Novel Schiff Base Derived from Amino Pyrene: Synthesis, Characterization, Crystal Structure Determination, and Anticancer Applications of the Ligand and Its Metal Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry and Characterizations
2.1.1. XRD Analysis of HL
2.1.2. NMR Spectral Studies (1H and 13C NMR Spectra)
2.1.3. Infrared Spectra and Coordination Mode
2.1.4. Magnetic Moments and Electronic Spectral Data
2.1.5. Mass Spectra
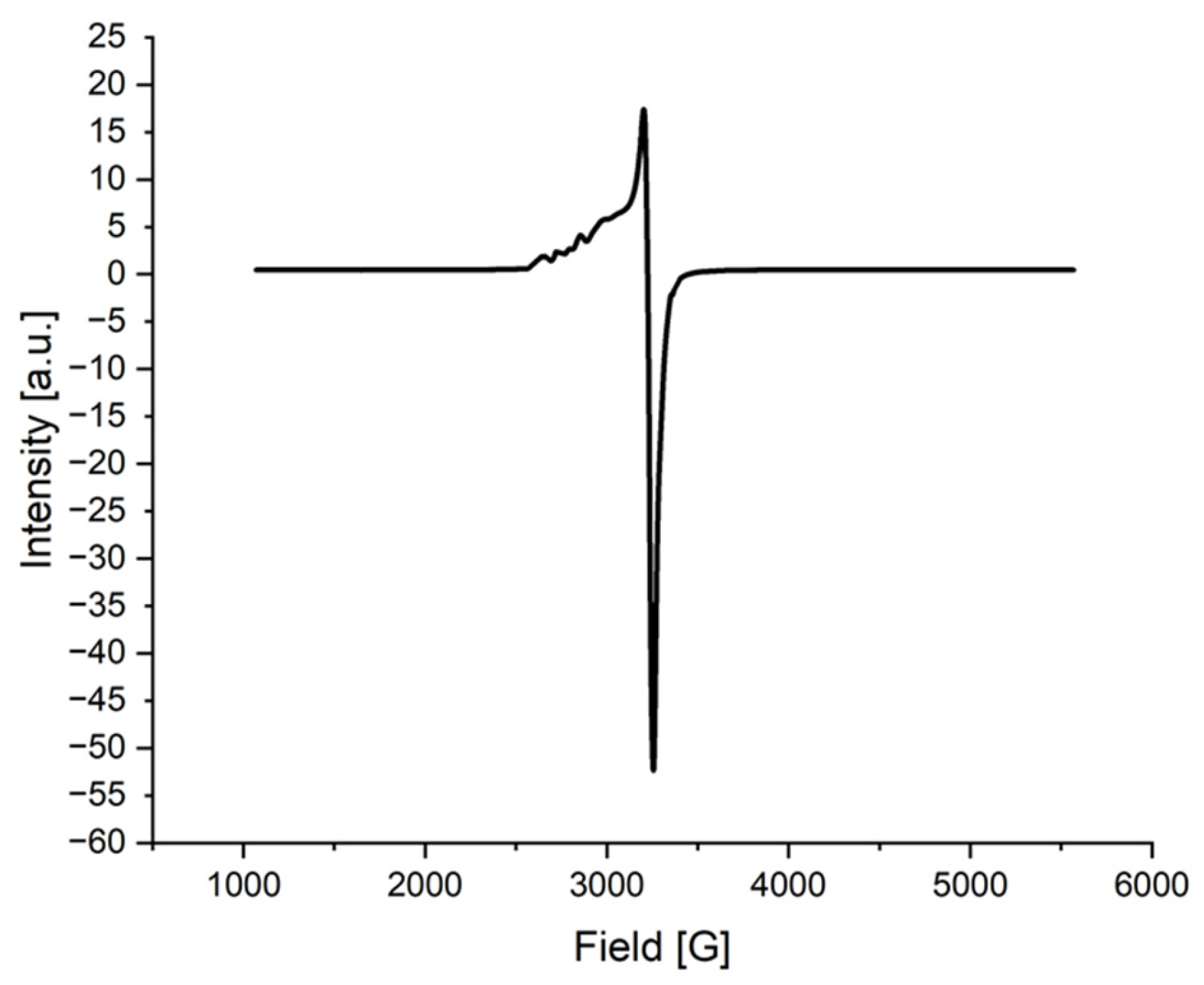
2.1.6. ESR Spectrum of the Cu(II) Complex
2.2. Cytotoxic Activity
3. Experimental Section
3.1. Materials and Instrumentation
3.2. Crystal Structure Determination of HL
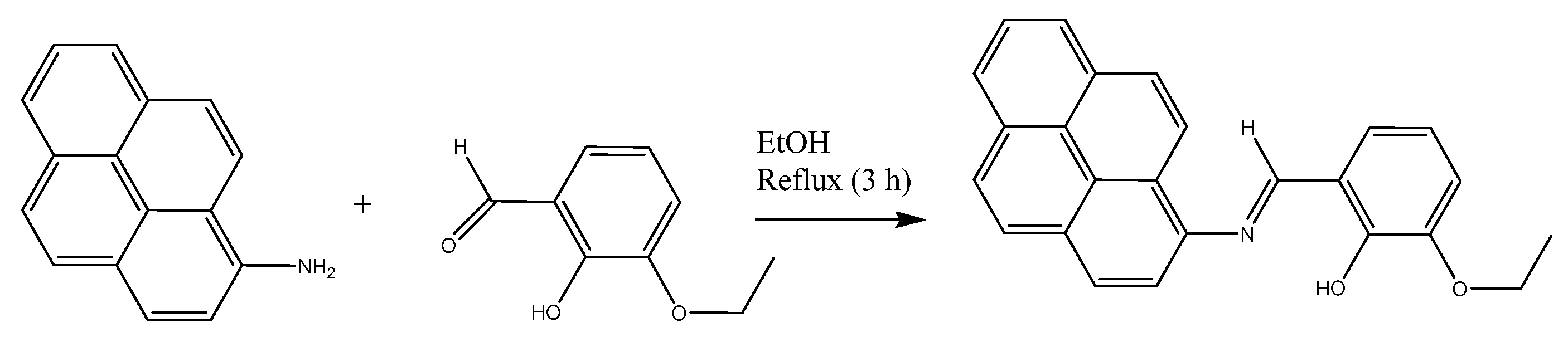
3.3. Synthesis of the Ligand (E)-2-Ethoxy-6-((pyren-1-ylimino)methyl)phenol (HL)
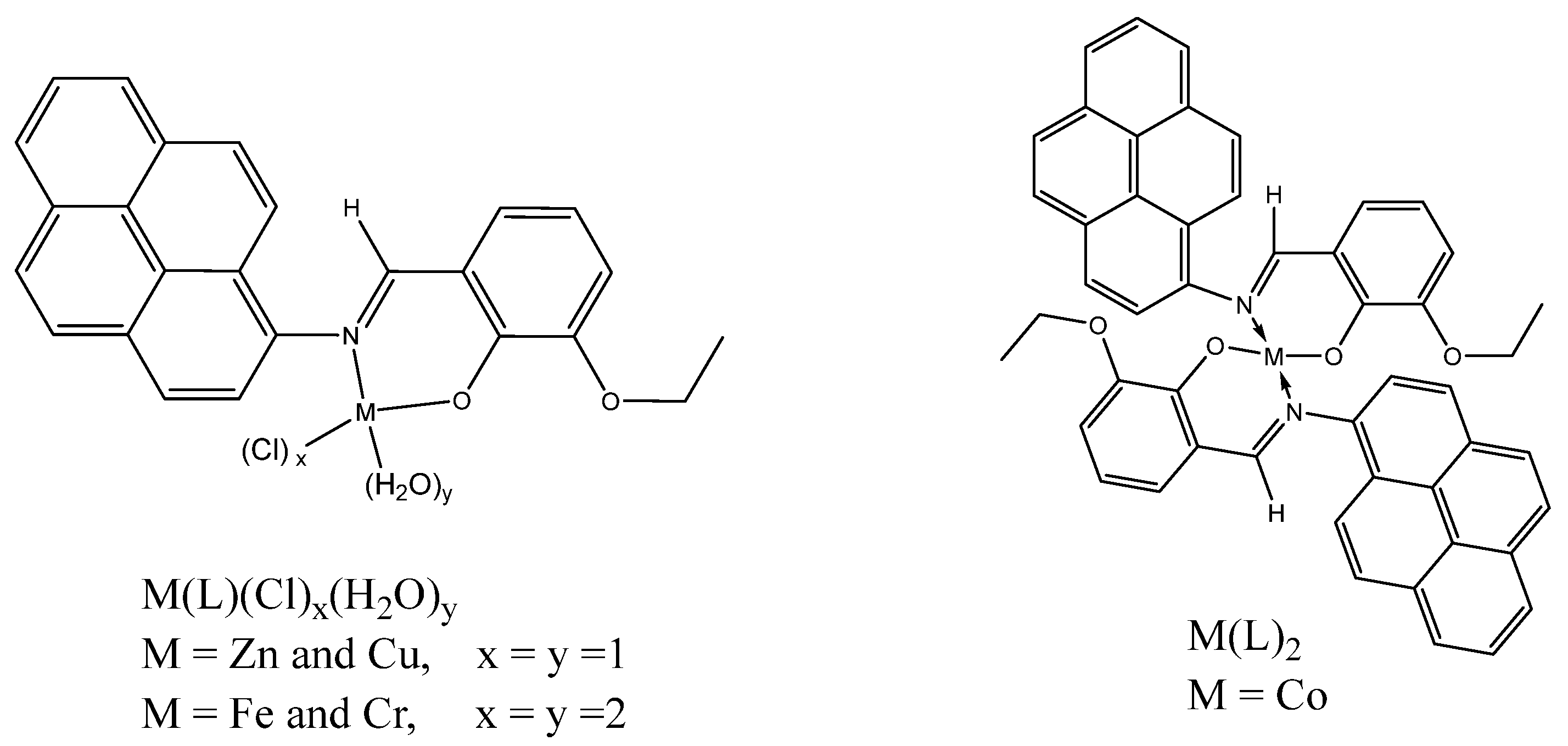
3.4. Synthesis of the Schiff Base Metal Complexes
- For [Cu(II)(L)(Cl)(H2O)], a black solid was obtained; m.p.: >250 °C with decomposition; yield (72.5%, 0.696 g).
- For [Co(II)(L)2], a dark red solid was obtained; m.p.: >250 °C with decomposition; yield (44.0%, 0.694 g).
- For [Cr(III)(L)(Cl)2(H2O)2], a pale brown solid was obtained; m.p.: >250 °C with decomposition; yield (16.9%, 0.090 g).
- For [Fe(III)(L)(Cl)2(H2O)], a black solid was obtained; m.p.: >250 °C with decomposition; yield (49.59%, 0.261 g).
3.5. Sample Preparation and Analysis for Mass Analysis
3.6. Cell Viability Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, Z.-Y.; Qiao, X.; Xie, C.-Z.; Shao, J.; Xu, J.-Y.; Qiang, Z.-Y.; Lou, J.-S. Activities of a novel Schiff Base copper (II) complex on growth inhibition and apoptosis induction toward MCF-7 human breast cancer cells via mitochondrial pathway. J. Inorg. Biochem. 2012, 117, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Darolia, P.J.; Chauhan, S.; Sindhu, M.; Verma, K.; Garg, S. Synthesis, Spectroscopic, Biological Activity, Molecular Docking and Density Functional Theoretical Investigations of Novel Tellurium (IV) Schiff Base Complexes. ChemistrySelect 2021, 6, 5778–5790. [Google Scholar] [CrossRef]
- El-Bindary, M.A.; El-Bindary, A.A. Synthesis, characterization, DNA binding, and biological action of dimedone arylhydrazone chelates. Appl. Organomet. Chem. 2022, 36, e6576. [Google Scholar] [CrossRef]
