidase Is the Primary Source of Superoxide Induced by NMDA Receptor Activation. Nat. Neurosci. 2009, 12, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Casado, M.; Isope, P.; Ascher, P. Involvement of Presynaptic N-Methyl-D-Aspartate Receptors in Cerebellar Long-Term Depression. Neuron 2002, 33, 123–130. [Google Scholar] [CrossRef]
- Sjöström, P.J.; Turrigiano, G.G.; Nelson, S.B. Neocortical LTD via Coincident Activation of Presynaptic NMDA and Cannabinoid Receptors. Neuron 2003, 39, 641–654. [Google Scholar] [CrossRef]
- Dawson, T.M.; Snyder, S.H. Gases as Biological Messengers: Nitric Oxide and Carbon Monoxide in the Brain. J. Neurosci. 1994, 14, 5147–5159. [Google Scholar] [CrossRef]
- Sato, Y.; Sagami, I.; Shimizu, T. Identification of Caveolin-1-Interacting Sites in Neuronal Nitric-Oxide Synthase: Molecular Mechanism for Inhibition of No Formation. J. Biol. Chem. 2004, 279, 8827–8836. [Google Scholar] [CrossRef]
- Hou, Q.; Huang, Y.; Amato, S.; Snyder, S.H.; Huganir, R.L.; Man, H.-Y. Regulation of AMPA Receptor Localization in Lipid Rafts. Mol. Cell Neurosci. 2008, 38, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.L. Molecular Pharmacology of Metabotropic Receptors Targeted by Neuropsychiatric Drugs. Nat. Struct. Mol. Biol. 2019, 26, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.; Goudet, C. Emerging Trends in Pain Modulation by Metabotropic Glutamate Receptors. Front. Mol. Neurosci. 2019, 11, 464. [Google Scholar] [CrossRef] [PubMed]
- Francesconi, A.; Kumari, R.; Zukin, R.S. Regulation of Group I Metabotropic Glutamate Receptor Trafficking and Signaling by the Caveolar/Lipid Raft Pathway. J. Neurosci. 2009, 29, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Reiner, A.; Levitz, J. Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S. Molecular Diversity of Glutamate Receptors and Implications for Brain Function. Science 1992, 258, 597–603. [Google Scholar] [CrossRef]
- Nakanishi, S.; Masu, M. Molecular Diversity and Functions of Glutamate Receptors. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 319–348. [Google Scholar] [CrossRef]
- Huh, E.; Agosto, M.A.; Wensel, T.G.; Lichtarge, O. Coevolutionary Signals in Metabotropic Glutamate Receptors Capture Residue Contacts and Long-Range Functional Interactions. J. Biol. Chem. 2023, 299, 103030. [Google Scholar] [CrossRef]
- Gandasi, N.R.; Arapi, V.; Mickael, M.E.; Belekar, P.A.; Granlund, L.; Kothegala, L.; Fredriksson, R.; Bagchi, S. Glutamine Uptake via SNAT6 and Caveolin Regulates Glutamine–Glutamate Cycle. Int. J. Mol. Sci. 2021, 22, 1167. [Google Scholar] [CrossRef]
- Mango, D.; Ledonne, A. Updates on the Physiopathology of Group I Metabotropic Glutamate Receptors (mGluRI)-Dependent Long-Term Depression. Cells 2023, 12, 1588. [Google Scholar] [CrossRef]
- Shigemoto, R.; Kinoshita, A.; Wada, E.; Nomura, S.; Ohishi, H.; Takada, M.; Flor, P.J.; Neki, A.; Abe, T.; Nakanishi, S.; et al. Differential Presynaptic Localization of Metabotropic Glutamate Receptor Subtypes in the Rat Hippocampus. J. Neurosci. 1997, 17, 7503–7522. [Google Scholar] [CrossRef] [PubMed]
- Grove-Strawser, D.; Boulware, M.I.; Mermelstein, P.G. Membrane Estrogen Receptors Activate the Metabotropic Glutamate Receptors mGluR5 and mGluR3 to Bidirectionally Regulate CREB Phosphorylation in Female Rat Striatal Neurons. Neuroscience 2010, 170, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Nakade, S.; Miyawaki, A.; Mikoshiba, K.; Ogawa, K. Localization of Inositol 1,4,5-Trisphosphate Receptor-like Protein in Plasmalemmal Caveolae. J. Cell Biol. 1992, 119, 1507–1513. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Miyawaki, A.; Mikoshiba, K. Inositol 1,4,5-Trisphosphate Receptor-like Protein in Plasmalemmal Caveolae Is Linked to Actin Filaments. J. Cell Sci. 1995, 108, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lockwich, T.P.; Liu, X.; Singh, B.B.; Jadlowiec, J.; Weiland, S.; Ambudkar, I.S. Assembly of Trp1 in a Signaling Complex Associated with Caveolin-Scaffolding Lipid Raft Domains. J. Biol. Chem. 2000, 275, 11934–11942. [Google Scholar] [CrossRef] [PubMed]
- Dunphy, J.T.; Greentree, W.K.; Linder, M.E. Enrichment of G-Protein Palmitoyltransferase Activity in Low Density Membranes: In Vitro Reconstitution of Gαi to These Domains Requires Palmitoyltransferase Activity. J. Biol. Chem. 2001, 276, 43300–43304. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Castillo, C.; Francesconi, A. Agonist-Dependent Signaling by Group I Metabotropic Glutamate Receptors Is Regulated by Association with Lipid Domains. J. Biol. Chem. 2013, 288, 32004–32019. [Google Scholar] [CrossRef]
