TMT-Based Proteomics Reveal the Mechanism of Action of Amygdalin against Rheumatoid Arthritis in a Rat Model through Regulation of Complement and Coagulation Cascades
Abstract
:1. Introduction
2. Results
2.1. Effects of Amygdalin on CFA-Induced Rats
2.1.1. Effects of Amygdalin on Histopathology in CFA-Induced Rats
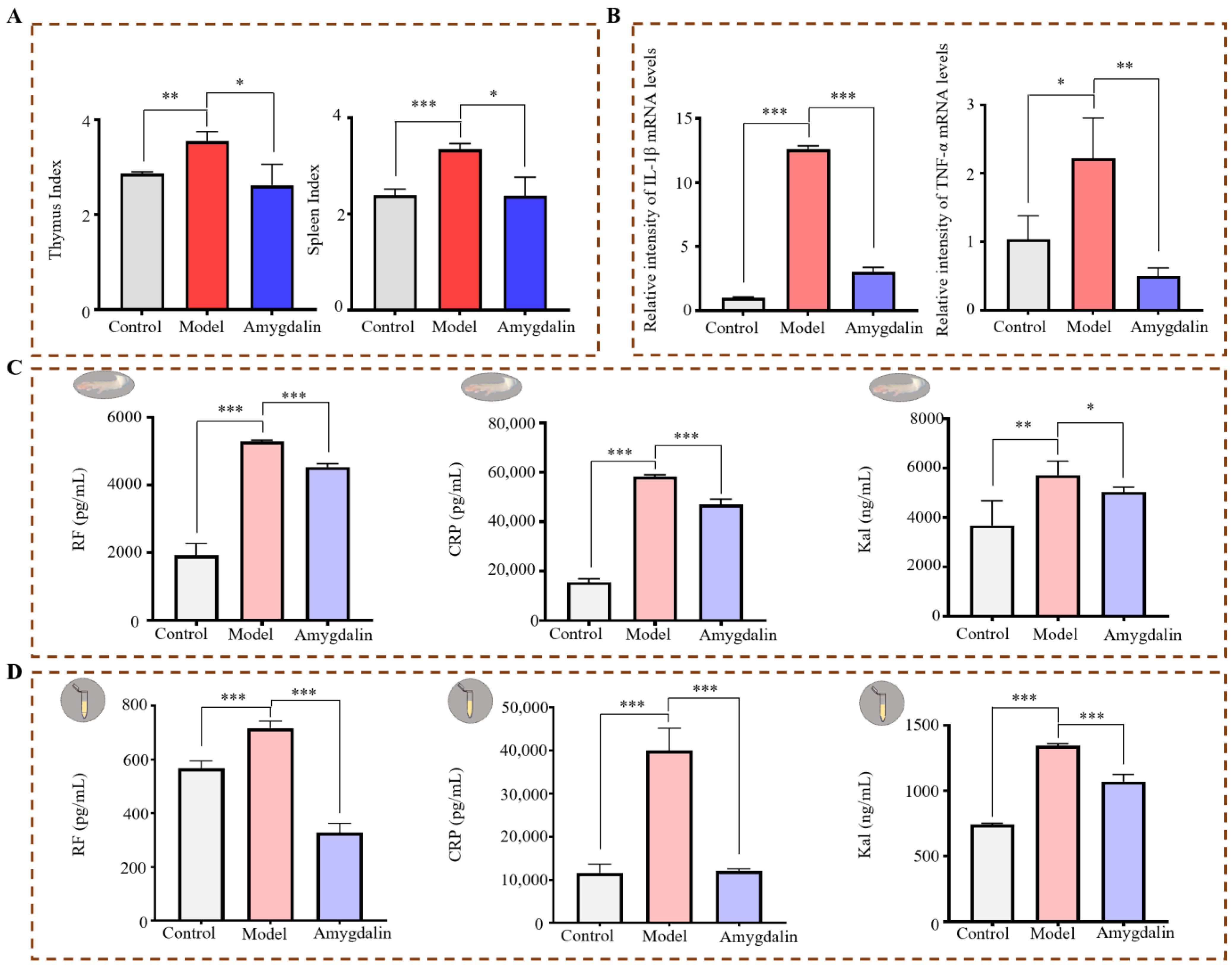
2.1.2. Effects of Amygdalin on CFA-Induced Cytokines in Rats
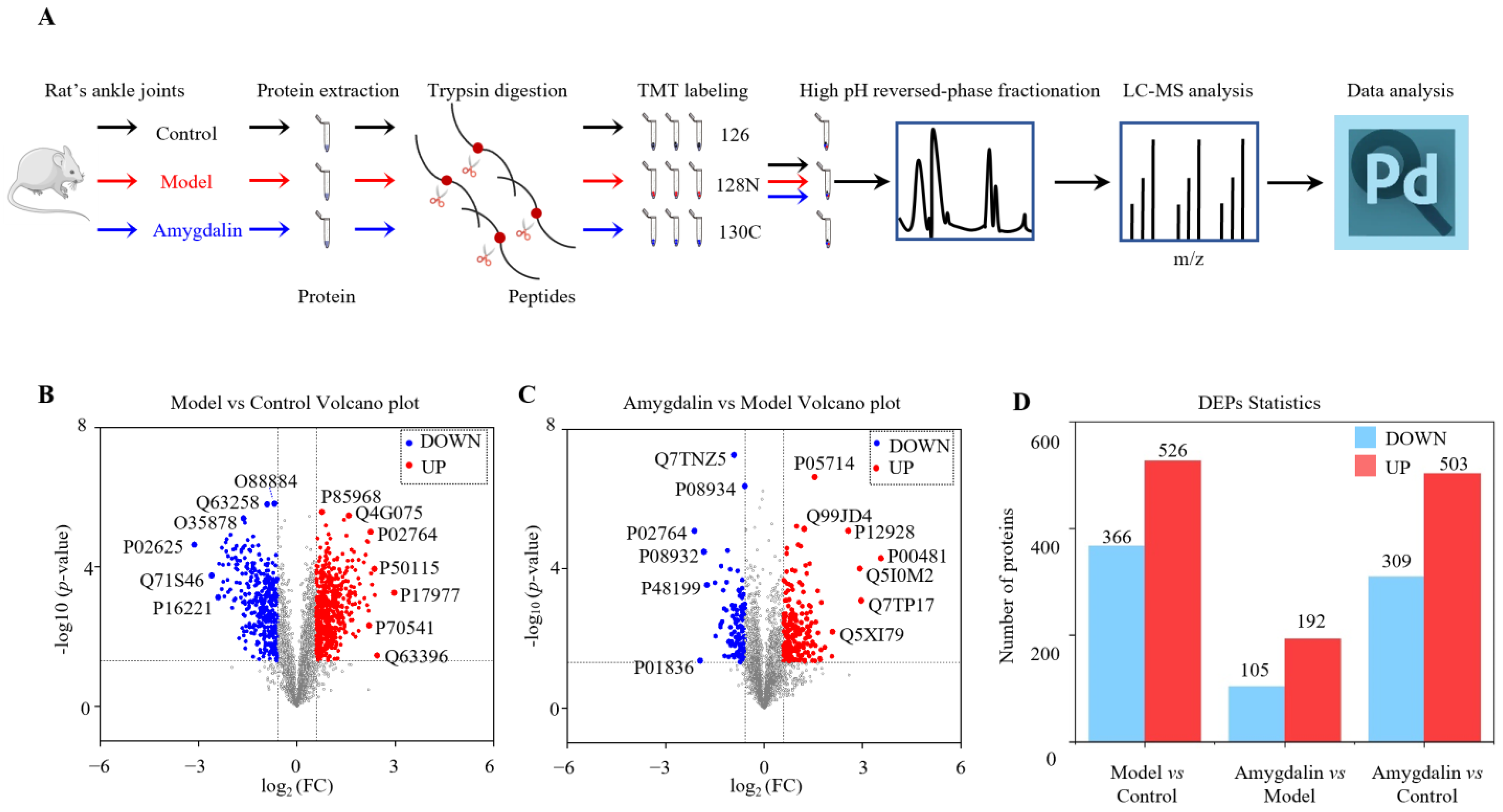
2.2. Identification of DAPs
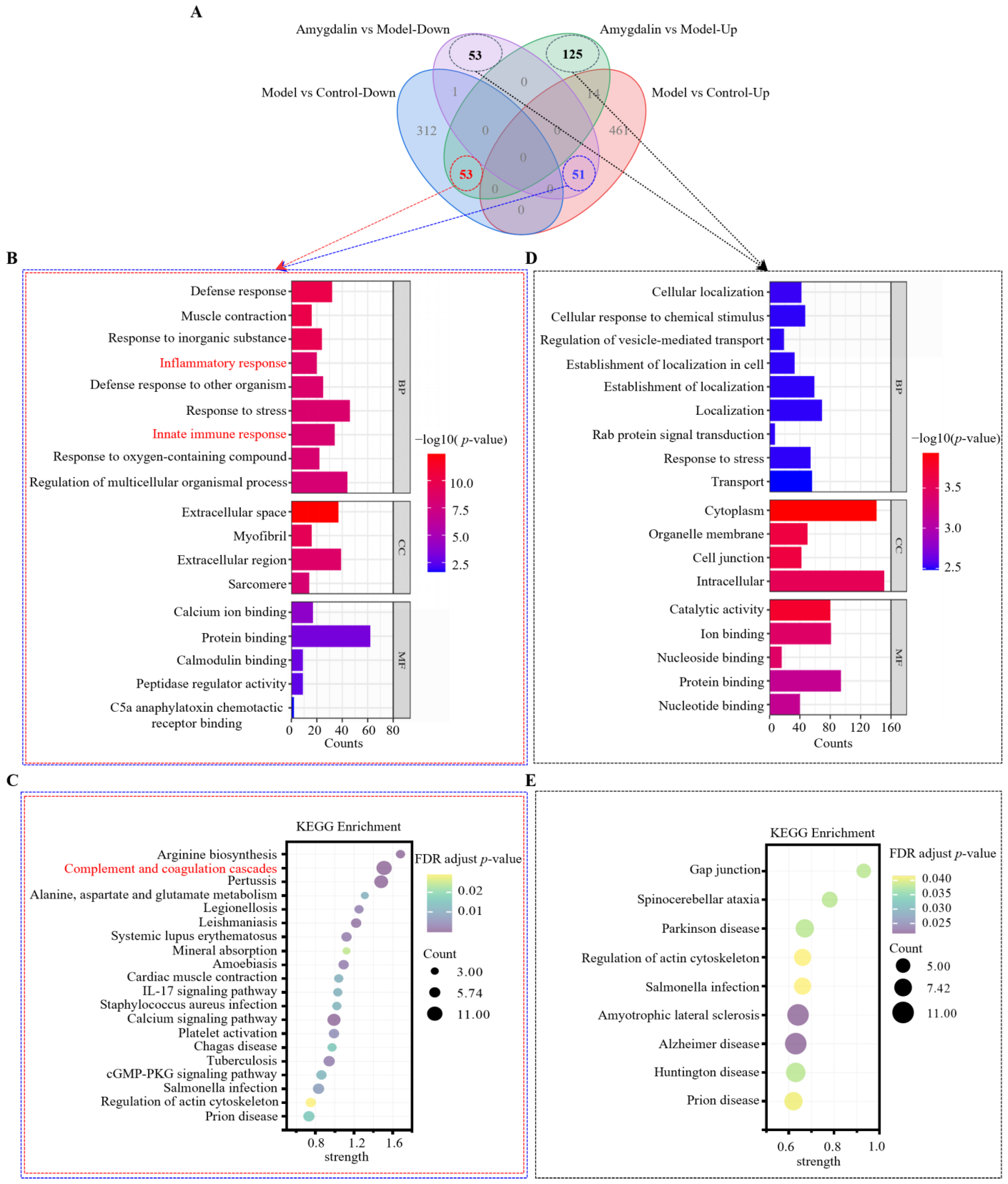
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses of the DAPs
2.4. Heatmap of the DAPs Related to Immune and Inflammation Pathways
2.5. Protein–Protein Interaction (PPI) Analyses and Verification of Proteomics
2.6. Verification of Proteomics and Molecular Docking
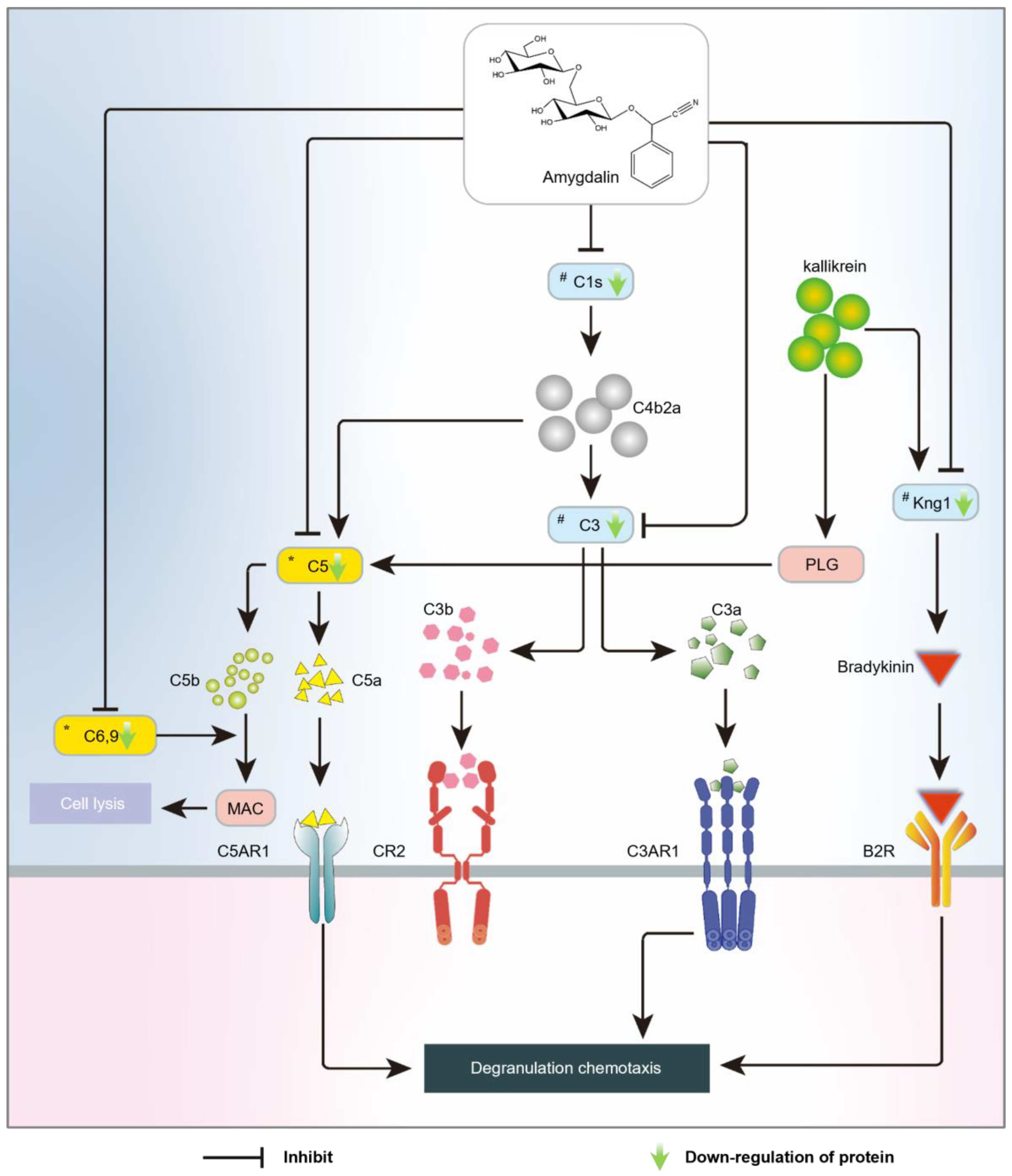
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. CFA-Induced Rat Model Preparation and Treatment
4.3. Measurement of Body Weight, Degree of Paw Swelling, and Arthritis Index Scores
4.4. Histopathological Observation
4.5. ELISA and Quantitative Polymerase Chain Reaction (qPCR) Analysis
4.6. TMT-Labeled Quantitative Proteomics
4.6.1. Peptide Preparation
4.6.2. TMT Labeling
4.6.3. High pH Reversed-Phase Fractionation
4.6.4. Nano LC-MS/MS Analysis and Proteomics Data Processing
4.6.5. Bioinformatics Annotation
4.7. Western Blotting Validation
4.8. Molecular Docking
4.9. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef] [PubMed]
- Radu, A.F.; Bungau, S.G. Management of rheumatoid arthritis: An overview. Cells 2021, 10, 2857. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, L.; Wu, H.; Zhang, Z.R.; Wang, W.Y.; Fu, J.; Deng, R.; Li, F.; Dai, X.J.; Zhan, X. Novel anti-inflammatory target of geniposide: Inhibiting Itgβ1/Ras-Erk1/2 signal pathway via the miRNA-124a in rheumatoid arthritis synovial fibroblasts. Int. Immunopharmacol. 