Cloning and Functional Characterization of NADPH-Cytochrome P450 Reductases in Aconitum vilmorinianum
Abstract
1. Introduction
2. Results
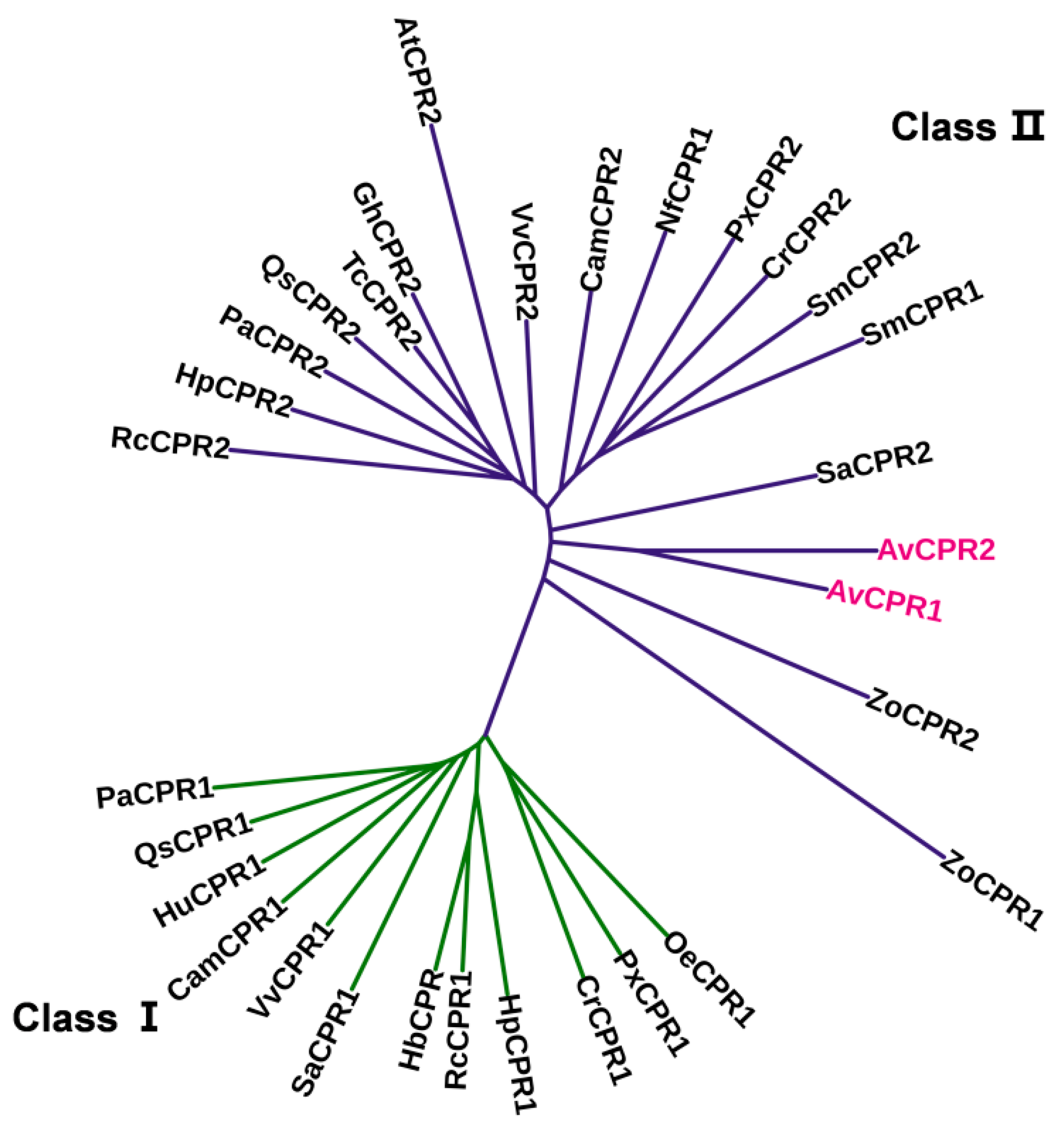
2.1. Cloning and Analysis of AvCPRs
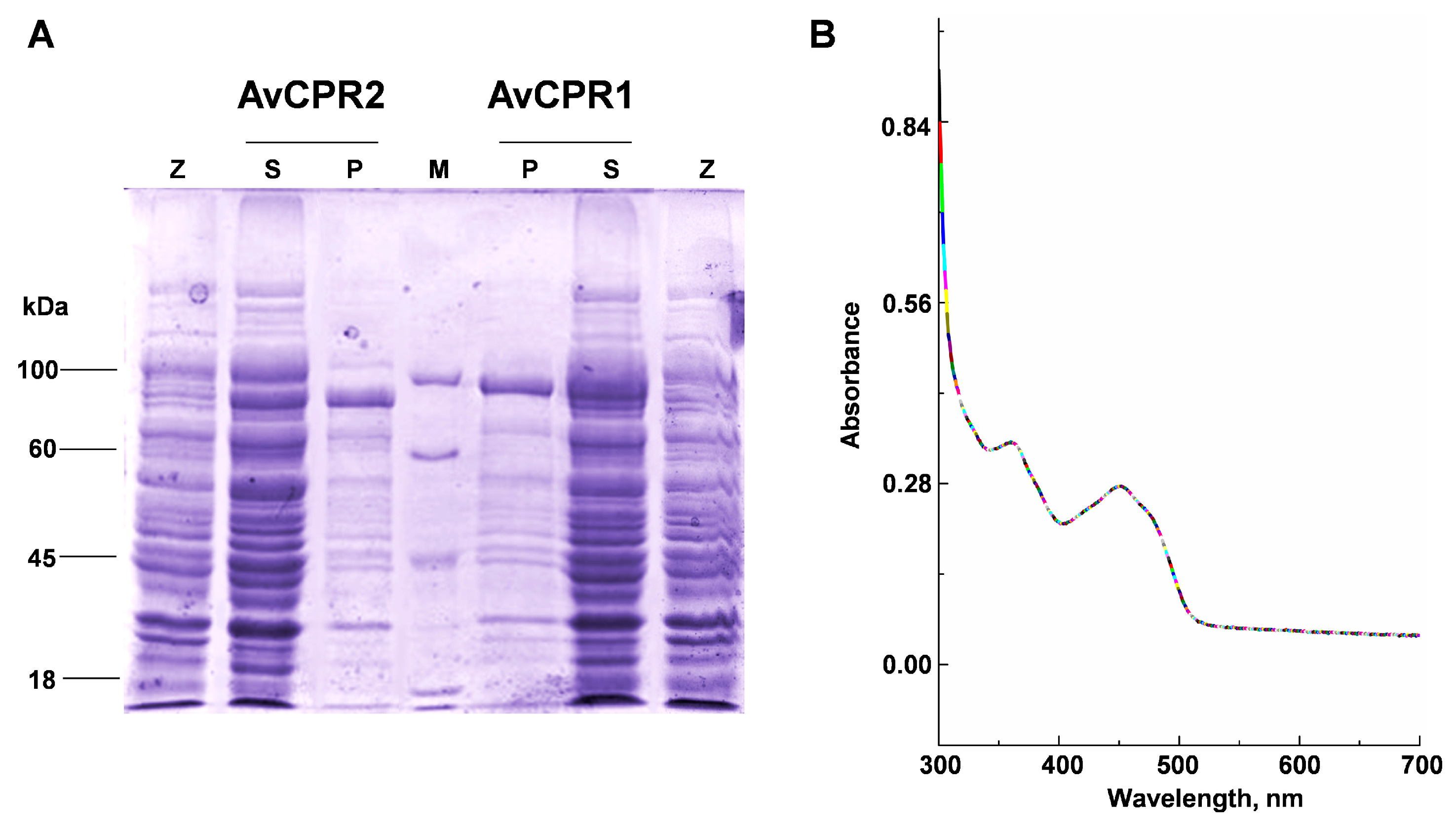
2.2. Catalytic Activities of Recombinant AvCPRs
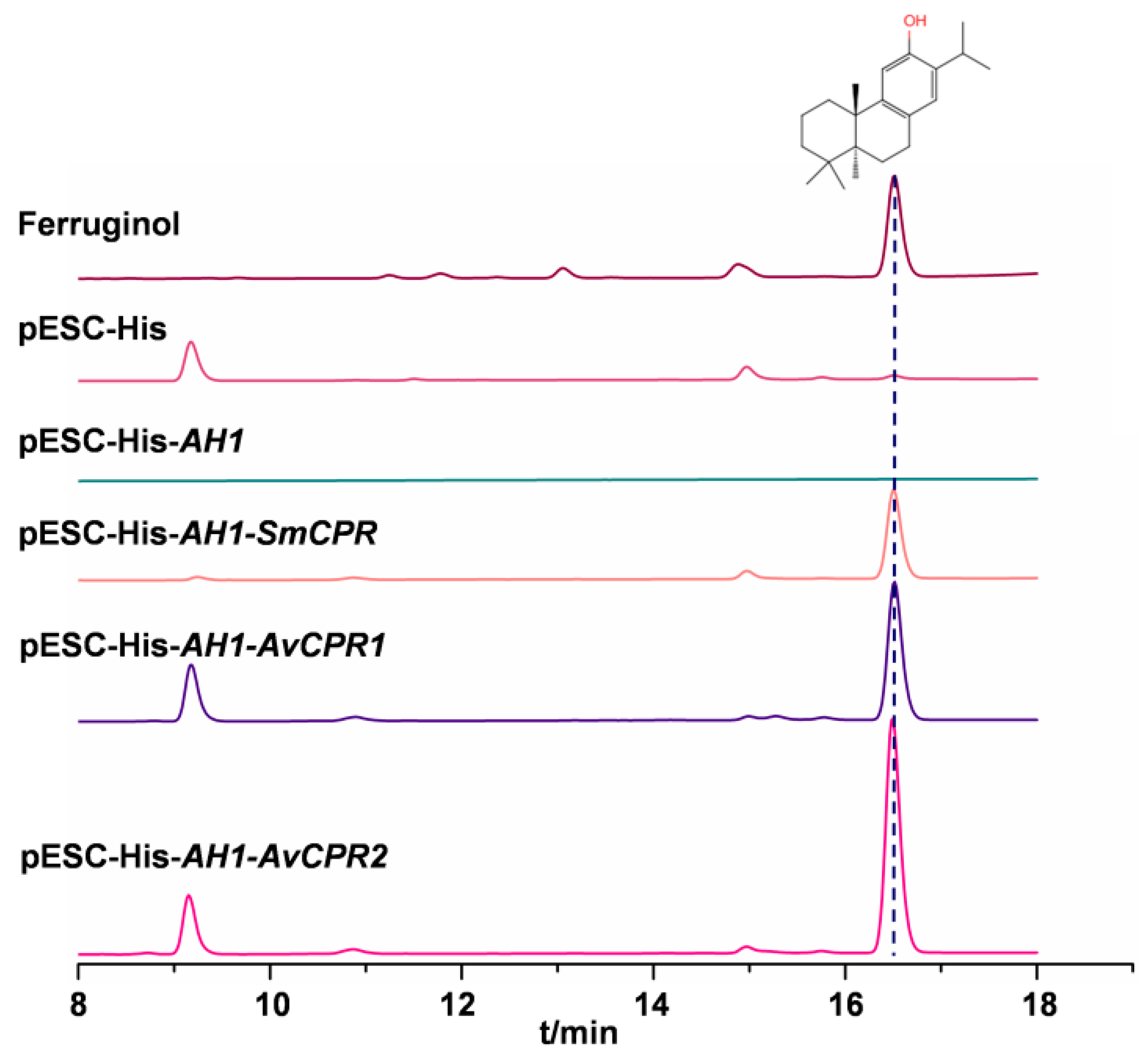
2.3. AvCPR1 and AvCPR2 Supported Heterogeneous P450 Monooxygenase Activity
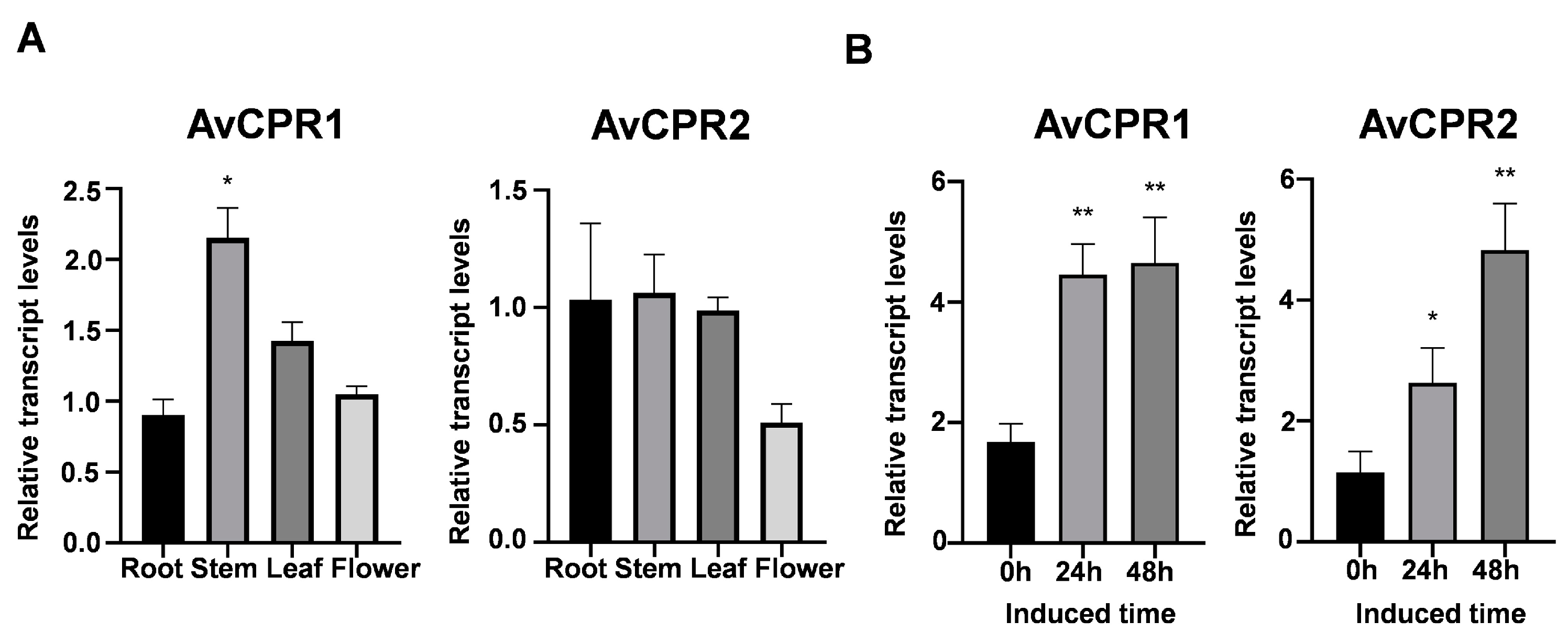
2.4. Expression Profiles of AvCPRs in A. vilmorinianum
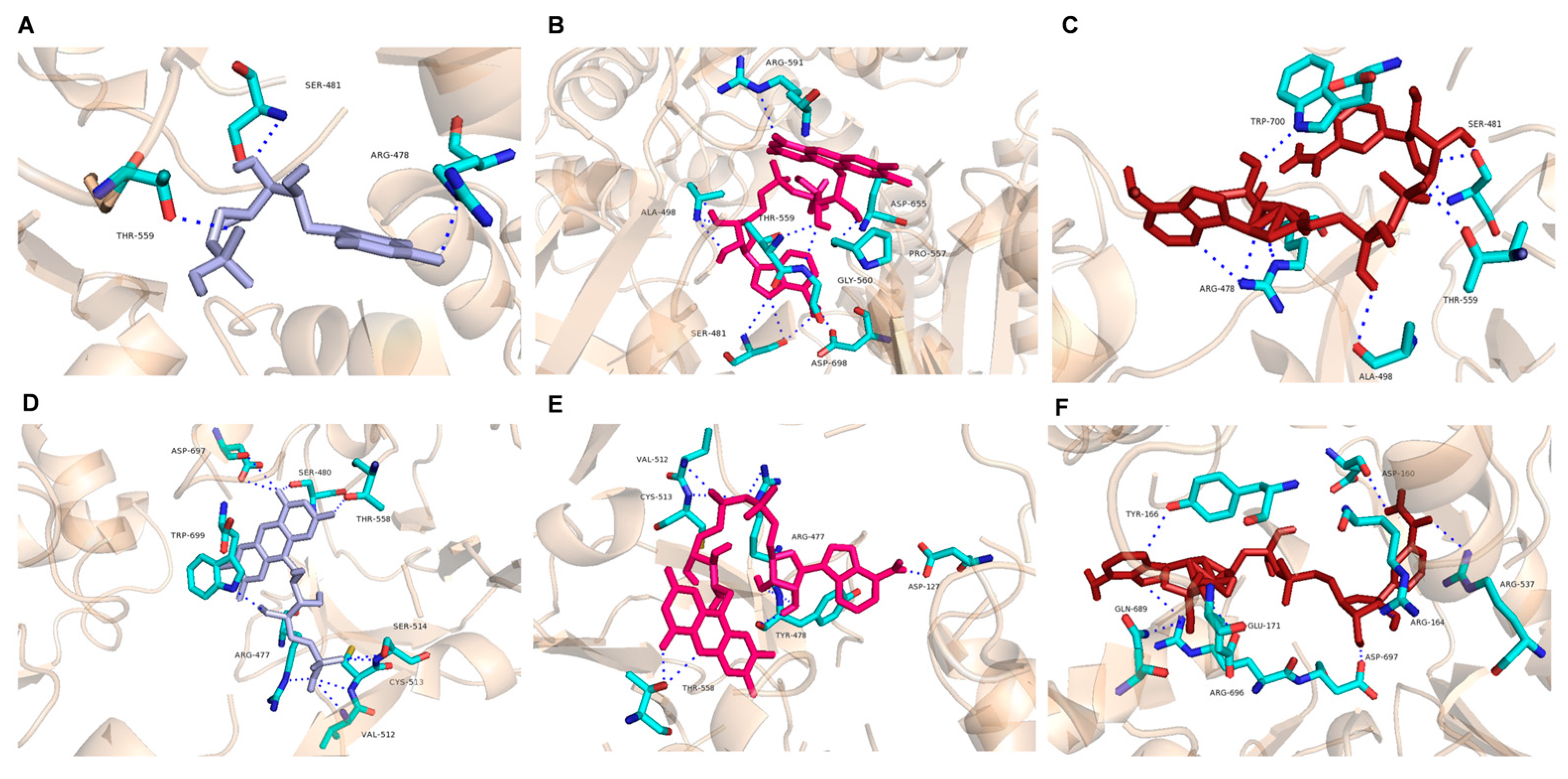
2.5. Docking Analysis of AvCPR1 and AvCPR2
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Total RNA Extraction and cDNA Synthesis
4.3. Molecular Cloning of AvCPRs
4.4. Bioinformatic Analysis of AvCPRs
4.5. Heterologous Expression of AvCPRs in E. coli
4.6. Enzymatic Activity of AvCPRs In Vitro
4.7. Heterologous Expression in Yeast
4.8. Expression Analysis of AvCPRs in A. vilmorinianum
4.9. Homology Modeling and Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flora of China Editorial Committee. Flora of China; Flora of China Editorial Committee: Beijing, China, 1979; Volume 27, p. 245. [Google Scholar]
- Liu, X.Y.; Ke, B.W.; Qin, Y. The diterpenoid alkaloids. Alkaloids Chem. Biol. 2022, 87, 1–360. [Google Scholar] [CrossRef]