- Al-Humaidi, J.Y. In situ alkaline media: Synthesis, spectroscopic, morphology and anticancer assignments of some transition metal ion complexes of 1-((2-aminophenylimino) methyl) naphthalen-2-ol Schiff base. J. Mol. Struct. 2019, 1183, 190–201. [Google Scholar] [CrossRef]
- Shabbir, M.; Akhter, Z.; Ismail, H.; Mirza, B. Synthetic bioactive novel ether based Schiff bases and their copper (II) complexes. J. Mol. Struct. 2017, 1146, 57–61. [Google Scholar] [CrossRef]
- El-Gammal, O.A.; El-Bindary, A.A.; Mohamed, F.S.; Rezk, G.N.; El-Bindary, M.A. Synthesis, characterization, design, molecular docking, anti COVID-19 activity, DFT calculations of novel Schiff base with some transition metal complexes. J. Mol. Liq. 2022, 346, 117850. [Google Scholar] [CrossRef]
- Bhorge, Y.R.; Tsai, H.-T.; Huang, K.-F.; Pape, A.J.; Janaki, S.N.; Yen, Y.-P. A new pyrene-based Schiff-base: A selective colorimetric and fluorescent chemosensor for detection of Cu (II) and Fe (III). Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 130, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Jhonsi, M.A.; Dhenadhayalan, N.; Lin, K.-C.; Ananth, D.A.; Sivasudha, T.; Narayanaswamy, R.; Kathiravan, A. Pyrene-based prospective biomaterial: In Vitro bioimaging, protein binding studies and detection of bilirubin and Fe3+. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 221, 117150. [Google Scholar] [CrossRef]
- Dalbera, S.; Kulovi, S.; Dalai, S. Pyrene-based Schiff Base as Selective Chemosensor for Copper (II) and Sulfide Ions. ChemistrySelect 2018, 3, 6561–6569. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, P.; Kumar, S.; Singhal, D.; Gupta, R. Turn-on fluorescent sensors for the selective detection of Al3+ (and Ga3+) and PPi ions. Inorg. Chem. 2019, 58, 10364–10376. [Google Scholar] [CrossRef]
- Yeldir, E.K.; Kaya, İ. Synthesis, characterization and investigation of fluorescent Sn2+ probe potential of pyrene-derived monomer and its oligo (azomethine) compound. Eur. Polym. J. 2022, 172, 111229. [Google Scholar] [CrossRef]
- Silva, C.S.; Moutinho, C.; Ferreira da Vinha, A.; Matos, C. Trace minerals in human health: Iron, zinc, copper, manganese and fluorine. Int. J. Sci. Res. Methodol. 2019, 13, 57–80. [Google Scholar]