2018, 65, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Firestein, G.S.; McInnes, I.B. Immunopathogenesis of rheumatoid arthritis. Immunity 2017, 46, 183–196. [Google Scholar] [CrossRef]
- Wasserman, A.M. Diagnosis and management of rheumatoid arthritis. Am. Fam. Physician 2011, 84, 1245–1252. [Google Scholar]
- Patel, J.P.; Konanur Srinivasa, N.K.; Gande, A.; Anusha, M.; Dar, H.; Baji, D.B. The role of biologics in rheumatoid arthritis: A narrative review. Cureus 2023, 15, e33293. [Google Scholar] [CrossRef]
- Bullock, J.; Rizvi, S.A.A.; Saleh, A.M.; Ahmed, S.S.; Do, D.P.; Ansari, R.A.; Ahmed, J. Rheumatoid arthritis: A brief overview of the treatment. Med. Princ. Pract. 2018, 27, 501–507. [Google Scholar] [CrossRef]
- Wei, Y.; Li, Y.; Wang, S.; Xiang, Z.; Li, X.; Wang, Q.; Dong, W.; Gao, P.; Dai, L. Phytochemistry and pharmacology of Armeniacae semen Amarum: A review. J. Ethnopharmacol. 2023, 308, 116265. [Google Scholar] [CrossRef]
- He, X.Y.; Wu, L.J.; Wang, W.X.; Xie, P.J.; Chen, Y.H.; Wang, F. Amygdalin–A pharmacological and toxicological review. J. Ethnopharmacol. 2020, 254, 112717. [Google Scholar] [CrossRef]
- Wang, L.; Pu, X.; Nie, X.; Wang, D.; Jiang, H.; Chen, Y.; Pang, L.; Wang, S.; Wang, X.; Xu, Z.; et al. Integrated serum pharmacochemistry and network pharmacological analysis used to explore possible anti-rheumatoid arthritis mechanisms of the Shentong-Zhuyu decoction. J. Ethnopharmacol. 2021, 273, 113988. [Google Scholar] [CrossRef]
- Pharmacopoeia Commission, N. Pharmacopoeia of the People’s Republic of China; China Pharmaceutical Science and Technology Press: Beijing, China, 2020. [Google Scholar]
