Inhibitory Mechanisms of Lekethromycin in Dog Liver Cytochrome P450 Enzymes Based on UPLC-MS/MS Cocktail Method
Abstract
1. Introduction
2. Results
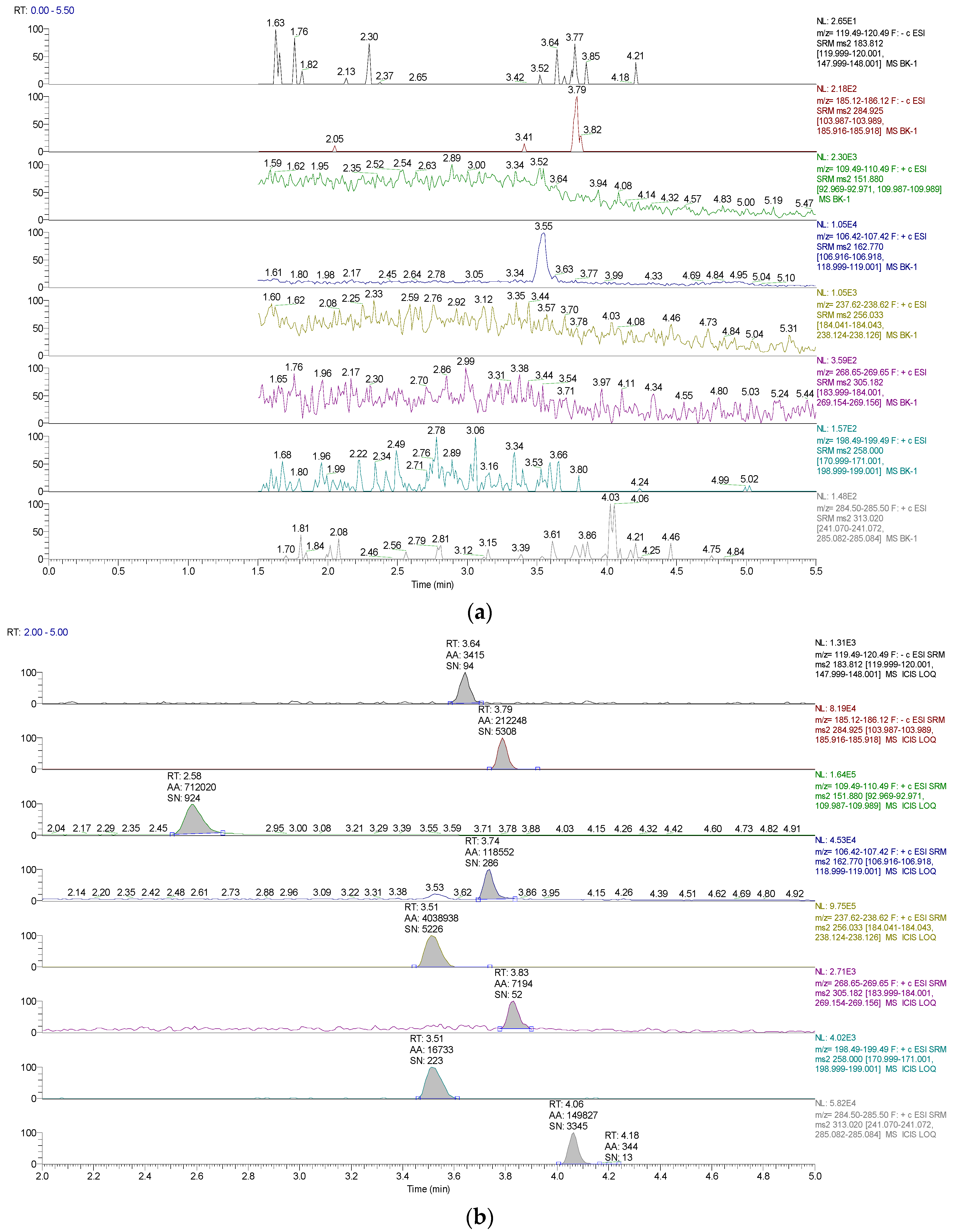
2.1. Methods Validation
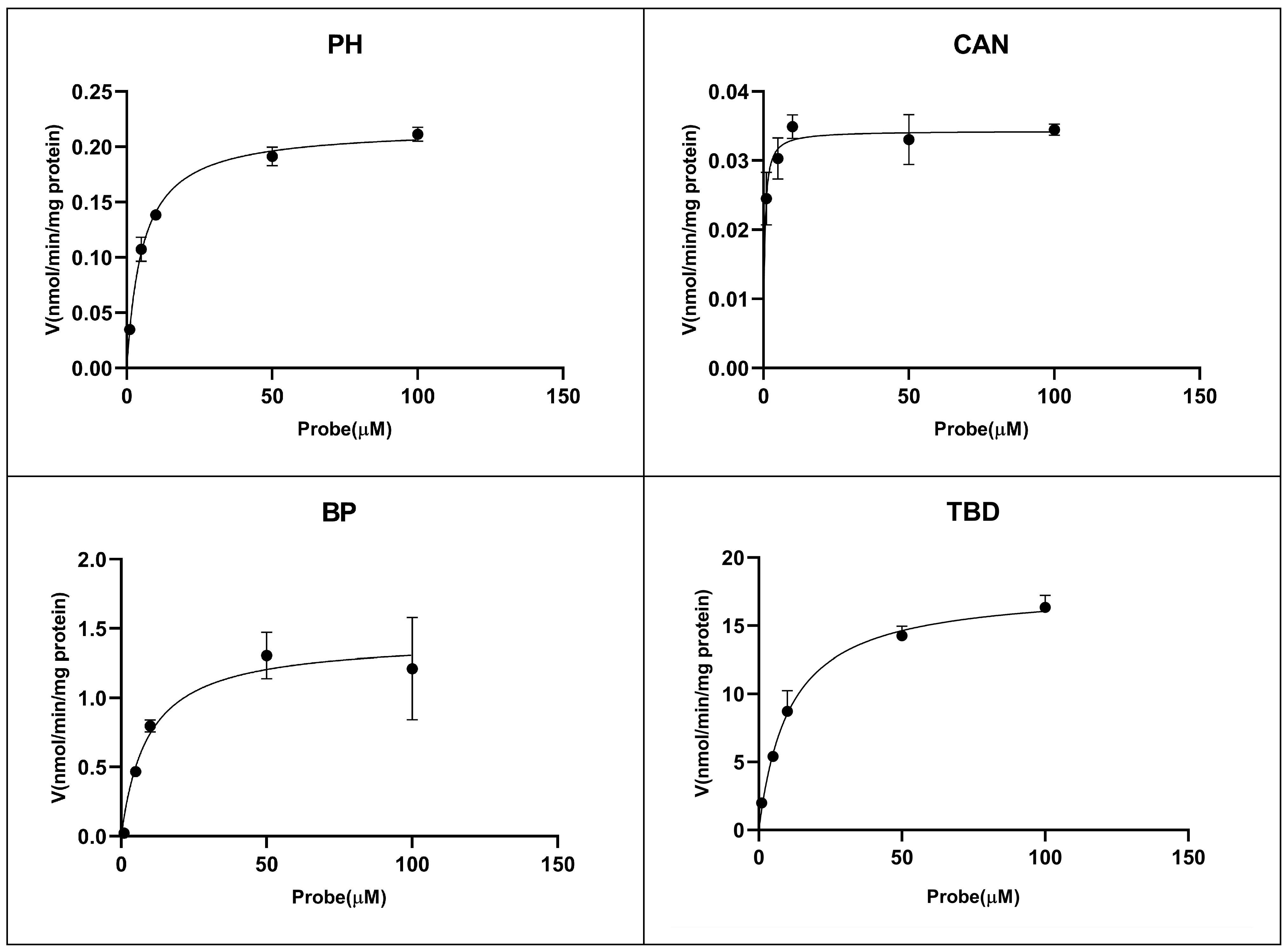
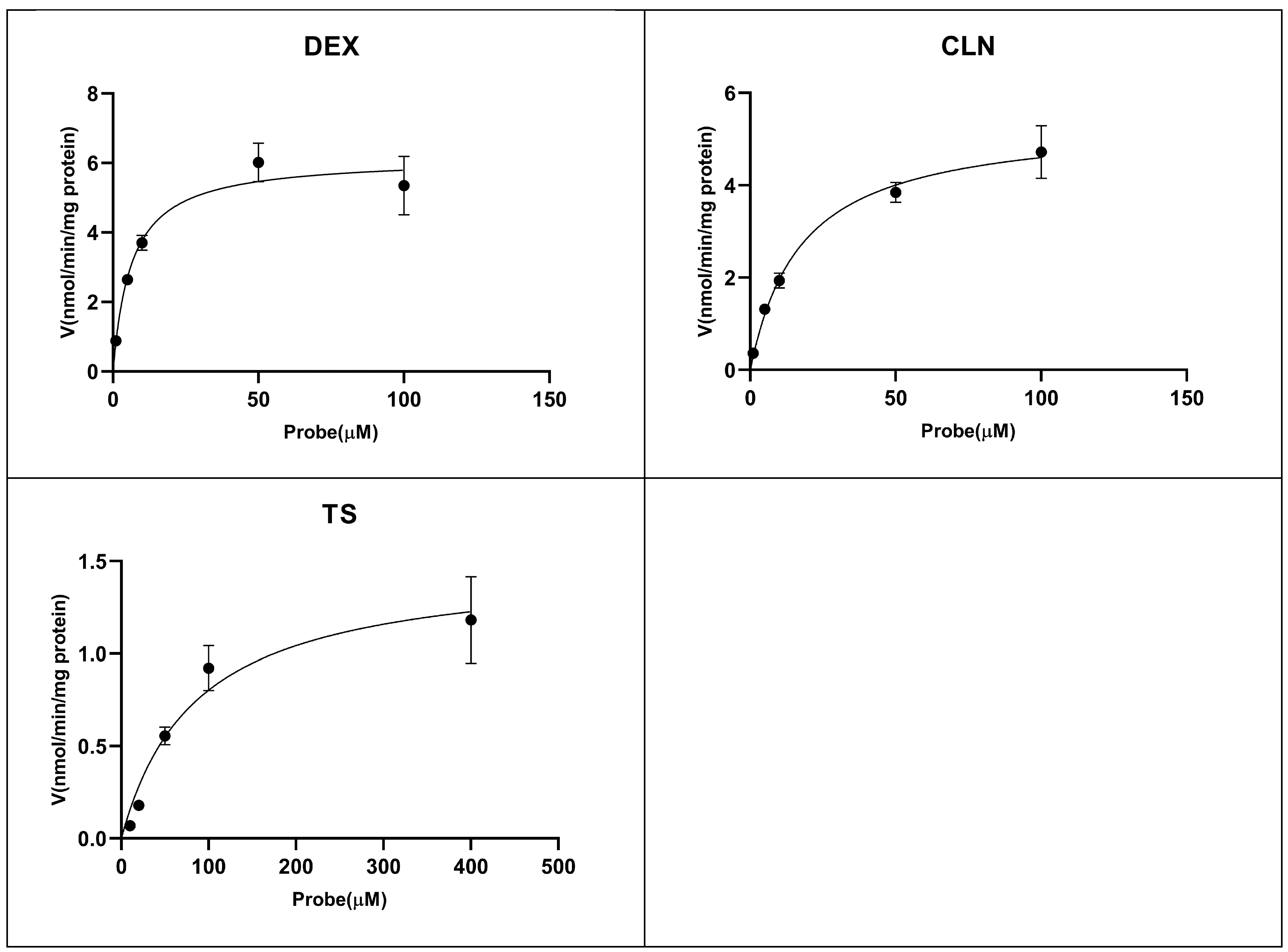
2.2. Enzyme Kinetic Study
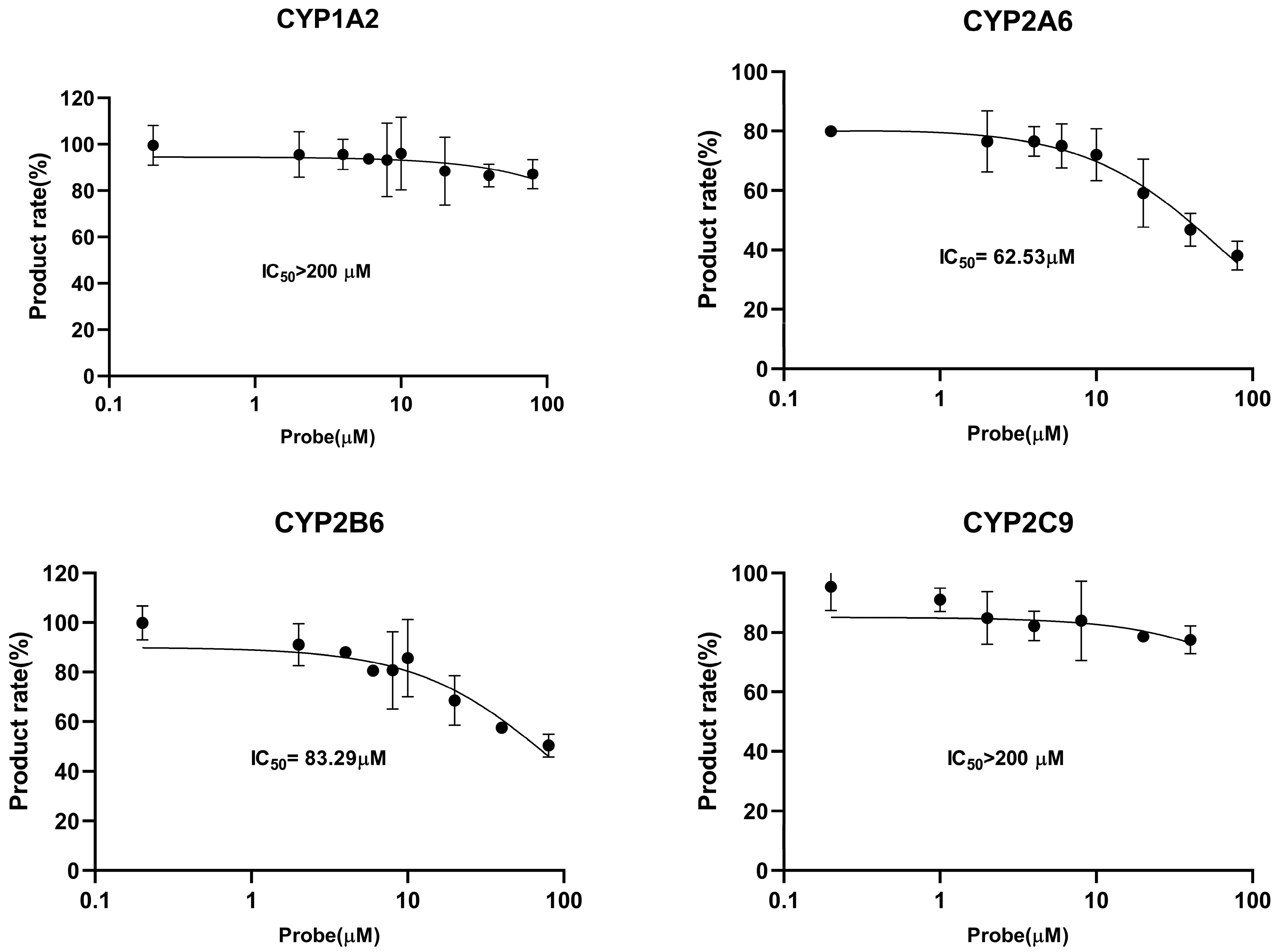
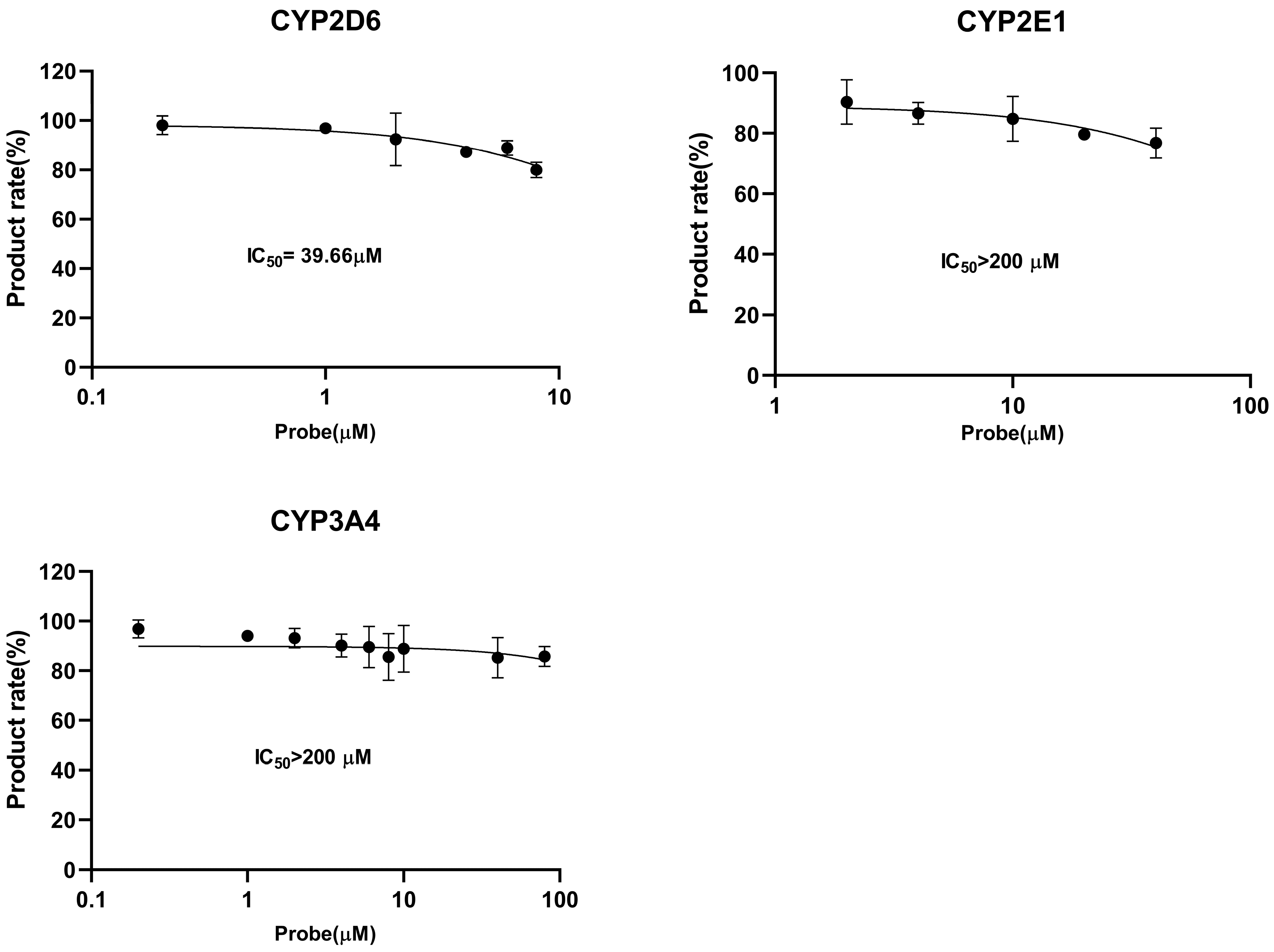
2.3. CYP450 Inhibition and Enzymatic Kinetic Study
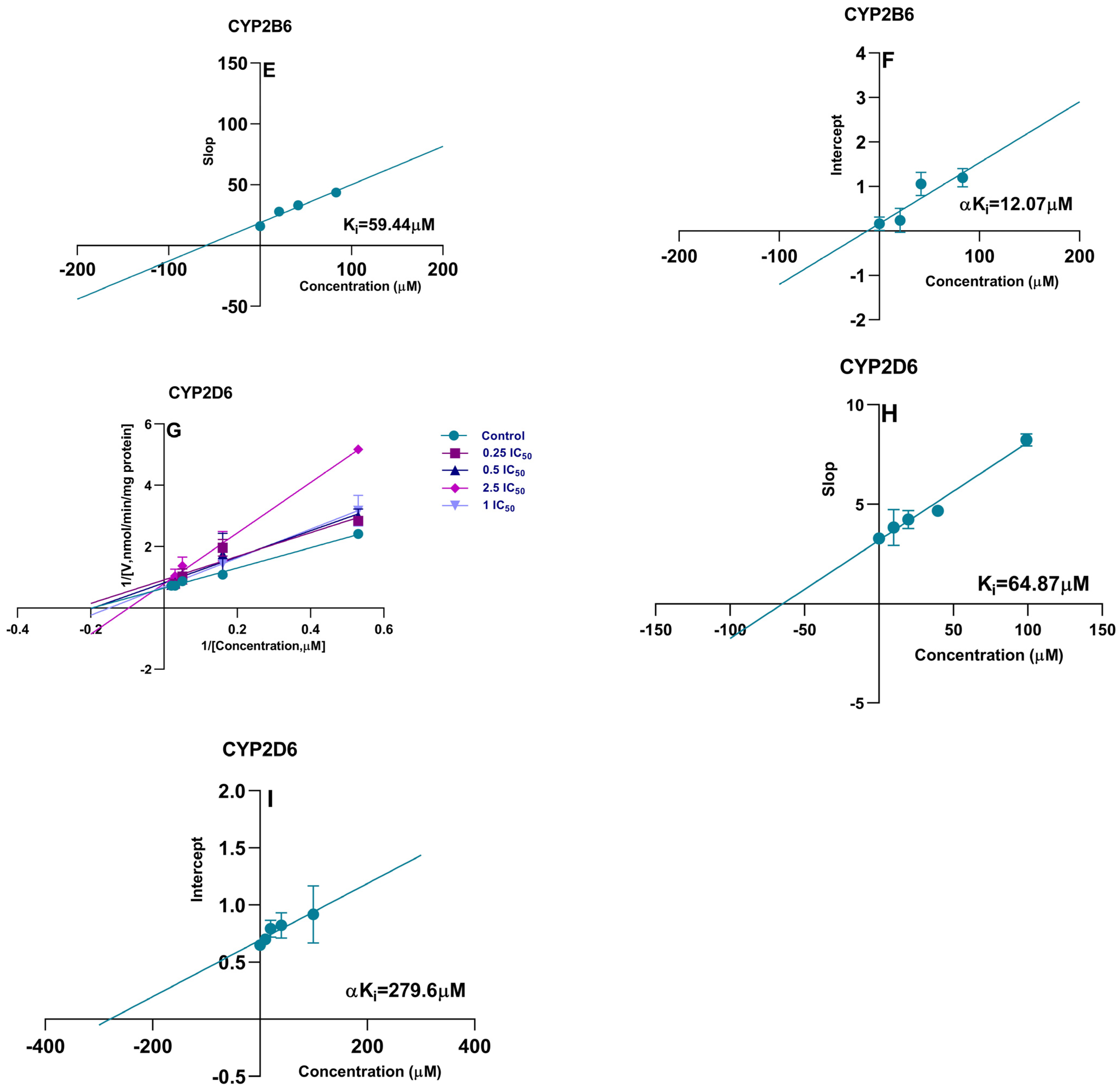
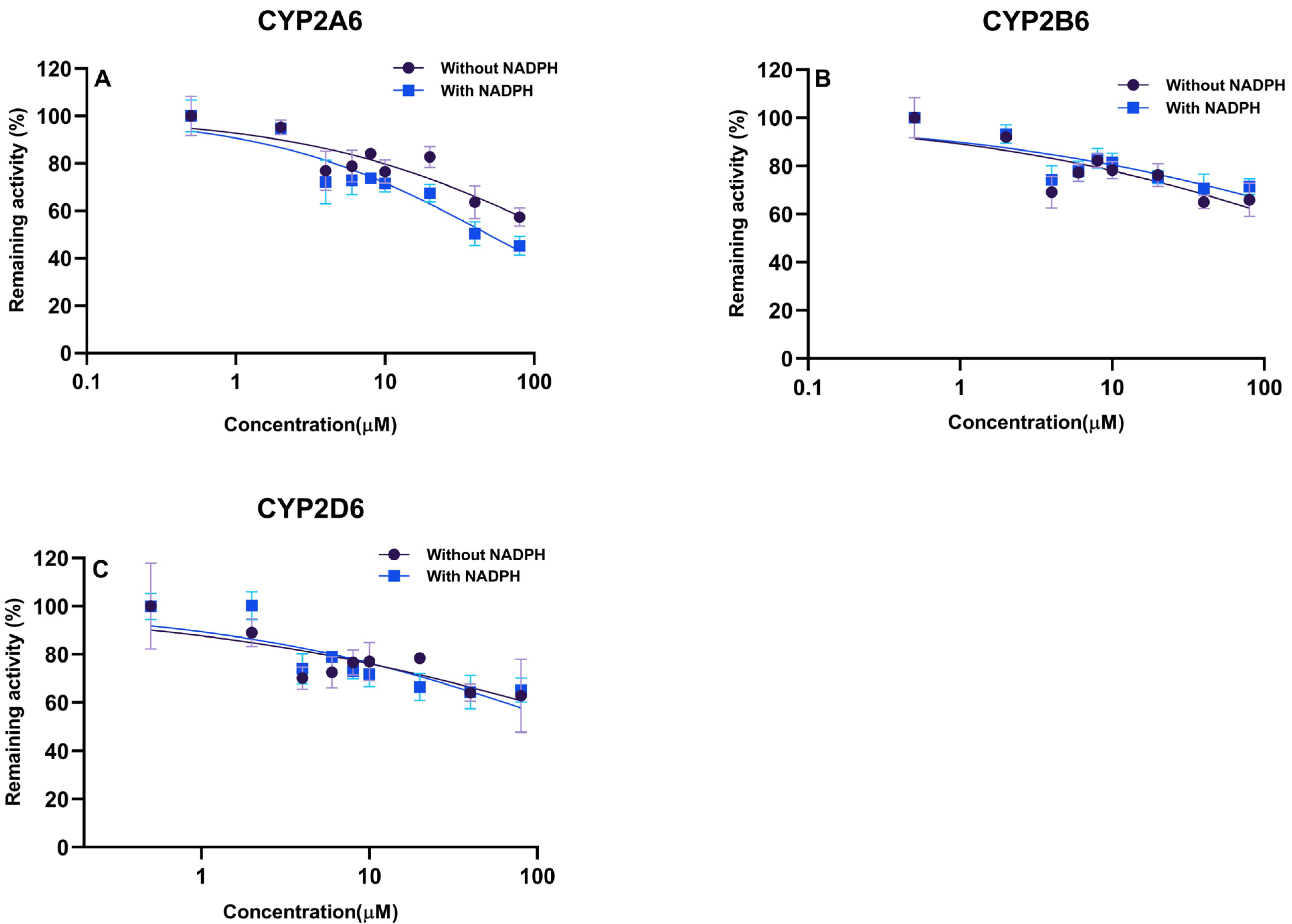
2.4. Irreversible Inhibition (IC50 Shift)
2.5. Chemical Inhibition Experiments