- El-Attar, M.; Aazam, E. Redox behavior, spectroscopic investigations, theoretical interpretation and biological effectiveness of some novel prepared bis-azomethine derivatives and their copper (II) complexes. J. Coord. Chem. 2021, 74, 779–803. [Google Scholar] [CrossRef]
- Aazam, E.S.; Zaki, M. Synthesis and characterization of Ni (II)/Zn (II) metal complexes derived from Schiff base and ortho-phenylenediamine: In vitro DNA binding, molecular modeling and RBC hemolysis. ChemistrySelect 2020, 5, 610–618. [Google Scholar] [CrossRef]
- Saritha, T.; Metilda, P. Synthesis, spectroscopic characterization and biological applications of some novel Schiff base transition metal (II) complexes derived from curcumin moiety. J. Saudi Chem. Soc. 2021, 25, 101245. [Google Scholar] [CrossRef]
- Aazam, E.S.; Büyükgüngör, O. 2-Amino-3-[(E)-(2-hydroxy-3-methylbenzylidene) amino] but-2-enedinitrile. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, o1406. [Google Scholar] [CrossRef] [PubMed]
- Hökelek, T.; Kılıç, Z.; Isıklan, M.; Toy, M. Intramolecular hydrogen bonding and tautomerism in Schiff bases. Part II. Structures of 1-[N-(2-pyridyl) aminomethylidene}-2 (1H)-naphtalenone (1) and bis [2-hydroxy-κO–N-(2-pyridyl)-1-naphthaldiminato-κN] zinc (II)(2). J. Mol. Struct. 2000, 523, 61–69. [Google Scholar] [CrossRef]
- Aazam, E.S.; Büyükgüngör, O. 7-[(3,5-Di-tert-butyl-2-hydroxybenzylidene) amino]-4-methyl-2H-chromen-2-one. Acta Crystallogr. Sect. E Struct. Rep. Online 2010, 66, o2587–o2588. [Google Scholar] [CrossRef]
- Durran, S.E.; Elsegood, M.R.; Hammond, S.R.; Smith, M.B. Coordination Studies of a New Unsymmetrical κ4-PNN′N″-Tetradentate Ligand: Stepwise Formation and Structural Characterization. Inorg. Chem. 2007, 46, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Mounika, K.; Pragathi, A.; Gyanakumari, C. Synthesis characterization and biological activity of a Schiff base derived from 3-ethoxy salicylaldehyde and 2-amino benzoic acid and its transition metal complexes. J. Sci. Res. 2010, 2, 513. [Google Scholar] [CrossRef]