- Li, X.P.; He, J.; He, S.L. Research Progress of Aconitum vilmorinianum. J. West China For. Sci. 2017, 46, 1–7. [Google Scholar] [CrossRef]
- Dong, S.; Cheng, C.L.; Zhu, P.F.; Zhou, Z.H.; Ma, X.X. HPLC Determination of Three Diester Diterpenoid Alkaloids of Wild and Cultivated Aconitum Vilmorinianum Radix from Different Producing Areas in Yunnan Province. Chin. J. Inf. TCM 2020, 27, 74–77. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Jeffreys, L.N.; Girvan, H.M.; McLean, K.J. Characterization of cytochrome P450 enzymes and their applications in synthetic biology. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2018; Volume 608, pp. 189–261. [Google Scholar] [CrossRef]
- Jensen, K.; Møller, B.L. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 2010, 71, 132–141. [Google Scholar] [CrossRef]
- Ro, D.K.; Ehlting, J.; Douglas, C.J. Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from Hybrid poplar. Plant Physiol. 2002, 130, 1837–1851. [Google Scholar] [CrossRef]
- Parage, C.; Foureau, E.; Kellner, F. Class II cytochrome P450 reductase governs the biosynthesis of alkaloids. Plant Physiol. 2016, 172, 1563–1577. [Google Scholar] [CrossRef]
- Qi, M.D.; Wang, J.; Ma, X.J. Cloning and functional identification of a new NADPH-cytochrome P450 reductase in Andrographis paniculata. China J. Chin. Mater. Medica 2018, 43, 309–315. [Google Scholar] [CrossRef]