- Hong, Y.H.; Kim, J.Y.; Lee, J.H.; Chae, H.G.; Jang, S.S.; Jeon, J.H.; Kim, C.H.; Kim, J.; Kim, S.J. Agonist-Induced Internalization of mGluR1α Is Mediated by Caveolin. J. Neurochem. 2009, 111, 61–71. [Google Scholar] [CrossRef]
- Hiester, B.G.; Bourke, A.M.; Sinnen, B.L.; Cook, S.G.; Gibson, E.S.; Smith, K.R.; Kennedy, M.J. L-Type Voltage-Gated Ca2+ Channels Regulate Synaptic-Activity-Triggered Recycling Endosome Fusion in Neuronal Dendrites. Cell Rep. 2017, 21, 2134–2146. [Google Scholar] [CrossRef]
- Davies, A.; Douglas, L.; Hendrich, J.; Wratten, J.; Tran Van Minh, A.; Foucault, I.; Koch, D.; Pratt, W.S.; Saibil, H.R.; Dolphin, A.C. The Calcium Channel A2δ-2 Subunit Partitions with CaV2.1 into Lipid Rafts in Cerebellum: Implications for Localization and Function. J. Neurosci. 2006, 26, 8748–8757. [Google Scholar] [CrossRef]
- Spencer, A.; Yu, L.; Guili, V.; Reynaud, F.; Ding, Y.; Ma, J.; Jullien, J.; Koubi, D.; Gauthier, E.; Cluet, D.; et al. Nerve Growth Factor Signaling from Membrane Microdomains to the Nucleus: Differential Regulation by Caveolins. Int. J. Mol. Sci. 2017, 18, 693. [Google Scholar] [CrossRef] [PubMed]
- Bilderback, T.R.; Gazula, V.R.; Lisanti, M.P.; Dobrowsky, R.T. Caveolin Interacts with Trk A and P75(NTR) and Regulates Neurotrophin Signaling Pathways. J. Biol. Chem. 1999, 274, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.S.; Zhou, J.; Feng, A.K.; Lynch, C.C.; Klumperman, J.; DeArmond, S.J.; Mobley, W.C. Nerve Growth Factor Signaling in Caveolae-like Domains at the Plasma Membrane. J. Biol. Chem. 1999, 274, 36707–36714. [Google Scholar] [CrossRef] [PubMed]
- Campbell, D.L.; Stamler, J.S.; Strauss, H.C. Redox Modulation of L-Type Calcium Channels in Ferret Ventricular Myocytes. Dual Mechanism Regulation by Nitric Oxide and S-Nitrosothiols. J. Gen. Physiol. 1996, 108, 277–293. [Google Scholar] [CrossRef]
- Toselli, M.; Biella, G.; Taglietti, V.; Cazzaniga, E.; Parenti, M. Caveolin-1 Expression and Membrane Cholesterol Content Modulate N-Type Calcium Channel Activity in NG108-15 Cells. Biophys. J. 2005, 89, 2443–2457. [Google Scholar] [CrossRef]
- Sepúlveda, M.R.; Berrocal-Carrillo, M.; Gasset, M.; Mata, A.M. The Plasma Membrane Ca2+-ATPase Isoform 4 Is Localized in Lipid Rafts of Cerebellum Synaptic Plasma Membranes. J. Biol. Chem. 2006, 281, 447–453. [Google Scholar] [CrossRef]
- Lopreiato, R.; Giacomello, M.; Carafoli, E. The Plasma Membrane Calcium Pump: New Ways to Look at an Old Enzyme. J. Biol. Chem. 2014, 289, 10261–10268. [Google Scholar] [CrossRef]
- Hirama, T.; Das, R.; Yang, Y.; Ferguson, C.; Won, A.; Yip, C.M.; Kay, J.G.; Grinstein, S.; Parton, R.G.; Fairn, G.D. Phosphatidylserine Dictates the Assembly and Dynamics of Caveolae in the Plasma Membrane. J. Biol. Chem. 2017, 292, 14292–14307. [Google Scholar] [CrossRef]
- Kagan, V.E.; Fabisiak, J.P.; Shvedova, A.A.; Tyurina, Y.Y.; Tyurin, V.A.; Schor, N.F.; Kawai, K. Oxidative Signaling Pathway for Externalization of Plasma Membrane Phosphatidylserine during Apoptosis. FEBS Lett. 2000, 477, 1–7. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, P.; Zhang, X. Phosphatidylserine Externalization in Caveolae Inhibits Ca2+ Efflux through Plasma Membrane Ca2+-ATPase in ECV304. Cell Calcium 2009, 45, 177–184. [Google Scholar] [CrossRef]
- Vacca, F.; Amadio, S.; Sancesario, G.; Bernardi, G.; Volonté, C. P2X3 Receptor Localizes into Lipid Rafts in Neuronal Cells. J. Neurosci. Res. 2004, 76, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Q.; Zhu, J.-X.; Wang, Y.; Zhang, X.; Bao, L. CaMKIIα and Caveolin-1 Cooperate to Drive ATP-Induced Membrane Delivery of the P2X3 Receptor. J. Mol. Cell Biol. 2014, 6, 140–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mojsilovic-Petrovic, J.; Jeong, G.-B.; Crocker, A.; Arneja, A.; David, S.; Russell, D.S.; Kalb, R.G. Protecting Motor Neurons from Toxic Insult by Antagonism of Adenosine A2a and Trk Receptors. J. Neurosci. 2006, 26, 9250–9263. [Google Scholar] [CrossRef]
- North, R.A. P2X3 Receptors and Peripheral Pain Mechanisms. J. Physiol. 2004, 554, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Wirkner, K.; Sperlagh, B.; Illes, P. P2X3 Receptor Involvement in Pain States. Mol. Neurobiol. 2007, 36, 165–183. [Google Scholar] [CrossRef]