- Dhahagani, K.; Kumar, S.M.; Chakkaravarthi, G.; Anitha, K.; Rajesh, J.; Ramu, A.; Rajagopal, G. Synthesis and spectral characterization of Schiff base complexes of Cu (II), Co (II), Zn (II) and VO (IV) containing 4-(4-aminophenyl) morpholine derivatives: Antimicrobial evaluation and anticancer studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 117, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Sunitha, M.; Jogi, P.; Ushaiah, B.; Kumari, C.G. Synthesis, characterization and antimicrobial activity of transition metal complexes of Schiff base ligand derived from 3-ethoxy salicylaldehyde and 2-(2-Aminophenyl) 1-H-benzimidazole. E J. Chem. 2012, 9, 2516–2523. [Google Scholar] [CrossRef]
- Durak, D.; Delikanlı, A.; Demetgül, C.; Kani, İ.; Serin, S. Crystal structure of an unsymmetrical Schiff base, immobilization of its cobalt and manganese complexes on a silica support, and catalytic studies. Transit. Met. Chem. 2013, 38, 199–206. [Google Scholar] [CrossRef]
- Pahonțu, E.; Ilieș, D.-C.; Shova, S.; Paraschivescu, C.; Badea, M.; Gulea, A.; Roșu, T. Synthesis, characterization, crystal structure and antimicrobial activity of copper (II) complexes with the Schiff base derived from 2-hydroxy-4-methoxybenzaldehyde. Molecules 2015, 20, 5771–5792. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.; El-Wakiel, N.; El-Ghamry, H. Synthesis, spectral, antitumor and antimicrobial studies on Cu (II) complexes of purine and triazole Schiff base derivatives. J. Mol. Struct. 2013, 1049, 326–335. [Google Scholar] [CrossRef]
- Joseyphus, R.S.; Nair, M.S. Synthesis, characterization and biological studies of some Co (II), Ni (II) and Cu (II) complexes derived from indole-3-carboxaldehyde and glycylglycine as Schiff base ligand. Arab. J. Chem. 2010, 3, 195–204. [Google Scholar] [CrossRef]
- Abdel-Rahman, L.H.; Abu-Dief, A.M.; Newair, E.F.; Hamdan, S.K. Some new nano-sized Cr (III), Fe (II), Co (II), and Ni (II) complexes incorporating 2-((E)-(pyridine-2-ylimino) methyl) napthalen-1-ol ligand: Structural characterization, electrochemical, antioxidant, antimicrobial, antiviral assessment and DNA interaction. J. Photochem. Photobiol. B Biol. 2016, 160, 18–31. [Google Scholar]
- Al-Fakeh, M.S.; Alsikhan, M.A.; Alnawmasi, J.S. Physico-Chemical Study of Mn (II), Co (II), Cu (II), Cr (III), and Pd (II) Complexes with Schiff-Base and Aminopyrimidyl Derivatives and Anti-Cancer, Antioxidant, Antimicrobial Applications. Molecules 2023, 28, 2555. [Google Scholar] [CrossRef]