- Lin, H.; Wang, J.; Qi, M. Molecular cloning and functional characterization of multiple NADPH-cytochrome P450 reductases from Andrographis paniculata. Int. J. Biol. Macromol. 2017, 102, 208–217. [Google Scholar] [CrossRef]
- Urban, P.; Mignotte, C.; Kazmaier, M.; Delorme, F.; Pompon, D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 1997, 272, 19176–19186. [Google Scholar] [CrossRef]
- Shet, M.S.; Sathasivan, K.; Arlotto, M.A. Purification, characterization, and cDNA cloning of an NADPH-cytochrome P450 reductase from mung bean. Proc. Natl. Acad. Sci. USA 1993, 90, 2890–2894. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kim, H.M.; Ma, S.H. Heterologous expression and functional characterization of the NADPH-cytochrome P450 reductase from Capsicum annuum. Plant Physiol. Biochem. 2014, 82, 116–122. [Google Scholar] [CrossRef]
- Yang, C.Q.; Lu, S.; Mao, Y.B. Characterization of two NADPH: Cytochrome P450 reductases from cotton (Gossypium hirsutum). Phytochemistry 2010, 71, 27–35. [Google Scholar] [CrossRef]
- Rana, S.; Lattoo, S.K.; Dhar, N. NADPH-cytochrome P450 reductase: Molecular cloning and functional characterization of two paralogs from Withania somnifera (L.) Dunal. PLoS ONE 2013, 8, 57–68. [Google Scholar] [CrossRef]