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Instruments and Chromatographic Conditions
4.3. Microsomal Incubation and Enzymatic Kinetic Study
4.4. CYP450 Inhibition Study
4.5. Irreversible Inhibition (IC50 Shift)
4.6. Chemical Inhibition Experiments
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cao, Y.; Sun, P.; Qiu, J.; Kong, J.; Yang, Y.; Liu, Y.; Zhou, D.; Wang, J.; Cao, X. Determination of lekethromycin in plasma and tissues of pneumonia-infected rats by ultra-high performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2023, 1227, 123811. [Google Scholar] [CrossRef]
- Sun, P.; Xiao, H.; Qiu, J.; Cao, Y.; Kong, J.; Zhang, S.; Cao, X. Plasma Protein Binding Rate and Pharmacokinetics of Lekethromycin in Rats. Antibiotics 2022, 11, 1241. [Google Scholar] [CrossRef]
- Xiao, H.; Sun, P.; Qiu, J.; Wang, J.; Yan, L.; Zhang, S.; Cao, X. Determination of lekethromycin, a novel macrolide lactone, in rat plasma by UPLC-MS/MS and its application to a pharmacokinetic study. Molecules 2020, 25, 4676. [Google Scholar] [CrossRef]
- Tang, D.-Q.; Li, Y.-J.; Li, Z.; Bian, T.-T.; Chen, K.; Zheng, X.-X.; Yu, Y.-Y.; Jiang, S.-S. Study on the interaction of plasma protein binding rate between edaravone and taurine in human plasma based on HPLC analysis coupled with ultrafiltration technique. Biomed. Chromatogr. 2015, 29, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Holbeck, S.L.; Camalier, R.; Crowell, J.A.; Govindharajulu, J.P.; Hollingshead, M.; Anderson, L.W.; Polley, E.; Rubinstein, L.; Srivastava, A.; Wilsker, D.; et al. The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity. Cancer Res. 2017, 77, 3564–3576. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. Clinical Drug Interaction Studies-Cytochrome P450 Enzyme-and Transporter-Mediated Drug Interactions Guidance for Industry. 2020. Available online: https://www.fda.gov/drugs/guidances-drugs/guidance-recap-podcast-clinical-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter. (accessed on 11 July 2023).