- Saghatforoush, L.A.; Aminkhani, A.; Chalabian, F. Iron (III) Schiff base complexes with asymmetric tetradentate ligands: Synthesis, spectroscopy, and antimicrobial properties. Transit. Met. Chem. 2009, 34, 899–904. [Google Scholar] [CrossRef]
- Capan, A.; Uruş, S.; Sönmez, M. Ru (III), Cr (III), Fe (III) complexes of Schiff base ligands bearing phenoxy Groups: Application as catalysts in the synthesis of vitamin K3. J. Saudi Chem. Soc. 2018, 22, 757–766. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Spek, A. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, J.M.; Armstrong, L.S.; Martinez, A.O. A rapid and simple MTT-based spectrophotometric assay for determining drug sensitivity in monolayer cultures. J. Tissue Cult. Methods 1988, 11, 15–17. [Google Scholar] [CrossRef]





| Compounds | Empirical Formula | Mol. wt. (g/mol) | Color | M.p. (°C) | Yield (%) | Elemental Found (calc.) | Ω (O hm−1 cm2 mol−1) | ||
|---|---|---|---|---|---|---|---|---|---|
| C | H | N | |||||||
| HL | C25H19NO2 | 365.4 | Orange | 105 | 96.69 | 82.3 (82.2) | 5.3 (5.2) | 3.9 (3.8) | - |
| Zn(II)(L)(Cl)(H2O) | C25H20ClNO3Zn | 481.0 | red | 215 | 22.93 | 61.9 (62.1) | 4.2 (4.2) | 3.1 (2.9) | 2.05 |
| Cu(II)(L)(Cl)(H2O) | C25H20ClNO3Cu | 480.0 | Black | >250 | 72.5 | 62.3 (62.4) | 3.9 (4.2) | 3.0 (2.9) | 5.53 |
| Co(II)(L)2 | C50H36N2O4Co | 787.8 | Dark red | >250 | 44.0 | 75.9 (76.2) | 4.5 (4.6) | 3.6 (3.6) | 12.4 |
| Cr(III)(L)(Cl)2(H2O)2 | C25H22Cl2NO4Cr | 533.0 | Pale Brown | >250 | 16.9 | 57.6 (57.4) | 4.2 (4.3) | 2.8 (2.7) | 15.15 |
| Fe(III)(L)(Cl)2(H2O)2 | C25H22Cl2NO4Fe | 526.0 | Black | >250 | 49.59 | 57.2 (57.0) | 4.1 (4.2) | 2.6 (2.7) | 10.15 |
| Crystal Data | |
|---|---|
| Chemical formula | C25H19NO2 |
| Mr | 365.41 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 150 |
| a, b, c (Å) | 7.0543 (13), 11.503 (2), 11.988 (2) |
| α, β, γ (°) | 104.225 (7), 97.308 (7), 102.335 (8) |
| V (Å3) | 904.4 (3) |
| Z | 2 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.09 |
| Crystal size (mm) | × × |
| Data collection | |
| Diffractometer | Bruker APEX-II CCD |
| Absorption correction | – |