- Liu, M.; Huang, W.; Wu, D.; Priestley, J.V. TRPV1, but Not P2X3, Requires Cholesterol for Its Function and Membrane Expression in Rat Nociceptors. Eur. J. Neurosci. 2006, 24, 1–6. [Google Scholar] [CrossRef]
- Frick, M.; Bright, N.A.; Riento, K.; Bray, A.; Merrified, C.; Nichols, B.J. Coassembly of Flotillins Induces Formation of Membrane Microdomains, Membrane Curvature, and Vesicle Budding. Curr. Biol. 2007, 17, 1151–1156. [Google Scholar] [CrossRef]
- Rivera-Milla, E.; Stuermer, C.A.O.; Málaga-Trillo, E. Ancient Origin of Reggie (Flotillin), Reggie-like, and Other Lipid-Raft Proteins: Convergent Evolution of the SPFH Domain. Cell Mol. Life Sci. 2006, 63, 343–357. [Google Scholar] [CrossRef]
- Bickel, P.E.; Scherer, P.E.; Schnitzer, J.E.; Oh, P.; Lisanti, M.P.; Lodish, H.F. Flotillin and Epidermal Surface Antigen Define a New Family of Caveolae-Associated Integral Membrane Proteins. J. Biol. Chem. 1997, 272, 13793–13802. [Google Scholar] [CrossRef]
- Morrow, I.C.; Rea, S.; Martin, S.; Prior, I.A.; Prohaska, R.; Hancock, J.F.; James, D.E.; Parton, R.G. Flotillin-1/Reggie-2 Traffics to Surface Raft Domains via a Novel Golgi-Independent Pathway. Identification of a Novel Membrane Targeting Domain and a Role for Palmitoylation. J. Biol. Chem. 2002, 277, 48834–48841. [Google Scholar] [CrossRef]
- Kokubo, H.; Helms, J.B.; Ohno-Iwashita, Y.; Shimada, Y.; Horikoshi, Y.; Yamaguchi, H. Ultrastructural Localization of Flotillin-1 to Cholesterol-Rich Membrane Microdomains, Rafts, in Rat Brain Tissue. Brain Res. 2003, 965, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, H.; Matsui, I. The Lipid Raft Markers Stomatin, Prohibitin, Flotillin, and HflK/C (SPFH)-Domain Proteins Form an Operon with NfeD Proteins and Function with Apolar Polyisoprenoid Lipids. Crit. Rev. Microbiol. 2020, 46, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Lapatsina, L.; Brand, J.; Poole, K.; Daumke, O.; Lewin, G.R. Stomatin-Domain Proteins. Eur. J. Cell Biol. 2012, 91, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Model, K.; Langer, T. Formation of Membrane-Bound Ring Complexes by Prohibitins in Mitochondria. Mol. Biol. Cell 2005, 16, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Daumke, O.; Lewin, G.R. SPFH Protein Cage—One Ring to Rule Them All. Cell Res. 2022, 32, 117–118. [Google Scholar] [CrossRef]
- Li, Y.; Martin, B.R.; Cravatt, B.F.; Hofmann, S.L. DHHC5 Protein Palmitoylates Flotillin-2 and Is Rapidly Degraded on Induction of Neuronal Differentiation in Cultured Cells. J. Biol. Chem. 2012, 287, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Strauss, K.; Goebel, C.; Runz, H.; Möbius, W.; Weiss, S.; Feussner, I.; Simons, M.; Schneider, A. Exosome Secretion Ameliorates Lysosomal Storage of Cholesterol in Niemann-Pick Type C Disease. J. Biol. Chem. 2010, 285, 26279–26288. [Google Scholar] [CrossRef]
- Solis, G.P.; Hoegg, M.; Munderloh, C.; Schrock, Y.; Malaga-Trillo, E.; Rivera-Milla, E.; Stuermer, C.A.O. Reggie/Flotillin Proteins Are Organized into Stable Tetramers in Membrane Microdomains. Biochem. J. 2007, 403, 313–322. [Google Scholar] [CrossRef]
- Riento, K.; Frick, M.; Schafer, I.; Nichols, B.J. Endocytosis of Flotillin-1 and Flotillin-2 Is Regulated by Fyn Kinase. J. Cell Sci. 2009, 122, 912–918. [Google Scholar] [CrossRef]
- Neumann-Giesen, C.; Fernow, I.; Amaddii, M.; Tikkanen, R. Role of EGF-Induced Tyrosine Phosphorylation of Reggie-1/Flotillin-2 in Cell Spreading and Signaling to the Actin Cytoskeleton. J. Cell Sci. 2007, 120, 395–406. [Google Scholar] [CrossRef]
- Swanwick, C.C.; Shapiro, M.E.; Vicini, S.; Wenthold, R.J. Flotillin-1 Promotes Formation of Glutamatergic Synapses in Hippocampal Neurons. Dev. Neurobiol. 2010, 70, 875–883. [Google Scholar] [CrossRef]
- Munderloh, C.; Solis, G.P.; Bodrikov, V.; Jaeger, F.A.; Wiechers, M.; Málaga-Trillo, E.; Stuermer, C.A.O. Reggies/Flotillins Regulate Retinal Axon Regeneration in the Zebrafish Optic Nerve and Differentiation of Hippocampal and N2a Neurons. J. Neurosci. 2009, 29, 6607–6615. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.C.; Solis, G.P.; Bodrikov, V.; Michel, U.; Haralampieva, D.; Shypitsyna, A.; Tönges, L.; Bähr, M.; Lingor, P.; Stuermer, C.A.O. Upregulation of Reggie-1/Flotillin-2 Promotes Axon Regeneration in the Rat Optic Nerve in Vivo and Neurite Growth in Vitro. Neurobiol. Dis. 2013, 51, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Stuermer, C.A.O. The Reggie/Flotillin Connection to Growth. Trends Cell Biol. 2010, 20, 6–13. [Google Scholar] [CrossRef]
