Nucleoside Analogs: A Review of Its Source and Separation Processes
Abstract
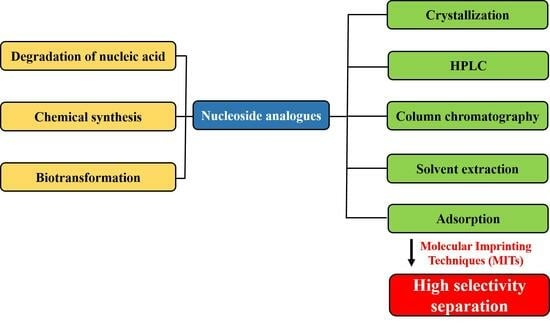
:1. Introduction
2. The Sources of Nucleoside Analogs
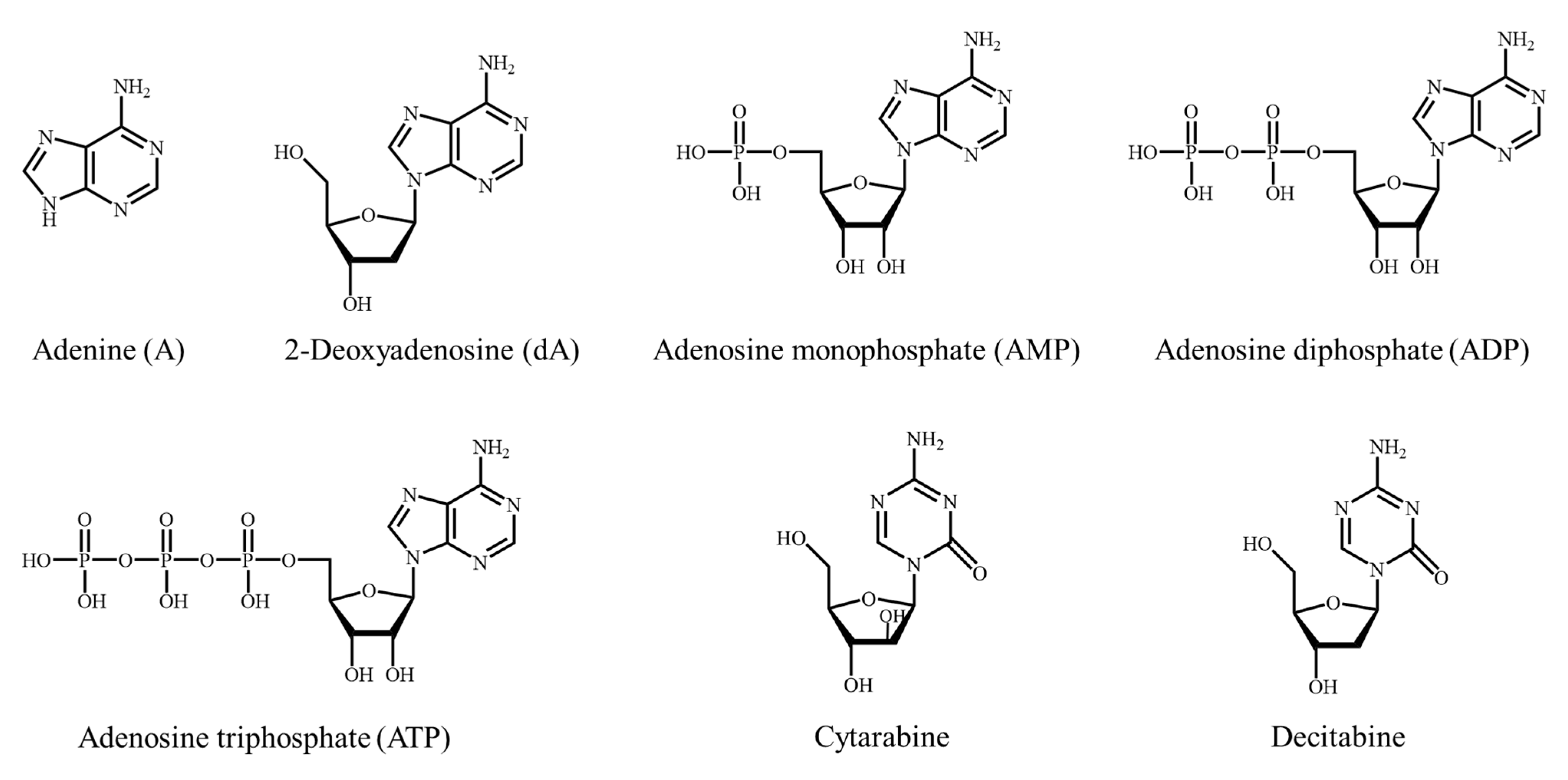
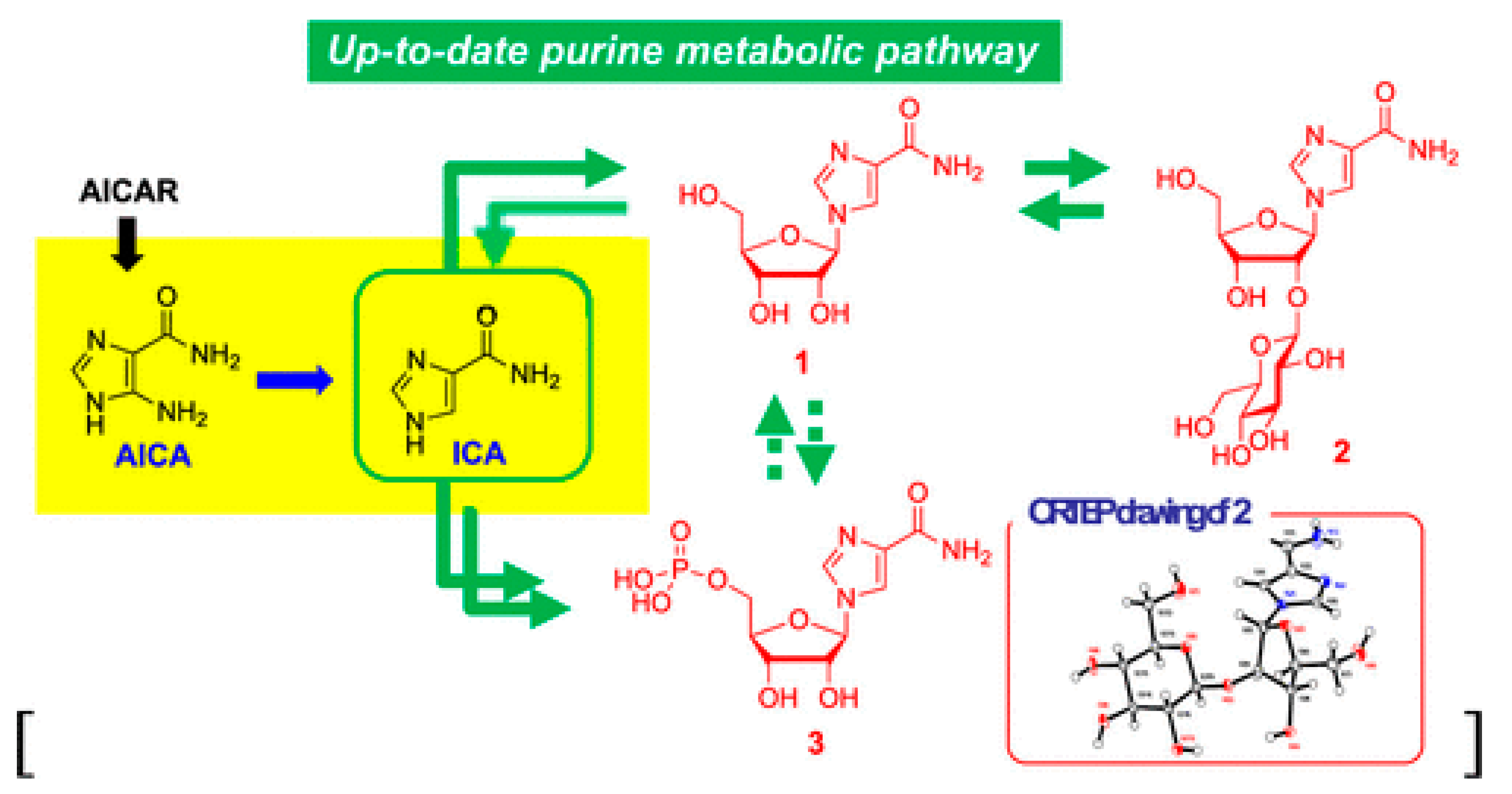
2.1. Degradation of Nucleic Acid
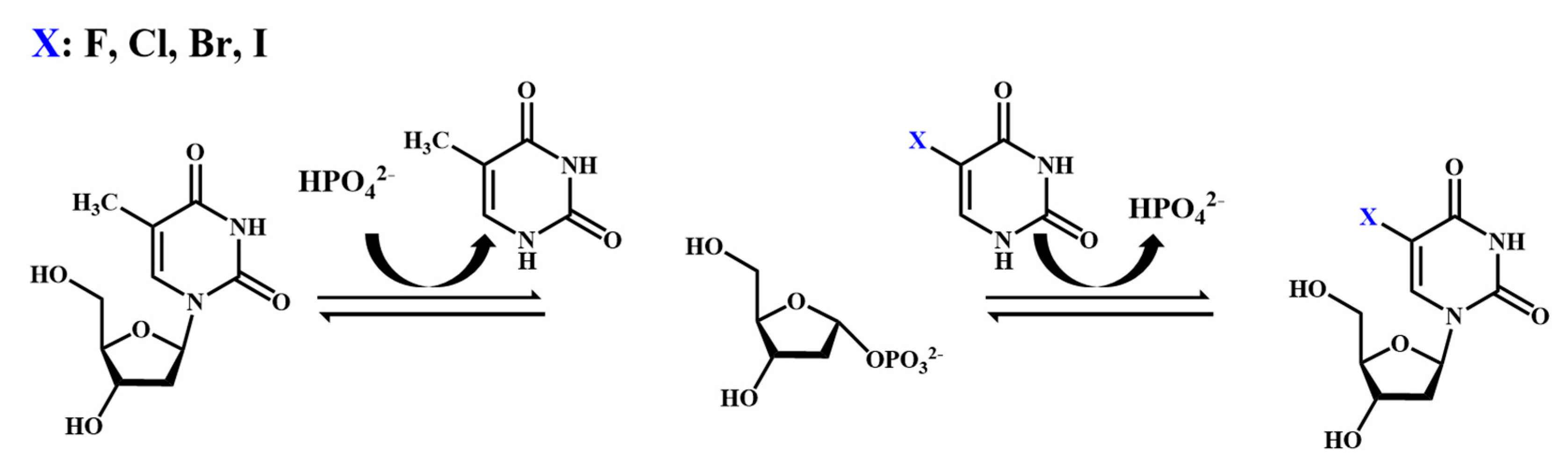
2.2. Chemical Synthesis
2.3. Biotransformation
3. Progress in the Separation of Nucleoside Analogs
3.1. Crystallization
3.2. High-Performance Liquid Chromatography
3.3. Column Chromatography
3.4. Solvent Extraction Method
3.5. Adsorption
4. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the Development of Nucleoside and Nucleotide Analogues for Cancer and Viral Diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef]
- Mashalidis, E.H.; Kaeser, B.; Terasawa, Y.; Katsuyama, A.; Kwon, D.Y.; Lee, K.; Hong, J.; Ichikawa, S.; Lee, S.Y. Chemical Logic of MraY Inhibition by Antibacterial Nucleoside Natural Products. Nat. Commun. 2019, 10, 2917. [Google Scholar] [CrossRef] [PubMed]
- Sampath, D.; Rao, V.A.; Plunkett, W. Mechanisms of Apoptosis Induction by Nucleoside Analogues. Oncogene 2003, 22, 9063–9074. [Google Scholar] [CrossRef] [PubMed]