- Food and Drug Administration. In Vitro Drug Interaction Studies-Cytochrome P450 Enzyme-and Transporter-Mediated Drug Interactions. 2020. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/in-vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions (accessed on 12 July 2023).
- Sun, L.; Mi, K.; Hou, Y.; Hui, T.; Zhang, L.; Tao, Y.; Liu, Z.; Huang, L. Pharmacokinetic and Pharmacodynamic Drug–Drug Interactions: Research Methods and Applications. Metabolites 2023, 13, 897. [Google Scholar] [CrossRef]
- Rendic, S.; Guengerich, F.P. Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Xenobiotic and Natural Chemicals. Chem. Res. Toxicol. 2015, 28, 38–42. [Google Scholar] [CrossRef]
- Ravindranath, V.; Strobel, H.W. Cytochrome P450-mediated metabolism in brain: Functional roles and their implications. Expert Opin. Drug Metab. Toxicol. 2013, 9, 551–558. [Google Scholar] [CrossRef]
- Seliskar, M.; Rozman, D. Mammalian cytochromes P450—Importance of tissue specificity. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2007, 1770, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Danielson, P.B. The Cytochrome P450 Superfamily: Biochemistry, Evolution and Drug Metabolism in Humans. Curr. Drug Metab. 2002, 3, 561–597. [Google Scholar] [CrossRef]
- Venkatakrishnan, K.; von Moltke, L.L.; Greenblatt, D.J. Human Drug Metabolism and the Cytochromes P450: Application and Relevance of In Vitro Models. J. Clin. Pharmacol. 2001, 41, 1149–1179. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M. Human drug metabolising cytochrome P450 enzymes: Properties and polymorphisms. Naunyn Schmiedebergs Arch. Pharmacol. 2004, 369, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Madani, S.; Wei, X.-X.; Reynolds, K.; Huang, S.-M. Evaluation of Cytochrome P450 Probe Substrates Commonly Used by the Pharmaceutical Industry to Study in Vitro Drug Interactions. Drug Metab. Dispos. 2002, 30, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, X.; Badawy, S.; Ihsan, A.; Liu, Z.; Xie, C.; Wang, X.; Tao, Y. A Review: Effects of Macrolides on CYP450 Enzymes. Curr. Drug Metab. 2020, 21, 928–937. [Google Scholar] [CrossRef]
- Shah, S.S.; Sasaki, K.; Hayashi, Y.; Motoyama, S.; Helmi, A.R.; Khalil, W.F.; Shimoda, M. Inhibitory effects of ketoconazole, cimetidine and erythromycin on hepatic CYP3A activities in cats. J. Vet. Med. Sci. 2009, 71, 1151–1159. [Google Scholar] [CrossRef]
- Wong, S.L.; Cao, G.; Mack, R.J.; Witt; Granneman, G.R. The Effect of Erythromycin on the CYP3A Component of Sertindole Clearance in Healthy Volunteers. J. Clin. Pharmacol. 1997, 37, 1056–1061. [Google Scholar] [CrossRef]
- Ostrowski, M.; Wilkowska, E.; Bączek, T. Impact of pharmaceutical dosage form on stability and dissolution of roxithromycin. Open Med. 2010, 5, 83–90. [Google Scholar] [CrossRef]
- Qin, Y.; Xu, W.; Mo, L.; Li, X.; Ge, B.; Xiong, J.; Gao, L.; Xu, P.; Xue, M. Comparison of Pharmacokinetics and Tissue Distribution Kinetics of Roxithromycin and Expression of CYP 3A1 between Pregnant Mice and Foetuses. Basic Clin. Pharmacol. Toxicol. 2017, 120, 146–151. [Google Scholar] [CrossRef]
- Bruce, M.A.; Hall, S.D.; Haehner-Daniels, B.D.; Gorski, J.C. In Vivo effect of clarithromycin on multiple cytochrome P450s. Drug Metab. Dispos. 2001, 29, 1023–1028. [Google Scholar]
- Togami, K.; Hayashi, Y.; Chono, S.; Morimoto, K. Involvement of intestinal permeability in the oral absorption of clarithromycin and telithromycin. Biopharm. Drug Dispos. 2014, 35, 321–329. [Google Scholar] [CrossRef]
- Grönlund, J.; Saari, T.; Hagelberg, N.; Martikainen, I.K.; Neuvonen, P.J.; Olkkola, K.T.; Laine, K. Effect of Telithromycin on the Pharmacokinetics and Pharmacodynamics of Oral Oxycodone. J. Clin. Pharmacol. 2010, 50, 101–108. [Google Scholar] [CrossRef]
- Nosaka, H.; Nadai, M.; Kato, M.; Yasui, K.; Yoshizumi, H.; Miyoshi, M.; Zhao, Y.L.; Baba, K.; Takagi, K.; Hasegawa, T. Effect of a newly developed ketolide antibiotic, telithromycin, on metabolism of theophylline and expression of cytochrome P450 in rats. Life Sci. 2006, 79, 50–56. [Google Scholar] [CrossRef]
- Lee, J.H.; Kang, H.E.; Lee, M.G. Pharmacokinetic interaction between telithromycin and metformin in diabetes mellitus rats. Xenobiotica 2010, 40, 217–224. [Google Scholar] [CrossRef]
- Guengerich, F.P. Inhibition of Cytochrome P450 Enzymes by Drugs-Molecular Basis and Practical Applications. Biomol. Ther. 2022, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, F.; Jiang, H.; Xu, J.; Meng, D.; Geng, P.; Dai, D.; Hu, J.; Zhou, Y.; Zhou, Q.; et al. Inhibition and Induction by Poziotinib of Different Rat Cytochrome P450 Enzymes In Vivo and in an In Vitro Cocktail Method. Front. Pharmacol. 2021, 11, 593518. [Google Scholar] [CrossRef] [PubMed]