- Huang, F.C.; Sung, P.H.; Do, Y.Y. Differential expression and functional characterization of the NADPH cytochrome P450 reductase genes from Nothapodytes foetida. Plant Sci. 2012, 190, 16–23. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, C.; Sun, W. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab. Eng. 2018, 45, 43–50. [Google Scholar] [CrossRef]
- Hubbard, P.A.; Shen, A.L.; Paschke, R. NADPH-cytochrome P450 oxidoreductase: Structural basis for hydride and electron transfer. J. Biol. Chem. 2001, 276, 29163–29170. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Gao, W.; Rong, Q. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc. 2012, 134, 3234–3241. [Google Scholar] [CrossRef]
- Chen, L.L.; Mao, L.Y.; Yin, B.W.; Tian, M.; Jin, B.L.; Wei, X.Y.; Huang, L.Q. Chemical constituents in different parts of seven species of Aconitum based on UHPLC-Q-TOF/MS. J. Pharm. Biomed. Anal. 2021, 193, 113713. [Google Scholar] [CrossRef]
- Shephard, E.A.; Phillips, I.R.; Bayney, R.M.; Pike, S.F.; Rabin, B.R. Quantification of NADPH: Cytochrome P-450 reductase in liver microsomes by a specific radioimmunoassay technique. Biochem. J. 1983, 211, 333–340. [Google Scholar] [CrossRef]
- Yabusaki, Y.; Murakami, H.; Ohkawa, H. Primary structure of Saccharomyces cerevisiae NADPH-cytochrome P450 reductase deduced from nucleotide sequence of its cloned gene. J. Biochem. 1988, 103, 1004–1010. [Google Scholar] [CrossRef]