- Madzharova, F.; Heiner, Z.; Guhlke, M.; Kneipp, J. Surface-Enhanced Hyper-Raman Spectra of Adenine, Guanine, Cytosine, Thymine, and Uracil. J. Phys. Chem. C 2016, 120, 15415–15423. [Google Scholar] [CrossRef]
- Shukla, M.K.; Leszczynski, J. TDDFT Investigation on Nucleic Acid Bases: Comparison with Experiments and Standard Approach. J. Comput. Chem. 2004, 25, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Stentoft, C.; Vestergaard, M.; Løvendahl, P.; Kristensen, N.B.; Moorby, J.M.; Jensen, S.K. Simultaneous Quantification of Purine and Pyrimidine Bases, Nucleosides and Their Degradation Products in Bovine Blood Plasma by High Performance Liquid Chromatography Tandem Mass Spectrometry. J. Chromatogr. A 2014, 1356, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.B. Enzymology of Purine and Pyrimidine Antimetabolites Used in the Treatment of Cancer. Chem. Rev. 2009, 109, 2880–2893. [Google Scholar] [CrossRef]
- Zhou, X.; Szeker, K.; Janocha, B.; Böhme, T.; Albrecht, D.; Mikhailopulo, I.A.; Neubauer, P. Recombinant Purine Nucleoside Phosphorylases from Thermophiles: Preparation, Properties and Activity towards Purine and Pyrimidine Nucleosides. FEBS J. 2013, 280, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Kuzuyama, T. Recent Advances in the Biosynthesis of Nucleoside Antibiotics. J. Antibiot. 