- Pouw, R.B.; Ricklin, D. Tipping the balance: Intricate roles of the complement system in disease and therapy. Semin. Immunopathol. 2021, 43, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.E.; Garbacki, N.; Molinaro, G.; Brown, N.J.; Marceau, F.; Adam, A. The kallikrein-kinin system: Current and future pharmacological targets. J. Pharmacol. Sci. 2005, 99, 6–38. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A.; van der Meer, J.W. Treating inflammation by blocking interleukin-1 in humans. Semin. Immunol. 2013, 25, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Teng, L.; Qu, Y.; Liu, J.; Zhu, X.; Chen, S.; Yang, L.; Huang, Y.; Song, Q.; Fu, Q. Anti-proliferation and anti-inflammation effects of corilagin in rheumatoid arthritis by downregulating NF-κB and MAPK signaling pathways. J. Ethnopharmacol. 2022, 284, 114791. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.; Guo, D.; Shi, Y.; Zhang, S.; Yang, R.; Cheng, J. The mechanism of action of paeoniae radix rubra-angelicae sinensis radix drug pair in the treatment of rheumatoid arthritis through PI3K/AKT/NF-κB signaling pathway. Front. Pharmacol. 2023, 14, 1113810. [Google Scholar] [CrossRef]
- Radner, H.; Aletaha, D. Anti-TNF in rheumatoid arthritis: An overview. Wien Med. Wochenschr. 2015, 165, 3–9. [Google Scholar] [CrossRef]
- Lin, Y.J.; Anzaghe, M.; Schülke, S. Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells 2020, 9, 880. [Google Scholar] [CrossRef]
- Scherer, H.U.; Häupl, T.; Burmester, G.R. The etiology of rheumatoid arthritis. J. Autoimmun. 2020, 110, 102400. [Google Scholar] [CrossRef]
- Pope, J.E.; Choy, E.H. C-reactive protein and implications in rheumatoid arthritis and associated comorbidities. Semin. Arthritis Rheum. 2021, 51, 219–229. [Google Scholar] [CrossRef]