- Porter, T.D.; Kasper, C.B. Coding nucleotide sequence of rat NADPH-cytochrome P-450 oxidoreductase cDNA and identification of flavin-binding domains. Proc. Natl. Acad. Sci. USA 1985, 82, 973–977. [Google Scholar] [CrossRef]
- Eberle, D.; Ullmann, P.; Werck-Reichhart, D.; Petersen, M. cDNA cloning and functional characterisation of CYP98A14 and NADPH: Cytochrome P450 reductase from Coleus blumei involved in rosmarinic acid biosynthesis. Plant Mol. Biol. 2009, 69, 239–253. [Google Scholar] [CrossRef]
- Rosco, A.; Pauli, H.H.; Priesner, W.; Kutchan, T.M. Cloning and heterologous expression of NADPH-cytochrome P450 reductases from the Papaveraceae. Arch. Biochem. Biophys. 1997, 348, 369–377. [Google Scholar] [CrossRef]
- Ponnamperuma, K.; Croteau, R. Purification and characterization of an NADPH-cytochrome P450 (cytochrome c) reductase from spearmint (Mentha spicata) glandular trichomes. Arch. Biochem. Biophys. 1996, 329, 9–16. [Google Scholar] [CrossRef]
- Jin, Z.; Cong, Y.; Zhu, S.; Xing, R.; Zhang, D.; Yao, X.; Yu, F. Two classes of cytochrome P450 reductase genes and their divergent functions in Camptotheca acuminata Decne. Int. J. Biol. Macromol. 2019, 138, 1098–1108. [Google Scholar] [CrossRef]