- Kwiatkowska, K.; Matveichuk, O.V.; Fronk, J.; Ciesielska, A. Flotillins: At the Intersection of Protein S-Palmitoylation and Lipid-Mediated Signaling. Int. J. Mol. Sci. 2020, 21, 2283. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, E.; Comunale, F.; Do Khoa, N.; Planchon, D.; Bodin, S.; Gauthier-Rouvière, C. Flotillin Microdomains Stabilize Cadherins at Cell-Cell Junctions. J. Cell Sci. 2013, 126, 5293–5304. [Google Scholar] [CrossRef]
- Bodin, S.; Planchon, D.; Rios Morris, E.; Comunale, F.; Gauthier-Rouvière, C. Flotillins in Intercellular Adhesion-from Cellular Physiology to Human Diseases. J. Cell Sci. 2014, 127, 5139–5147. [Google Scholar] [CrossRef]
- Seong, E.; Yuan, L.; Arikkath, J. Cadherins and Catenins in Dendrite and Synapse Morphogenesis. Cell Adhes. Migr. 2015, 9, 202–213. [Google Scholar] [CrossRef]
- Meister, M.; Tikkanen, R. Endocytic Trafficking of Membrane-Bound Cargo: A Flotillin Point of View. Membranes 2014, 4, 356–371. [Google Scholar] [CrossRef]
- Otto, G.P.; Nichols, B.J. The Roles of Flotillin Microdomains--Endocytosis and Beyond. J. Cell Sci. 2011, 124, 3933–3940. [Google Scholar] [CrossRef]
- Hu, J.; Gao, Y.; Huang, Q.; Wang, Y.; Mo, X.; Wang, P.; Zhang, Y.; Xie, C.; Li, D.; Yao, J. Flotillin-1 Interacts With and Sustains the Surface Levels of TRPV2 Channel. Front. Cell Dev. Biol. 2021, 9, 634160. [Google Scholar] [CrossRef]
- Dietzen, D.J.; Hastings, W.R.; Lublin, D.M. Caveolin Is Palmitoylated on Multiple Cysteine Residues: Palmitoylation IS Not Necessary for Localization of Caveolin to Caveolae. J. Biol. Chem. 1995, 270, 6838–6842. [Google Scholar] [CrossRef]
- Parat, M.-O.; Fox, P.L. Palmitoylation of Caveolin-1 in Endothelial Cells Is Post-Translational but Irreversible. J. Biol. Chem. 2001, 276, 15776–15782. [Google Scholar] [CrossRef] [PubMed]
- Südhof, T.C.; Rizo, J. Synaptic Vesicle Exocytosis. Cold Spring Harb. Perspect. Biol. 2011, 3, a005637. [Google Scholar] [CrossRef] [PubMed]
- Ogunmowo, T.H.; Jing, H.; Raychaudhuri, S.; Kusick, G.F.; Imoto, Y.; Li, S.; Itoh, K.; Ma, Y.; Jafri, H.; Dalva, M.B.; et al. Membrane Compression by Synaptic Vesicle Exocytosis Triggers Ultrafast Endocytosis. Nat. Commun. 2023, 14, 2888. [Google Scholar] [CrossRef]
- Braun, J.E.A.; Madison, D.V. A Novel SNAP25–Caveolin Complex Correlates with the Onset of Persistent Synaptic Potentiation. J. Neurosci. 2000, 20, 5997–6006. [Google Scholar] [CrossRef] [PubMed]
- Pombo, I.; Rivera, J.; Blank, U. Munc18-2/Syntaxin3 Complexes Are Spatially Separated from Syntaxin3-Containing SNARE Complexes. FEBS Lett. 2003, 550, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tu, J.; Zhang, S.; Cai, B.; Liu, Z.; Hou, S.; Zhong, Q.; Hu, X.; Liu, W.; Li, G.; et al. Different Regions of Synaptic Vesicle Membrane Regulate VAMP2 Conformation for the SNARE Assembly. Nat. Commun. 2020, 11, 1531. [Google Scholar] [CrossRef]
- Glebov, O.O.; Bright, N.A.; Nichols, B.J. Flotillin-1 Defines a Clathrin-Independent Endocytic Pathway in Mammalian Cells. Nat. Cell Biol. 2006, 8, 46–54. [Google Scholar] [CrossRef]
- Taverna, E.; Saba, E.; Rowe, J.; Francolini, M.; Clementi, F.; Rosa, P. Role of Lipid Microdomains in P/Q-Type Calcium Channel (Cav2.1) Clustering and Function in Presynaptic Membranes. J. Biol. Chem. 2004, 279, 5127–5134. [Google Scholar] [CrossRef]
- Davies, A.; Kadurin, I.; Alvarez-Laviada, A.; Douglas, L.; Nieto-Rostro, M.; Bauer, C.S.; Pratt, W.S.; Dolphin, A.C. The A2δ Subunits of Voltage-Gated Calcium Channels Form GPI-Anchored Proteins, a Posttranslational Modification Essential for Function. Proc. Natl. Acad. Sci. USA 2010, 107, 1654–1659. [Google Scholar] [CrossRef]
- Swanwick, C.C.; Shapiro, M.E.; Yi, Z.; Chang, K.; Wenthold, R.J. NMDA Receptors Interact with Flotillin-1 and -2, Lipid Raft-Associated Proteins. FEBS Lett. 2009, 583, 1226–1230. [Google Scholar] [CrossRef]