- Fukami, Y.; Koike, H.; Iijima, M.; Mouri, N.; Nishi, R.; Katsuno, M. Role of complement components in vasculitic neuropathy associated with systemic lupus erythematosus and rheumatoid arthritis. Muscle Nerve 2022, 66, 175–182. [Google Scholar] [CrossRef]
- Banda, N.K.; Deane, K.D.; Bemis, E.A.; Strickland, C.; Seifert, J.; Jordan, K.; Goldman, K.; Morgan, B.P.; Moreland, L.W.; Lewis, M.J.; et al. Analysis of complement gene expression, clinical associations, and biodistribution of complement proteins in the synovium of early rheumatoid arthritis patients reveals unique pathophysiologic features. J. Immunol. 2022, 208, 2482–2496. [Google Scholar] [CrossRef] [PubMed]
- Biró, E.; Nieuwland, R.; Tak, P.P.; Pronk, L.M.; Schaap, M.C.; Sturk, A.; Hack, C.E. Activated complement components and complement activator molecules on the surface of cell-derived microparticles in patients with rheumatoid arthritis and healthy individuals. Ann. Rheum. Dis. 2007, 66, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Holers, V.M.; Banda, N.K. Complement in the initiation and evolution of rheumatoid arthritis. Front. Immunol. 2018, 9, 1057. [Google Scholar] [CrossRef]
- Ammitzboll, C.G.; Thiel, S.; Ellingsen, T.; Deleuran, B.; Jorgensen, A.; Jensenius, J.C.; Stengaard-Pedersen, K. Levels of lectin pathway proteins in plasma and synovial fluid of rheumatoid arthritis and osteoarthritis. Rheumatol. Int. 2012, 32, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, L.; La Torre, F.; Soresina, A.; Federici, S.; Cancrini, C.; Castagnoli, R.; Cinicola, B.L.; Corrente, S.; Giardino, G.; Lougaris, V.; et al. Inherited defects in the complement system. Pediatr. Allergy Immunol. 2022, 33 (Suppl. 27), 73–76. [Google Scholar] [CrossRef] [PubMed]