- Koopmann, E.; Hahlbrock, K. Differentially regulated NADPH: Cytochrome P450 oxidoreductases in parsley. Proc. Natl. Acad. Sci. USA 1997, 94, 14954–14959. [Google Scholar] [CrossRef]
- Liao, J.; Xie, L.; Shi, H.; Cui, S.; Lan, F.; Luo, Z.; Ma, X. Development of an efficient transient expression system for Siraitia grosvenorii fruit and functional characterization of two NADPH-cytochrome P450 reductases. Phytochemistry 2021, 189, 112824. [Google Scholar] [CrossRef]
- Hamdane, D.; Xia, C.; Im, S.C.; Zhang, H.; Kim, J.J.P.; Waskell, L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009, 284, 11374–11384. [Google Scholar] [CrossRef]
- Hong, Y.; Li, H.; Yuan, Y.C.; Chen, S. Sequence-function correlation of aromatase and its interaction with reductase. J. Steroid Biochem. Mol. Biol. 2010, 118, 203–206. [Google Scholar] [CrossRef][Green Version]
- Nadler, S.G.; Strobel, H.W. Role of electrostatic interactions in the reaction of NADPH-cytochrome P-450 reductase with cytochromes P-450. Arch. Biochem. Biophys. 1988, 261, 418–429. [Google Scholar] [CrossRef]
- Zhao, Q.; Modi, S.; Smith, G.; Paine, M.; McDONAGH, P.D.; Wolf, C.R.; Driessen, H.P. Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 Å resolution. Protein Sci. 1999, 8, 298–306. [Google Scholar] [CrossRef]
- Wang, M.; Roberts, D.L.; Paschke, R.; Shea, T.M.; Masters, B.S.S.; Kim, J.J.P. Three-dimensional structure of NADPH-cytochrome P450 reductase: Prototype for FMN-and FAD-containing enzymes. Proc. Natl. Acad. Sci. USA 1997, 94, 8411–8416. [Google Scholar] [CrossRef]