- Simons, K.; Ikonen, E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Abulrob, A.; Tauskela, J.S.; Mealing, G.; Brunette, E.; Faid, K.; Stanimirovic, D. Protection by Cholesterol-Extracting Cyclodextrins: A Role for N-Methyl-d-Aspartate Receptor Redistribution. J. Neurochem. 2005, 92, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Arenas, F.; Garcia-Ruiz, C.; Fernandez-Checa, J.C. Intracellular Cholesterol Trafficking and Impact in Neurodegeneration. Front. Mol. Neurosci. 2017, 10, 382. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Nelson, A.R.; Sagare, A.P.; Zlokovic, B.V. Impaired Vascular-Mediated Clearance of Brain Amyloid Beta in Alzheimer’s Disease: The Role, Regulation and Restoration of LRP1. Front. Aging Neurosci. 2015, 7, 136. [Google Scholar] [CrossRef]
- Beffert, U.; Stolt, P.C.; Herz, J. Functions of Lipoprotein Receptors in Neurons. J. Lipid Res. 2004, 45, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, C.; Kulik, A.; Frotscher, M.; Herz, J.; Schäfer, M.; Bock, H.H.; May, P. Low Density Lipoprotein Receptor-Related Protein 1 (LRP1) Modulates N-Methyl-D-Aspartate (NMDA) Receptor-Dependent Intracellular Signaling and NMDA-Induced Regulation of Postsynaptic Protein Complexes. J. Biol. Chem. 2013, 288, 21909–21923. [Google Scholar] [CrossRef]
- Bodrikov, V.; Pauschert, A.; Kochlamazashvili, G.; Stuermer, C.A.O. Reggie-1 and Reggie-2 (Flotillins) Participate in Rab11a-Dependent Cargo Trafficking, Spine Synapse Formation and LTP-Related AMPA Receptor (GluA1) Surface Exposure in Mouse Hippocampal Neurons. Exp. Neurol. 2017, 289, 31–45. [Google Scholar] [CrossRef]
- Rajakulendran, S.; Hanna, M.G. The Role of Calcium Channels in Epilepsy. Cold Spring Harb. Perspect. Med. 2016, 6, a022723. [Google Scholar] [CrossRef]
- Arikkath, J.; Campbell, K.P. Auxiliary Subunits: Essential Components of the Voltage-Gated Calcium Channel Complex. Curr. Opin. Neurobiol. 2003, 13, 298–307. [Google Scholar] [CrossRef]
- Schlick, B.; Flucher, B.E.; Obermair, G.J. Voltage-Activated Calcium Channel Expression Profiles in Mouse Brain and Cultured Hippocampal Neurons. Neuroscience 2010, 167, 786–798. [Google Scholar] [CrossRef]
- Catterall, W.A. Voltage-Gated Calcium Channels. Cold Spring Harb. Perspect. Biol. 2011, 3, a003947. [Google Scholar] [CrossRef]
- Ilic, K.; Lin, X.; Malci, A.; Stojanović, M.; Puljko, B.; Rožman, M.; Vukelić, Ž.; Heffer, M.; Montag, D.; Schnaar, R.L.; et al. Plasma Membrane Calcium ATPase-Neuroplastin Complexes Are Selectively Stabilized in GM1-Containing Lipid Rafts. Int. J. Mol. Sci. 2021, 22, 13590. [Google Scholar] [CrossRef]
- Jiang, L.; Fernandes, D.; Mehta, N.; Bean, J.L.; Michaelis, M.L.; Zaidi, A. Partitioning of the Plasma Membrane Ca2+-ATPase into Lipid Rafts in Primary Neurons: Effects of Cholesterol Depletion. J. Neurochem. 2007, 102, 378–388. [Google Scholar] [CrossRef]
- Stuermer, C.A.; Lang, D.M.; Kirsch, F.; Wiechers, M.; Deininger, S.O.; Plattner, H. Glycosylphosphatidyl Inositol-Anchored Proteins and Fyn Kinase Assemble in Noncaveolar Plasma Membrane Microdomains Defined by Reggie-1 and -2. Mol. Biol. Cell 2001, 12, 3031–3045. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, D.N.; Min, W.; Gong, Y.; Heng, Y.M.; Boggs, J.M. Two Types of Detergent-Insoluble, Glycosphingolipid/Cholesterol-Rich Membrane Domains from Isolated Myelin. J. Neurochem. 2005, 94, 1696–1710. [Google Scholar] [CrossRef] [PubMed]
- del Toro, D.; Xifró, X.; Pol, A.; Humbert, S.; Saudou, F.; Canals, J.M.; Alberch, J. Altered Cholesterol Homeostasis Contributes to Enhanced Excitotoxicity in Huntington’s Disease. J. Neurochem. 2010, 115, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Sonnino, S.; Chiricozzi, E.; Grassi, S.; Mauri, L.; Prioni, S.; Prinetti, A. Chapter Three-Gangliosides in Membrane Organization. In Progress in Molecular Biology and Translational Science; Schnaar, R.L., Lopez, P.H.H., Eds.; Gangliosides in Health and Disease; Academic Press: Cambridge, MA, USA, 2018; Volume 156, pp. 83–120. [Google Scholar]
- Prinetti, A.; Chigorno, V.; Tettamanti, G.; Sonnino, S. Sphingolipid-Enriched Membrane Domains from Rat Cerebellar Granule Cells Differentiated in Culture: A Compositional Study. J. Biol. Chem. 2000, 275, 11658–11665. [Google Scholar] [CrossRef] [PubMed]
- Vyas, K.A.; Patel, H.V.; Vyas, A.A.; Schnaar, R.L. Segregation of Gangliosides GM1 and GD3 on Cell Membranes, Isolated Membrane Rafts, and Defined Supported Lipid Monolayers. Biol. Chem. 2001, 382, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J. Lipid Rafts and Human Diseases: Why We Need to Target Gangliosides. FEBS Open Bio 2023, 13, 1636–1650. [Google Scholar] [CrossRef]