- Defendi, F.; Thielens, N.M.; Clavarino, G.; Cesbron, J.Y.; Dumestre-Pérard, C. The immunopathology of complement proteins and innate immunity in autoimmune disease. Clin. Rev. Allergy Immunol. 2020, 58, 229–251. [Google Scholar] [CrossRef]
- Geisbrecht, B.V.; Lambris, J.D.; Gros, P. Complement component C3: A structural perspective and potential therapeutic implications. Semin. Immunol. 2022, 59, 101627. [Google Scholar] [CrossRef]
- Zarantonello, A.; Revel, M.; Grunenwald, A.; Roumenina, L.T. C3-dependent effector functions of complement. Immunol. Rev. 2023, 313, 120–138. [Google Scholar] [CrossRef]
- Ikeda, Z.; Kamei, T.; Sasaki, Y.; Reynolds, M.; Sakai, N.; Yoshikawa, M.; Tawada, M.; Morishita, N.; Dougan, D.R.; Chen, C.H.; et al. Discovery of a novel series of potent, selective, orally available, and brain-penetrable C1s inhibitors for modulation of the complement pathway. J. Med. Chem. 2023, 66, 6354–6371. [Google Scholar] [CrossRef]
- Barnum, S.R.; Bubeck, D.; Schein, T.N. Soluble membrane attack complex: Biochemistry and immunobiology. Front. Immunol. 2020, 11, 585108. [Google Scholar] [CrossRef]
- Laursen, N.S.; Magnani, F.; Gottfredsen, R.H.; Petersen, S.V.; Andersen, G.R. Structure, function and control of complement C5 and its proteolytic fragments. Curr. Mol. Med. 2012, 12, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Jore, M.M.; Johnson, S.; Sheppard, D.; Barber, N.M.; Li, Y.I.; Nunn, M.A.; Elmlund, H.; Lea, S.M. Structural basis for therapeutic inhibition of complement C5. Nat. Struct. Mol. Biol. 2016, 23, 378–386. [Google Scholar] [CrossRef]
- Banda, N.K.; Kraus, D.; Vondracek, A.; Huynh, L.H.; Bendele, A.; Holers, V.M.; Arend, W.P. Mechanisms of effects of complement inhibition in murine collagen-induced arthritis. Arthritis Rheum. 2002, 46, 3065–3075. [Google Scholar] [CrossRef] [PubMed]