- Sane, R.S.; Ramsden, D.; Sabo, J.P.; Cooper, C.; Rowland, L.; Ting, N.; Whitcher-Johnstone, A.; Tweedie, D.J. Contribution of Major Metabolites toward Complex Drug-Drug Interactions of Deleobuvir: In Vitro Predictions and In Vivo Outcomes. Drug Metab. Dispos. 2016, 44, 466–475. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Wang, S.; Zhou, Q.; Dai, D.; Shi, J.; Xu, X.; Luo, Q. Cytochrome P450-Based Drug-Drug Interactions of Vonoprazan In Vitro and In Vivo. Front. Pharmacol. 2020, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Hisaka, A.; Ohno, Y.; Yamamoto, T.; Suzuki, H. Prediction of pharmacokinetic drug–drug interaction caused by changes in cytochrome P450 activity using in vivo information. Pharmacol. Ther. 2010, 125, 230–248. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Wong, Y. Enzyme Kinetics for Clinically Relevant CYP Inhibition. Curr. Drug Metab. 2005, 6, 241–257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kozakai, K.; Yamada, Y.; Oshikata, M.; Kawase, T.; Suzuki, E.; Haramaki, Y.; Taniguchi, H. Cocktail-substrate Approach-based High-throughput Assay for Evaluation of Direct and Time-dependent Inhibition of Multiple Cytochrome P450 Isoforms. Drug Metab. Pharmacokinet. 2014, 29, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Uchida, S.; Tanaka, S.; Kamiya, C.; Katayama, N.; Hakamata, A.; Odagiri, K.; Inui, N.; Kawakami, J.; Watanabe, H.; et al. Verification of a cocktail approach for quantitative drug–drug interaction assessment: A comparative analysis between the results of a single drug and a cocktail drug. Xenobiotica 2021, 51, 404–412. [Google Scholar] [CrossRef]
- Adiraju, S.K.S.; Shekar, K.; Fraser, J.F.; Smith, M.T.; Ghassabian, S. An improved LC–MS/MS method for simultaneous evaluation of CYP2C9, CYP2C19, CYP2D6 and CYP3A4 activity. Bioanalysis 2018, 10, 1577–1590. [Google Scholar] [CrossRef]
- Showande, S.J.; Fakeye, T.O.; Kajula, M.; Hokkanen, J.; Tolonen, A. Potential inhibition of major human cytochrome P450 isoenzymes by selected tropical medicinal herbs-Implication for herb-drug interactions. Food Sci. Nutr. 2019, 7, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Fliszár-Nyúl, E.; Ungvári, O.; Dombi, Á.; Özvegy-Laczka, C.; Poór, M. Interactions of Mycotoxin Alternariol with Cytochrome P450 Enzymes and OATP Transporters. Metabolites 2022, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Kamble, S.H.; Sharma, A.; King, T.I.; Berthold, E.C.; León, F.; Meyer, P.K.L.; Kanumuri, S.R.R.; McMahon, L.R.; McCurdy, C.R.; Avery, B.A. Exploration of cytochrome P450 inhibition mediated drug-drug interaction potential of kratom alkaloids. Toxicol. Lett. 2020, 319, 148–154. [Google Scholar] [CrossRef]
- Amaeze, O.; Eng, H.; Horlbogen, L.; Varma, M.V.S.; Slitt, A. Cytochrome P450 Enzyme Inhibition and Herb-Drug Interaction Potential of Medicinal Plant Extracts Used for Management of Diabetes in Nigeria. Eur. J. Drug Metab. Pharmacokinet. 2021, 46, 437–450. [Google Scholar] [CrossRef]
- Or, P.M.; Lam, F.F.; Kwan, Y.; Cho, C.; Yu, H.; Lin, G.; Lau, C.B.; Fung, K.; Leung, P.; Yeung, J.H. Effects of Radix Astragali and Radix Rehmanniae, the components of an anti-diabetic foot ulcer herbal formula, on metabolism of model CYP1A2, CYP2C9, CYP2D6, CYP2E1 and CYP3A4 probe substrates in pooled human liver microsomes and specific CYP isoforms. Phytomedicine 2012, 19, 535–544. [Google Scholar] [CrossRef]
- Witt, L.; Suzuki, Y.; Hohmann, N.; Mikus, G.; Haefeli, W.E.; Burhenne, J. Ultrasensitive quantification of the CYP2E1 probe chlorzoxazone and its main metabolite 6-hydroxychlorzoxazone in human plasma using ultra performance liquid chromatography coupled to tandem mass spectrometry after chlorzoxazone microdosing. J. Chromatogr. B 2016, 1027, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z.-Y.; Lu, S.; Powers, D.; Kansra, V.; Wang, X. Effects of rolapitant administered orally on the pharmacokinetics of dextromethorphan (CYP2D6), tolbutamide (CYP2C9), omeprazole (CYP2C19), efavirenz (CYP2B6), and repaglinide (CYP2C8) in healthy subjects. Support. Care Cancer 2019, 27, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Khayyat, M.H.; Mashhadian, N.V.; Eghbal, S.; Jalali, N. Inter-individual Variability of Coumarin 7-hydroxylation (CYP2A6 activity) in an Iranian Population. Iran J. Basic Med. Sci. 2013, 16, 610–614. [Google Scholar]
- Gao, N.; Qi, B.; Liu, F.-J.; Fang, Y.; Zhou, J.; Jia, L.-J.; Qiao, H.-L. Inhibition of Baicalin on Metabolism of Phenacetin, a Probe of CYP1A2, in Human Liver Microsomes and in Rats. PLoS ONE 2014, 9, e89752. [Google Scholar] [CrossRef]
- Nirogi, R.; Palacharla, R.C.; Mohammed, A.R.; Manoharan, A.; Ponnamaneni, R.K.; Bhyrapuneni, G. Evaluation of metabolism dependent inhibition of CYP2B6 mediated bupropion hydroxylation in human liver microsomes by monoamine oxidase inhibitors and prediction of potential as perpetrators of drug interaction. Chem. Biol. Interact. 2015, 230, 9–20. [Google Scholar] [CrossRef]