- Díaz, M.; de Pablo, D.P.; Valdés-Baizabal, C.; Santos, G.; Marin, R. Molecular and Biophysical Features of Hippocampal “Lipid Rafts Aging” Are Modified by Dietary N-3 Long-chain Polyunsaturated Fatty Acids. Aging Cell 2023, 22, e13867. [Google Scholar] [CrossRef] [PubMed]
- Azzaz, F.; Chahinian, H.; Yahi, N.; Fantini, J.; Di Scala, C. AmyP53 Prevents the Formation of Neurotoxic β-Amyloid Oligomers through an Unprecedent Mechanism of Interaction with Gangliosides: Insights for Alzheimer’s Disease Therapy. Int. J. Mol. Sci. 2023, 24, 1760. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Fantini, J. Deciphering the Glycolipid Code of Alzheimer’s and Parkinson’s Amyloid Proteins Allowed the Creation of a Universal Ganglioside-Binding Peptide. PLoS ONE 2014, 9, e104751. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Yahi, N. The Driving Force of Alpha-Synuclein Insertion and Amyloid Channel Formation in the Plasma Membrane of Neural Cells: Key Role of Ganglioside- and Cholesterol-Binding Domains. Adv. Exp. Med. Biol. 2013, 991, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Mahfoud, R.; Garmy, N.; Maresca, M.; Yahi, N.; Puigserver, A.; Fantini, J. Identification of a Common Sphingolipid-Binding Domain in Alzheimer, Prion, and HIV-1 Proteins. J. Biol. Chem. 2002, 277, 11292–11296. [Google Scholar] [CrossRef]
- Di Scala, C.; Yahi, N.; Boutemeur, S.; Flores, A.; Rodriguez, L.; Chahinian, H.; Fantini, J. Common Molecular Mechanism of Amyloid Pore Formation by Alzheimer’s β-Amyloid Peptide and α-Synuclein. Sci. Rep. 2016, 6, 28781. [Google Scholar] [CrossRef]
- Sahu, S.K.; Saxena, R.; Chattopadhyay, A. Cholesterol Depletion Modulates Detergent Resistant Fraction of Human serotonin1A Receptors. Mol. Membr. Biol. 2012, 29, 290–298. [Google Scholar] [CrossRef]
- Gupta, V.K.; Sharma, N.S.; Kesh, K.; Dauer, P.; Nomura, A.; Giri, B.; Dudeja, V.; Banerjee, S.; Bhattacharya, S.; Saluja, A.; et al. Metastasis and Chemoresistance in CD133 Expressing Pancreatic Cancer Cells Are Dependent on Their Lipid Raft Integrity. Cancer Lett. 2018, 439, 101–112. [Google Scholar] [CrossRef]
- Brügger, B.; Glass, B.; Haberkant, P.; Leibrecht, I.; Wieland, F.T.; Kräusslich, H.-G. The HIV Lipidome: A Raft with an Unusual Composition. Proc. Natl. Acad. Sci. USA 2006, 103, 2641–2646. [Google Scholar] [CrossRef]
- Mergia, A. The Role of Caveolin 1 in HIV Infection and Pathogenesis. Viruses 2017, 9, 129. [Google Scholar] [CrossRef]
- Sahay, B.; Mergia, A. The Potential Contribution of Caveolin 1 to HIV Latent Infection. Pathogens 2020, 9, 896. Available online: https://www.mdpi.com/2076-0817/9/11/896 (accessed on 26 July 2023). [CrossRef]
- Gisslén, M.; Hagberg, L.; Norkrans, G.; Lekman, A.; Fredman, P. Increased Cerebrospinal Fluid Ganglioside GM1 Concentrations Indicating Neuronal Involvement in All Stages of HIV-1 Infection. J. Neurovirol 1997, 3, 148–152. [Google Scholar] [CrossRef]
- Ledeen, R.W.; Wu, G. Ganglioside Function in Calcium Homeostasis and Signaling. Neurochem. Res. 2002, 27, 637–647. [Google Scholar] [CrossRef]
- Jiang, L.; Bechtel, M.D.; Bean, J.L.; Winefield, R.; Williams, T.D.; Zaidi, A.; Michaelis, E.K.; Michaelis, M.L. Effects of Gangliosides on the Activity of the Plasma Membrane Ca2+-ATPase. Biochim. Biophys. Acta 2014, 1838, 1255–1265. [Google Scholar] [CrossRef]
- Zhao, Y.; Fan, X.; Yang, F.; Zhang, X. Gangliosides Modulate the Activity of the Plasma Membrane Ca2+-ATPase from Porcine Brain Synaptosomes. Arch. Biochem. Biophys. 2004, 427, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Bechtel, M.D.; Galeva, N.A.; Williams, T.D.; Michaelis, E.K.; Michaelis, M.L. Decreases in Plasma Membrane Ca2+-ATPase in Brain Synaptic Membrane Rafts from Aged Rats. J. Neurochem. 2012, 123, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Colina, C.; Cervino, V.; Benaim, G. Ceramide and Sphingosine Have an Antagonistic Effect on the Plasma-Membrane Ca2+-ATPase from Human Erythrocytes. Biochem. J. 2002, 362, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, Y.; Duan, J.; Yang, F.; Zhang, X. Gangliosides Activate the Phosphatase Activity of the Erythrocyte Plasma Membrane Ca2+-ATPase. Arch. Biochem. Biophys. 2005, 444, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R.O.; Masco, D.; Brooker, G.; Spiegel, S. Endogenous Ganglioside GM1 Modulates L-Type Calcium Channel Activity in N18 Neuroblastoma Cells. J. Neurosci. 1994, 14, 2272–2281. [Google Scholar] [CrossRef]