| No. of measured, independent, and observed [I > 2σ(I)] reflections | 20,633, 3078, 2033 |
| Rint | 0.110 |
| (sin θ/λ)max (Å−1) | 0.588 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.060, 0.132, 1.08 |
| No. of reflections | 3078 |
| No. of parameters | 256 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.22, −0.20 |
| Bond Lengths (Å) | Bond Lengths (Å) | ||
|---|---|---|---|
| O1-C11 | 1.372 (3) | C11-O1-C12 | 117.5 (2) |
| O1-C12 | 1.443 (3) | C8-N1-C7 | 122.0 (2) |
| O2-C10 | 1.353 (3) | C6-C7-N1 | 117.3 (2) |
| N1-C7 | 1.415 (3) | C19-C7-N1 | 122.6 (2) |
| N1-C8 | 1.282 (3) | N1-C8-C9 | 121.2 (3) |
| O1-C11 | 1.372 (3) | O2-C10-C9 | 122.3 (2) |
| O1-C12 | 1.443 (3) | O2-C10-C11 | 117.7 (2) |
| N1-C8 | 1.282 (3) | O1-C12-C13 | 106.9 (2) |
| C11-C14 | 1.374 (4) | ||
| C12-C13 | 1.503 (4) | ||
| C14-C15 | 1.407 (4) | ||
| Chemical Shift (δ, ppm) | Multiplicity | Number of Protons | Functional Group Assigned |
|---|---|---|---|
| 13.61 | Singlet | 1 | -OH |
| 9.28 | Singlet | 1 | -CH=N- |
| 8.49–8.12 | Multiplet | 9 | Pyrene protons |
| 7.42–6.99 | Doublet | 3 | Aromatic protons |
| 4.14 | Quartet | 2 | -OCH2 |
| 1.43 | Triplet | 3 | -CH3 |
| Compound | vOH/ vH2O | vC=N | vC=C | vC-O | vC-N | vM-N | vM-O |
|---|---|---|---|---|---|---|---|
| HL | 3431 | 1605 | 1459 | 1252 | 1186 | - | - |
| Zn(II)(L)(Cl)(H2O) | 3415 | 1631 | 1492 | 1235 | 1118 | 843 | 608 |
| Cu(II)(L)(Cl)(H2O) | 3416 | 1596 | 1365 | 1208 | 1132 | 844 | 613 |
| Co(II)(L)2 | 3415 | 1631 | 1492 | 1234 | 1118 | 843 | 626 |
| Cr(III)(L)(Cl)2(H2O)2 | 3420 | 1738 | 1365 | 1228 | 1216 | 846 | 527 |
| Fe(III)(L)(Cl)2(H2O)2 | 3433 | 1629 | 1482 | 1238 | 1189 | 845 | 607 |
| Ligand and Complexes | Vmax (nm) | µeff. (B.M) | Geometry | dn |
|---|---|---|---|---|
| HL | 292, 370, 425 | - | - | - |
| Zn(II)(L)(Cl)(H2O) | 295, 380, 430 | Diamagnetic | Tetrahedral | d10 |
| Cu(II)(L)(Cl)(H2O) | 275, 347, 425, 700 | 1.70 | Distorted tetrahedral | d9 |
| Co(II)(L)2 | 293, 390, 410, 675 | 4.16 | Tetrahedral | d7 |
| Cr(III)(L)(Cl)2(H2O)2 | 300, 399, 625, 725 | 3.61 | Distorted octahedral | d3 |
| Fe(III)(L)(Cl)2(H2O)2 | 311, 385, 695 | 5.91 | Distorted octahedral | d5 |
| Compound | Expected m/z | Found m/z | Peak Assigned |
|---|---|---|---|