- Bogaards, J.J.P.; Bertrand, M.; Jackson, P.; Oudshoorn, M.J.; Weaver, R.J.; Van Bladeren, P.J.; Walther, B. Determining the best animal model for human cytochrome P450 activities: A comparison of mouse, rat, rabbit, dog, micropig, monkey and man. Xenobiotica 2000, 30, 1131–1152. [Google Scholar] [CrossRef]
- Polasek, T.M.; Miners, J.O. Quantitative prediction of macrolide drug-drug interaction potential from in vitro studies using testosterone as the human cytochrome P4503A substrate. Eur. J. Clin. Pharmacol. 2006, 62, 203–208. [Google Scholar] [CrossRef]
- Polasek, T.M.; Miners, J.O. Macrolide-theophylline interactions: No role for the inhibition of cytochrome P4501A2. Br. J. Clin. Pharmacol. 2008, 66, 898–900. [Google Scholar] [CrossRef]
- Ring, B.; Wrighton, S.A.; Mohutsky, M. Reversible Mechanisms of Enzyme Inhibition and Resulting Clinical Significance. Methods Mol. Biol. 2014, 1113, 37–56. [Google Scholar] [CrossRef]
- Carrão, D.B.; Habenchus, M.D.; de Albuquerque, N.C.P.; da Silva, R.M.; Lopes, N.P.; de Oliveira, A.R.M. In Vitro inhibition of human CYP2D6 by the chiral pesticide fipronil and its metabolite fipronil sulfone: Prediction of pesticide-drug interactions. Toxicol. Lett. 2019, 313, 196–204. [Google Scholar] [CrossRef]
- Shou, M.; Lin, Y.; Lu, P.; Tang, C.; Mei, Q.; Cui, D.; Tang, W.; Ngui, J.S.; Lin, C.C.; Singh, R.; et al. Enzyme Kinetics of Cytochrome P450-Mediated Reactions. Curr. Drug Metab. 2012, 2, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Stresser, D.M.; Mao, J.; Kenny, J.R.; Jones, B.C.; Grime, K. Exploring concepts of in vitro time-dependent CYP inhibition assays. Expert Opin. Drug Metab. Toxicol. 2014, 10, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Bao, S.; Li, Y.; Lin, Q.; Yang, S.; He, J. Inhibitory Mechanisms of Myricetin on Human and Rat Liver Cytochrome P450 Enzymes. Eur. J. Drug Metab. Pharmacokinet. 2019, 44, 611–618. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, A.M.; Dang, C.H. Basic Review of the Cytochrome P450 System. J. Adv. Pract. Oncol. 2013, 4, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.; Magro, L.; Moretti, U.; Cutroneo, P.; Moschini, M.; Motola, D.; Tuccori, M.; Conforti, A. Identifying Adverse Drug Reactions Associated with Drug-Drug Interactions. Drug Saf. 2010, 33, 667–675. [Google Scholar] [CrossRef]
- Wu, J.; Guan, X.; Dai, Z.; He, R.; Ding, X.; Yang, L.; Ge, G. Molecular probes for human cytochrome P450 enzymes: Recent progress and future perspectives. Coord. Chem. Rev. 2021, 427, 213600. [Google Scholar] [CrossRef]
- Mo, S.-L.; Liu, Y.-H.; Duan, W.; Wei, M.; Kanwar, J.; Zhou, S.-F. Substrate Specificity, Regulation, and Polymorphism of Human Cytochrome P450 2B6. Curr. Drug Metab. 2009, 10, 730–753. [Google Scholar] [CrossRef]
- Patki, K.C.; von Moltke, L.L.; Greenblatt, D.J. In Vitro Metabolism of Midazolam, Triazolam, Nifedipine, and Testosterone by Human Liver Microsomes and Recombinant Cytochromes P450: Role of CYP3A4 and CYP3A5. Drug Metab. Dispos. 2003, 31, 938–944. [Google Scholar] [CrossRef]
- VandenBrink, B.M.; Foti, R.S.; Rock, D.A.; Wienkers, L.C.; Wahlstrom, J.L. Prediction of CYP2D6 Drug Interactions from In Vitro Data: Evidence for Substrate-Dependent Inhibition. Drug Metab. Dispos. 2012, 40, 47–53. [Google Scholar] [CrossRef]
- Tang, L.; Ye, L.; Lv, C.; Zheng, Z.; Gong, Y.; Liu, Z. Involvement of CYP3A4/5 and CYP2D6 in the metabolism of aconitine using human liver microsomes and recombinant CYP450 enzymes. Toxicol. Lett. 2011, 202, 47–54. [Google Scholar] [CrossRef]
- Perloff, E.S.; Mason, A.K.; Dehal, S.S.; Blanchard, A.P.; Morgan, L.; Ho, T.; Dandeneau, A.; Crocker, R.M.; Chandler, C.M.; Boily, N.; et al. Validation of cytochrome P450 time-dependent inhibition assays: A two-time point IC50 shift approach facilitates kinact assay design. Xenobiotica 2009, 39, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Berry, L.; Zhao, Z. An Examination of IC50 and IC50-Shift Experiments in Assessing Time-Dependent Inhibition of CYP3A4, CYP2D6 and CYP2C9 in Human Liver Microsomes. Drug Metab. Lett. 2008, 2, 51–59. [Google Scholar] [CrossRef]
- Berry, L.M.; Zhao, Z.; Lin, M.-H.J. Dynamic Modeling of Cytochrome P450 Inhibition In Vitro: Impact of Inhibitor Depletion on IC50 Shift. Drug Metab. Dispos. 2013, 41, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Su, L.; Ji, D.; Gu, W.; Mao, C. Interaction between CYP450 enzymes and metabolism of traditional Chinese medicine as well as enzyme activity assay. Zhongguo Zhong Yao Za Zhi 2015, 40, 3524–3529. [Google Scholar] [PubMed]
- Periti, P.; Mazzei, T.; Mini, E.; Novelli, A. Pharmacokinetic Drug Interactions of Macrolides. Clin. Pharmacokinet. 1992, 23, 106–131. [Google Scholar] [CrossRef]
- Pessayre, D.; Larrey, D.; Funck-Brentano, C.; Benhamou, J.P. Drug interactions and hepatitis produced by some macrolide antibiotics. J. Antimicrob. Chemother. 1985, 16 (Suppl. A), 181–194. [Google Scholar] [CrossRef]