- Frank, C.; Giammarioli, A.M.; Pepponi, R.; Fiorentini, C.; Rufini, S. Cholesterol Perturbing Agents Inhibit NMDA-Dependent Calcium Influx in Rat Hippocampal Primary Culture. FEBS Lett. 2004, 566, 25–29. [Google Scholar] [CrossRef]
- de Erausquin, G.A.; Manev, H.; Guidotti, A.; Costa, E.; Brooker, G. Gangliosides Normalize Distorted Single-Cell Intracellular Free Ca2+ Dynamics after Toxic Doses of Glutamate in Cerebellar Granule Cells. Proc. Natl. Acad. Sci. USA 1990, 87, 8017–8021. [Google Scholar] [CrossRef] [PubMed]
- Manev, H.; Favaron, M.; Vicini, S.; Guidotti, A.; Costa, E. Glutamate-Induced Neuronal Death in Primary Cultures of Cerebellar Granule Cells: Protection by Synthetic Derivatives of Endogenous Sphingolipids. J. Pharmacol. Exp. Ther. 1990, 252, 419–427. [Google Scholar] [PubMed]
- Costa, E.; Armstrong, D.M.; Guidotti, A.; Kharlamov, A.; Kiedrowski, L.; Manev, H.; Polo, A.; Wroblewski, J.T. Gangliosides in the Protection against Glutamate Excitotoxicity. Prog. Brain Res. 1994, 101, 357–373. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.A.; Dosemeci, A.; Reese, T.S. Co-Segregation of AMPA Receptors with GM1 Ganglioside in Synaptosomal Membrane Sub-Fractions. Biochem. J. 2010, 427, 535–540. [Google Scholar] [CrossRef]
- Kasahara, K.; Watanabe, Y.; Yamamoto, T.; Sanai, Y. Association of Src Family Tyrosine Kinase Lyn with Ganglioside GD3 in Rat Brain. Possible Regulation of Lyn by Glycosphingolipid in Caveolae-like Domains. J. Biol. Chem. 1997, 272, 29947–29953. [Google Scholar] [CrossRef]
- Hayashi, T.; Huganir, R.L. Tyrosine Phosphorylation and Regulation of the AMPA Receptor by SRC Family Tyrosine Kinases. J. Neurosci. 2004, 24, 6152–6160. [Google Scholar] [CrossRef]
- Goncalves, J.; Bartol, T.M.; Camus, C.; Levet, F.; Menegolla, A.P.; Sejnowski, T.J.; Sibarita, J.-B.; Vivaudou, M.; Choquet, D.; Hosy, E. Nanoscale Co-Organization and Coactivation of AMPAR, NMDAR, and mGluR at Excitatory Synapses. Proc. Natl. Acad. Sci. USA 2020, 117, 14503–14511. [Google Scholar] [CrossRef]
- Li, S.; Raychaudhuri, S.; Lee, S.A.; Brockmann, M.M.; Wang, J.; Kusick, G.; Prater, C.; Syed, S.; Falahati, H.; Ramos, R.; et al. Asynchronous Release Sites Align with NMDA Receptors in Mouse Hippocampal Synapses. Nat. Commun. 2021, 12, 677. [Google Scholar] [CrossRef]
- Prendergast, J.; Umanah, G.K.E.; Yoo, S.-W.; Lagerlöf, O.; Motari, M.G.; Cole, R.N.; Huganir, R.L.; Dawson, T.M.; Dawson, V.L.; Schnaar, R.L. Ganglioside Regulation of AMPA Receptor Trafficking. J. Neurosci. 2014, 34, 13246–13258. [Google Scholar] [CrossRef]
- Wu, G.; Ledeen, R.W. Gangliosides as Modulators of Neuronal Calcium. Prog. Brain Res. 1994, 101, 101–112. [Google Scholar] [CrossRef]
- Fang, Y.; Wu, G.; Xie, X.; Lu, Z.H.; Ledeen, R.W. Endogenous GM1 Ganglioside of the Plasma Membrane Promotes Neuritogenesis by Two Mechanisms. Neurochem. Res. 2000, 25, 931–940. [Google Scholar] [CrossRef]
- Wu, G.; Lu, Z.-H.; Nakamura, K.; Spray, D.C.; Ledeen, R.W. Trophic Effect of Cholera Toxin B Subunit in Cultured Cerebellar Granule Neurons: Modulation of Intracellular Calcium by GM1 Ganglioside. J. Neurosci. Res. 1996, 44, 243–254. [Google Scholar] [CrossRef]
- Milani, D.; Minozzi, M.C.; Petrelli, L.; Guidolin, D.; Skaper, S.D.; Spoerri, P.E. Interaction of Ganglioside GM1 with the B Subunit of Cholera Toxin Modulates Intracellular Free Calcium in Sensory Neurons. J. Neurosci. Res. 1992, 33, 466–475. [Google Scholar] [CrossRef]
- Ando, S.; Tanaka, Y.; Waki, H.; Kon, K.; Iwamoto, M.; Fukui, F. Gangliosides and Sialylcholesterol as Modulators of Synaptic Functionsa. Ann. N. Y. Acad. Sci. 1998, 845, 232–239. [Google Scholar] [CrossRef]
- Berg, L.K.; Larsson, M.; Morland, C.; Gundersen, V. Pre- and Postsynaptic Localization of NMDA Receptor Subunits at Hippocampal Mossy Fibre Synapses. Neuroscience 2013, 230, 139–150. [Google Scholar] [CrossRef]
- Nosov, G.; Kahms, M.; Klingauf, J. The Decade of Super-Resolution Microscopy of the Presynapse. Front. Synaptic Neurosci. 2020, 12, 32. [Google Scholar] [CrossRef]
- Nakatani, Y.; Hotta, S.; Utsunomiya, I.; Tanaka, K.; Hoshi, K.; Ariga, T.; Yu, R.K.; Miyatake, T.; Taguchi, K. Cav2.1 Voltage-Dependent Ca2+ Channel Current Is Inhibited by Serum from Select Patients with Guillain-Barré Syndrome. Neurochem. Res. 2009, 34, 149–157. [Google Scholar] [CrossRef]