- Park, J.W.; Reed, J.R.; Brignac-Huber, L.M.; Backes, W.L. Cytochrome P450 system proteins reside in different regions of the endoplasmic reticulum. Biochem. J. 2014, 464, 241–249. [Google Scholar] [CrossRef]
- Waskell, L.; Kim, J.-J. Electron transfer partners of cytochrome P450. In Cytochrome P450; Springer: Boston, MA, USA, 2015; pp. 33–68. [Google Scholar]
- Lv, Y.; Marsafari, M.; Koffas, M.; Zhou, J.; Xu, P. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef]
- Istiandari, P.; Yasumoto, S.; Srisawat, P.; Tamura, K.; Chikugo, A.; Suzuki, H.; Muranaka, T. Comparative Analysis of NADPH-Cytochrome P450 Reductases from Legumes for Heterologous Production of Triterpenoids in Transgenic Saccharomyces cerevisiae. Front. Plant Sci. 2021, 12, 762546. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, Y.J.; Hillwig, M.L. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12108–12113. [Google Scholar] [CrossRef]
- Zeng, L.F.; Li, G.D.; Wang, B.J. Selection of optimal qRT-PCR reference genes for Aconitum vilmorinianum. China J. Chin. Mater. Medica 2021, 46, 3116–3122. [Google Scholar] [CrossRef]

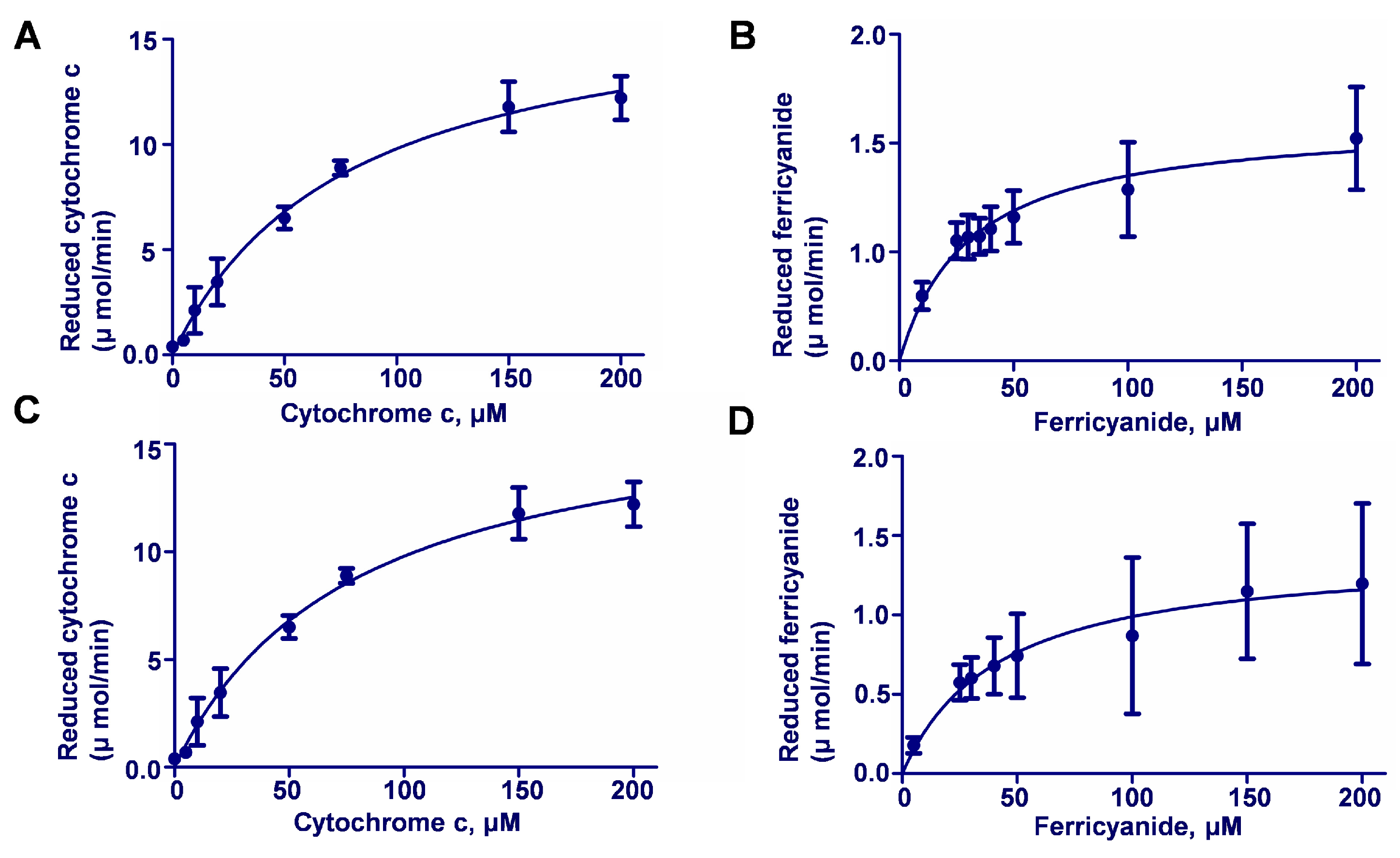
| Vmax (μmol·min−1·mg−1) | Km (μmol·L−1) | K cat (min−1) | ||