- Golikova, M.V.; Strukova, E.N.; Portnoy, Y.A.; Dovzhenko, S.A.; Kobrin, M.B.; Zinner, S.H.; Firsov, A.A. Resistance studies with Streptococcus pneumoniae using an in vitro dynamic model: Amoxicillin versus azithromycin at clinical exposures. J. Chemother. 2019, 31, 252–260. [Google Scholar] [CrossRef]
- Obach, R.S.; Walsky, R.L.; Venkatakrishnan, K.; Gaman, E.A.; Houston, J.B.; Tremaine, L.M. The Utility of in Vitro Cytochrome P450 Inhibition Data in the Prediction of Drug-Drug Interactions. J. Pharmacol. Exp. Ther. 2006, 316, 336–348. [Google Scholar] [CrossRef]
- Kapelyukh, Y.; Wolf, R. CYP2D6 substrates and drug metabolism. In CYP2D6: Genetics, Pharmacology and Clinical Relevance; Future Medicine Ltd.: London, UK, 2014; pp. 80–100. [Google Scholar] [CrossRef]
- de Groot, M.J.; Wakenhut, F.; Whitlock, G.; Hyland, R. Understanding CYP2D6 interactions. Drug Discov. Today 2009, 14, 964–972. [Google Scholar] [CrossRef]
- Qiu, F.; Zhang, R.; Sun, J.; A, J.; Hao, H.; Peng, Y.; Ai, H.; Wang, G. Inhibitory Effects of Seven Components of Danshen Extract on Catalytic Activity of Cytochrome P450 Enzyme in Human Liver Microsomes. Drug Metab. Dispos. 2008, 36, 1308–1314. [Google Scholar] [CrossRef]


| Analyte | Concentration (ng/mL) | Recovery (%) | Intra-Day RSD (%) | Inter-Day RSD (%) |
|---|---|---|---|---|
| ACE | 10 | 104.80 ± 6.36 | 6.07 | 5.73 |
| 50 | 101.66 ± 2.00 | 1.96 | 4.19 | |
| 200 | 101.92 ± 1.44 | 1.41 | 2.69 | |
| HCAN | 10 | 99.86 ± 8.06 | 8.07 | 7.29 |
| 50 | 99.39 ± 7.04 | 7.09 | 7.24 | |
| 200 | 99.90 ± 6.47 | 6.48 | 4.93 | |
| HBP | 10 | 104.86 ± 8.55 | 8.16 | 7.84 |
| 50 | 103.64 ± 3.43 | 3.31 | 5.56 | |
| 200 | 101.29 ± 3.18 | 3.14 | 3.82 | |
| HTBD | 10 | 105.04 ± 11.72 | 11.15 | 10.04 |
| 50 | 102.01 ± 9.83 | 9.64 | 8.48 | |
| 200 | 95.53 ± 1.30 | 1.36 | 2.04 | |
| HDEX | 10 | 109.42 ± 1.8 | 12.14 | 10.84 |
| 50 | 106.93 ± 4.05 | 1.68 | 6.67 | |
| 200 | 97.79 ± 3.71 | 3.79 | 6.34 | |
| HCLN | 10 | 97.35 ± 8.65 | 8.88 | 9.31 |
| 50 | 101.99 ± 11.41 | 11.18 | 9.81 | |
| 200 | 97.63 ± 1.48 | 1.52 | 7.14 | |
| HTS | 10 | 95.63 ± 6.89 | 7.20 | 9.08 |
| 50 | 97.40 ± 1.76 | 1.80 | 4.59 | |
| 200 | 101.89 ± 2.45 | 2.40 | 3.96 |
| Isoforms | Reactions | Time (min) | Protein Concentration (mg/mL) | Km (μM) | Vmax (nmol/min/mg protein) | IC50 (μM) |
|---|---|---|---|---|---|---|
| CYP1A2 | Phenacetin- O-deethylation | 15 | 0.2 | 5.39 | 0.22 | >200 |
| CYP2A6 | Coumarin 5-hydroxylation | 15 | 0.2 | 0.41 | 0.034 | 62.53 |
| CYP2B6 | Bupropion 1-hydroxylation | 15 | 0.2 | 9.15 | 1.43 | 83.29 |
| CYP2C9 | Tolbutamide 4-hydroxylation | 15 | 0.2 | 10.76 | 17.78 | >200 |
| CYP2D6 | Dextromethorphan O-demethylation | 15 | 0.2 | 6.33 | 6.16 | 39.66 |
| CYP2E1 | Chlorzoxazone 1-hydroxylation | 15 | 0.2 | 17.42 | 5.4 | >200 |
| CYP3A4 | Testosterone 6β-hydroxylation | 15 | 0.2 | 85.21 | 1.49 | >200 |
| CYP450 Isoform | Shift Ratio (−NADPH/+NADPH) | Ki (μM) | αKi (μM) |
|---|---|---|---|
| CYP2A6 | 1.29 | 135.6 | 227.6 |
| CYP2D6 | 1.10 | 64.87 | 279.6 |
| CYP2B6 | 0.88 | 59.44 | 12.07 |
| CYP450 Isoform | Substrates | Metabolites | Inhibitors |
|---|---|---|---|
| CYP1A2 | PH | ACE | α-Naphthoflavone |
| CYP2A6 | CAN | HCAN | Pilocarpine |
| CYP2B6 | BP | HBP | Thio-TEPA |
| CYP2C9 | TBD | HTBD | Sulfaphenazole |
| CYP2D6 | DEX | HDEX | Quinidine |
| CYP2E1 | CLN | HCLN | Sodium diethyldithiocarbamate |
| CYP3A4 | TS | HTS | Ketoconazole |
| Substrates | Quantitative Ion | Collision Energy (eV) | Ion Source |
|---|---|---|---|
| PH | 180 > 110 | 20 | ESI+ |
| CAN | 147 > 110 | 19 | ESI+ |
| BP | 240 > 184 | 18 | ESI+ |
| TBD | 271 > 91 | 31 | ESI+ |
| DEX | 272 > 215 | 23.5 | ESI+ |
| CLN | 168 > 132 | 20 | ESI− |
| TS | 289 > 97 | 22 | ESI+ |
| ACE | 152 > 110 | 16 | ESI+ |
| HCAN | 163 > 119 | 34 | ESI+ |
| HBP | 256 > 184 | 15 | ESI+ |
| HTBD | 285 > 186 | 17 | ESI− |
| HDEX | 258 > 171 | 30 | ESI+ |
| HCLN | 184 > 120 | 15 | ESI− |
| HTS | 305 > 184 | 10 | ESI+ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, P.; Cao, Y.; Qiu, J.; Kong, J.; Zhang, S.; Cao, X. Inhibitory Mechanisms of Lekethromycin in Dog Liver Cytochrome P450 Enzymes Based on UPLC-MS/MS Cocktail Method. Molecules 2023, 28, 7193. https://doi.org/10.3390/molecules28207193
Sun P, Cao Y, Qiu J, Kong J, Zhang S, Cao X. Inhibitory Mechanisms of Lekethromycin in Dog Liver Cytochrome P450 Enzymes Based on UPLC-MS/MS Cocktail Method. Molecules. 2023; 28(20):7193. https://doi.org/10.3390/molecules28207193
Chicago/Turabian StyleSun, Pan, Yuying Cao, Jicheng Qiu, Jingyuan Kong, Suxia Zhang, and Xingyuan Cao. 2023. "Inhibitory Mechanisms of Lekethromycin in Dog Liver Cytochrome P450 Enzymes Based on UPLC-MS/MS Cocktail Method" Molecules 28, no. 20: 7193. https://doi.org/10.3390/molecules28207193
APA StyleSun, P., Cao, Y., Qiu, J., Kong, J., Zhang, S., & Cao, X. (2023). Inhibitory Mechanisms of Lekethromycin in Dog Liver Cytochrome P450 Enzymes Based on UPLC-MS/MS Cocktail Method. Molecules, 28(20), 7193. https://doi.org/10.3390/molecules28207193