- Taylor, C.P.; Garrido, R. Immunostaining of Rat Brain, Spinal Cord, Sensory Neurons and Skeletal Muscle for Calcium Channel Alpha2-Delta (Alpha2-Delta) Type 1 Protein. Neuroscience 2008, 155, 510–521. [Google Scholar] [CrossRef]
- Bauer, C.S.; Nieto-Rostro, M.; Rahman, W.; Tran-Van-Minh, A.; Ferron, L.; Douglas, L.; Kadurin, I.; Sri Ranjan, Y.; Fernandez-Alacid, L.; Millar, N.S.; et al. The Increased Trafficking of the Calcium Channel Subunit Alpha2delta-1 to Presynaptic Terminals in Neuropathic Pain Is Inhibited by the Alpha2delta Ligand Pregabalin. J. Neurosci. 2009, 29, 4076–4088. [Google Scholar] [CrossRef]
- Zhang, J.; Diamond, J.S. Distinct Perisynaptic and Synaptic Localization of NMDA and AMPA Receptors on Ganglion Cells in Rat Retina. J. Comp. Neurol. 2006, 498, 810–820. [Google Scholar] [CrossRef]
- Zhang, J.; Diamond, J.S. Subunit- and Pathway-Specific Localization of NMDA Receptors and Scaffolding Proteins at Ganglion Cell Synapses in Rat Retina. J. Neurosci. 2009, 29, 4274–4286. [Google Scholar] [CrossRef]
- Lu, C.-R.; Hwang, S.J.; Phend, K.D.; Rustioni, A.; Valtschanoff, J.G. Primary Afferent Terminals That Express Presynaptic NR1 in Rats Are Mainly from Myelinated, Mechanosensitive Fibers. J. Comp. Neurol. 2003, 460, 191–202. [Google Scholar] [CrossRef]
- Grant, S.G.N. Synapse Signalling Complexes and Networks: Machines Underlying Cognition. Bioessays 2003, 25, 1229–1235. [Google Scholar] [CrossRef]
- Henley, J.; Poo, M. Guiding Neuronal Growth Cones Using Ca2+ Signals. Trends Cell Biol. 2004, 14, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.Q.; Poo, M.-M. Calcium Signaling in Neuronal Motility. Annu. Rev. Cell Dev. Biol. 2007, 23, 375–404. [Google Scholar] [CrossRef]
- Davies, S.N.; Lester, R.A.; Reymann, K.G.; Collingridge, G.L. Temporally Distinct Pre- and Post-Synaptic Mechanisms Maintain Long-Term Potentiation. Nature 1989, 338, 500–503. [Google Scholar] [CrossRef]
- Brown, T.C.; Correia, S.S.; Petrok, C.N.; Esteban, J.A. Functional Compartmentalization of Endosomal Trafficking for the Synaptic Delivery of AMPA Receptors during Long-Term Potentiation. J. Neurosci. 2007, 27, 13311–13315. [Google Scholar] [CrossRef]
- Petralia, R.S.; Wang, Y.-X.; Wenthold, R.J. Internalization at Glutamatergic Synapses during Development. Eur. J. Neurosci. 2003, 18, 3207–3217. [Google Scholar] [CrossRef]
- Vulchanova, L.; Riedl, M.S.; Shuster, S.J.; Buell, G.; Surprenant, A.; North, R.A.; Elde, R. Immunohistochemical Study of the P2X2 and P2X3 Receptor Subunits in Rat and Monkey Sensory Neurons and Their Central Terminals. Neuropharmacology 1997, 36, 1229–1242. [Google Scholar] [CrossRef]
- Llewellyn-Smith, I.J.; Burnstock, G. Ultrastructural Localization of P2X3 Receptors in Rat Sensory Neurons. Neuroreport 1998, 9, 2545–2550. [Google Scholar] [CrossRef]
- Cook, S.P.; Vulchanova, L.; Hargreaves, K.M.; Elde, R.; McCleskey, E.W. Distinct ATP Receptors on Pain-Sensing and Stretch-Sensing Neurons. Nature 1997, 387, 505–508. [Google Scholar] [CrossRef]
- Souslova, V.; Cesare, P.; Ding, Y.; Akopian, A.N.; Stanfa, L.; Suzuki, R.; Carpenter, K.; Dickenson, A.; Boyce, S.; Hill, R.; et al. Warm-Coding Deficits and Aberrant Inflammatory Pain in Mice Lacking P2X3 Receptors. Nature 2000, 407, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- Westover, E.J.; Covey, D.F. The Enantiomer of Cholesterol. J. Membr. Biol. 2004, 202, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Gorospe, B.; Moura, J.J.G.; Gutierrez-Merino, C.; Samhan-Arias, A.K. Biochemical and Biophysical Characterization of the Caveolin-2 Interaction with Membranes and Analysis of the Protein Structural Alteration by the Presence of Cholesterol. Int. J. Mol. Sci. 2022, 23, 15203. [Google Scholar] [CrossRef] [PubMed]
- Levental, I.; Grzybek, M.; Simons, K. Greasing Their Way: Lipid Modifications Determine Protein Association with Membrane Rafts. Biochemistry 2010, 49, 6305–6316. [Google Scholar] [CrossRef]
- Levental, I.; Lingwood, D.; Grzybek, M.; Coskun, U.; Simons, K. Palmitoylation Regulates Raft Affinity for the Majority of Integral Raft Proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 22050–22054. [Google Scholar] [CrossRef]
- Song, Y.; Kenworthy, A.K.; Sanders, C.R. Cholesterol as a Co-Solvent and a Ligand for Membrane Proteins. Protein Sci. 2014, 23, 1–22. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).