- Irmscher, S.; Döring, N.; Halder, L.D.; Jo, E.A.H.; Kopka, I.; Dunker, C.; Jacobsen, I.D.; Luo, S.; Slevogt, H.; Lorkowski, S.; et al. Kallikrein cleaves C3 and activates complement. J. Innate Immun. 2018, 10, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Bekassy, Z.; Lopatko Fagerström, I.; Bader, M.; Karpman, D. Crosstalk between the renin-angiotensin, complement and kallikrein-kinin systems in inflammation. Nat. Rev. Immunol. 2022, 22, 411–428. [Google Scholar] [CrossRef]
- Dobó, J.; Major, B.; Kékesi, K.A.; Szabó, I.; Megyeri, M.; Hajela, K.; Juhász, G.; Závodszky, P.; Gál, P. Cleavage of kininogen and subsequent bradykinin release by the complement component: Mannose-binding lectin-associated serine protease (MASP)-1. PLoS ONE 2011, 6, e20036. [Google Scholar] [CrossRef]
- Yang, A.; Zhou, J.; Wang, B.; Dai, J.; Colman, R.W.; Song, W.; Wu, Y. A critical role for plasma kallikrein in the pathogenesis of autoantibody-induced arthritis. FASEB J. 2017, 31, 5419–5431. [Google Scholar] [CrossRef]
- Choudhary, N.; Bhatt, L.K.; Prabhavalkar, K.S. Experimental animal models for rheumatoid arthritis. Immunopharmacol. Immunotoxicol. 2018, 40, 193–200. [Google Scholar] [CrossRef]
- Luo, Y.; Shen, Y.; Zong, L.; Xie, J.; Dai, L.; Luo, X. Anti-rheumatoid arthritis potential of Rhododendron molle G. Don leaf extract in adjuvant induced arthritis rats. J. Ethnopharmacol. 2023, 307, 116175. [Google Scholar] [CrossRef]
- Gu, W.Z.; Brandwein, S.R. Inhibition of type II collagen-induced arthritis in rats by triptolide. Int. J. Immunopharmacol. 1998, 20, 389–400. [Google Scholar] [CrossRef]