|---|---|---|---|---|
| Cytochrome c | AvCPR1 | 24.31 ± 3.55 | 344.20 ± 69.68 | 607.00 ± 88.75 |
| AvCPR2 | 17.44 ± 0.86 | 78.11 ± 9.15 | 174.4 ± 8.60 | |
| K3Fe(CN)6 | AvCPR1 | 1.19 ± 0.11 | 35.68 ± 9.31 | 29.75 ± 2.75 |
| AvCPR2 | 1.37 ± 0.08 | 38.75 ± 5.57 | 13.70 ± 0.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Li, G.; Wang, X.; Yang, C.; Xu, F.; Qian, Z.; Ma, X. Cloning and Functional Characterization of NADPH-Cytochrome P450 Reductases in Aconitum vilmorinianum. Molecules 2023, 28, 7409. https://doi.org/10.3390/molecules28217409
Cheng J, Li G, Wang X, Yang C, Xu F, Qian Z, Ma X. Cloning and Functional Characterization of NADPH-Cytochrome P450 Reductases in Aconitum vilmorinianum. Molecules. 2023; 28(21):7409. https://doi.org/10.3390/molecules28217409
Chicago/Turabian StyleCheng, Jingping, Guodong Li, Xue Wang, Congwei Yang, Furong Xu, Zigang Qian, and Xiaohui Ma. 2023. "Cloning and Functional Characterization of NADPH-Cytochrome P450 Reductases in Aconitum vilmorinianum" Molecules 28, no. 21: 7409. https://doi.org/10.3390/molecules28217409
APA StyleCheng, J., Li, G., Wang, X., Yang, C., Xu, F., Qian, Z., & Ma, X. (2023). Cloning and Functional Characterization of NADPH-Cytochrome P450 Reductases in Aconitum vilmorinianum. Molecules, 28(21), 7409. https://doi.org/10.3390/molecules28217409