| HL | 365 | 366 | [M+1]+ |
| Zn(II)(L)(Cl)(H2O) | 481 | 482 | [M+1]+ |
| Cu(II)(L)(Cl)(H2O) | 480 | 519 | [M-Cl+DMF]+ |
| Co(II)(L)2 | 787 | 788 | [M+1]+ |
| Cr(III)(L)(Cl)2(H2O)2 | 522 | 432 | [M+1-2Cl-H2O]+ |
| Fe(III)(L)(Cl)2(H2O)2 | 526 | 527 | [M+1]+ |
| Sample | 100 μg | 25 μg | 6.3 μg | 1.6 μg | 0.4 μg |
|---|---|---|---|---|---|
| HL | 46.88 | 56.88 | 64.25 | 76.44 | 83.44 |
| Zn(II)(L)(Cl)(H2O) | 38.03 | 47.13 | 52.75 | 64.16 | 68.00 |
| Cu(II)(L)(Cl)(H2O) | 29.44 | 40.36 | 49.30 | 59.01 | 68.46 |
| Co(II)(L)2 | 35.06 | 49.50 | 62.81 | 72.06 | 79.94 |
| Cr(III)(L)(Cl)2(H2O)2 | 45.81 | 56.54 | 65.30 | 75.32 | 84.14 |
| Fe(III)(L)(Cl)2(H2O)2 | 38.65 | 46.52 | 56.04 | 66.85 | 76.13 |
| 5FU b | 37.63 | 47.40 | 59.44 | 66.00 | 76.15 |
| Compound | Cytotoxicity IC50 a μg/mL | SD ± |
|---|---|---|
| HL | 63.901 | 3.67 |
| Zn(II)(L)(Cl)(H2O) | 12.742 | 0.73 |
| Cu(II)(L)(Cl)(H2O) | 5.661 | 0.33 |
| Co(II)(L)2 | 21.141 | 1.21 |
| Cr(III)(L)(Cl)2(H2O)2 | 58.708 | 3.37 |
| Fe(III)(L)(Cl)2(H2O)2 | 16.895 | 0.97 |
| 5FU b | 18.047 | 1.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aazam, E.S.; Majrashi, M.A. Novel Schiff Base Derived from Amino Pyrene: Synthesis, Characterization, Crystal Structure Determination, and Anticancer Applications of the Ligand and Its Metal Complexes. Molecules 2023, 28, 7352. https://doi.org/10.3390/molecules28217352
Aazam ES, Majrashi MA. Novel Schiff Base Derived from Amino Pyrene: Synthesis, Characterization, Crystal Structure Determination, and Anticancer Applications of the Ligand and Its Metal Complexes. Molecules. 2023; 28(21):7352. https://doi.org/10.3390/molecules28217352
Chicago/Turabian StyleAazam, Elham S., and Maryam A. Majrashi. 2023. "Novel Schiff Base Derived from Amino Pyrene: Synthesis, Characterization, Crystal Structure Determination, and Anticancer Applications of the Ligand and Its Metal Complexes" Molecules 28, no. 21: 7352. https://doi.org/10.3390/molecules28217352
APA StyleAazam, E. S., & Majrashi, M. A. (2023). Novel Schiff Base Derived from Amino Pyrene: Synthesis, Characterization, Crystal Structure Determination, and Anticancer Applications of the Ligand and Its Metal Complexes. Molecules, 28(21), 7352. https://doi.org/10.3390/molecules28217352