- Bao, Y.; Sun, Y.W.; Ji, J.; Gan, L.; Zhang, C.F.; Wang, C.Z.; Yuan, C.S. Genkwanin ameliorates adjuvant-induced arthritis in rats through inhibiting JAK/STAT and NF-kappaB signaling pathways. Phytomedicine 2019, 63, 153036. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]



| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| IL-1β | AGAACATAAGCCAACAAGTGGT | ACAGGTATAGATTCTTCCCCTT |
| TNF-α | CCTCTTCTCATTCCTGCTCGT | TCCTCCTTGTTGGGACCGAT |
| β-actin | CGGGACCTGACAGACTACCTC | AAGTCTAGGGCAACATAGCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Chai, J.-H.; Zhang, Y.; Jing, X.-J.; Kong, X.-W.; Liang, J.; Xia, Y.-G. TMT-Based Proteomics Reveal the Mechanism of Action of Amygdalin against Rheumatoid Arthritis in a Rat Model through Regulation of Complement and Coagulation Cascades. Molecules 2023, 28, 7126. https://doi.org/10.3390/molecules28207126
Zhou L, Chai J-H, Zhang Y, Jing X-J, Kong X-W, Liang J, Xia Y-G. TMT-Based Proteomics Reveal the Mechanism of Action of Amygdalin against Rheumatoid Arthritis in a Rat Model through Regulation of Complement and Coagulation Cascades. Molecules. 2023; 28(20):7126. https://doi.org/10.3390/molecules28207126
Chicago/Turabian StyleZhou, Lan, Jun-Hong Chai, Yi Zhang, Xiao-Jie Jing, Xiang-Wen Kong, Jun Liang, and Yong-Gang Xia. 2023. "TMT-Based Proteomics Reveal the Mechanism of Action of Amygdalin against Rheumatoid Arthritis in a Rat Model through Regulation of Complement and Coagulation Cascades" Molecules 28, no. 20: 7126. https://doi.org/10.3390/molecules28207126
APA StyleZhou, L., Chai, J.-H., Zhang, Y., Jing, X.-J., Kong, X.-W., Liang, J., & Xia, Y.-G. (2023). TMT-Based Proteomics Reveal the Mechanism of Action of Amygdalin against Rheumatoid Arthritis in a Rat Model through Regulation of Complement and Coagulation Cascades. Molecules, 28(20), 7126. https://doi.org/10.3390/molecules28207126