2019, 72, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K. Organometallic nucleosides-Synthesis, Transformations, and Applications. Coord. Chem. Rev. 2021, 432, 213705. [Google Scholar] [CrossRef]
- Lopez-Gomez, C.; Levy, R.J.; Sanchez-Quintero, M.J.; Juanola-Falgarona, M.; Barca, E.; Garcia-Diaz, B.; Tadesse, S.; Garone, C.; Hirano, M. Deoxycytidine and Deoxythymidine Treatment for Thymidine Kinase 2 Deficiency. Ann. Neurol. 2017, 81, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Zhao, Y.; Yamagata, Y.; Hua, Y.; Yang, W. Watching DNA Polymerase η Make a Phosphodiester Bond. Nature 2012, 487, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Moretti, J.E.; Müller, U.F. A Ribozyme That Triphosphorylates RNA 5′-Hydroxyl Groups. Nucleic Acids Res. 2014, 42, 4767–4778. [Google Scholar] [CrossRef]
- Degrève, B.; De Clercq, E.; Balzarini, J. Bystander Effect of Purine Nucleoside Analogues in HSV-1tk Suicide Gene Therapy Is Superior to that of Pyrimidine Nucleoside Analogues. Gene Ther. 1999, 6, 162–170. [Google Scholar] [CrossRef]
- Damaraju, V.L.; Damaraju, S.; Young, J.D.; Baldwin, S.A.; Mackey, J.; Sawyer, M.B.; Cass, C.E. Nucleoside Anticancer Drugs: The Role of Nucleoside Transporters in Resistance to Cancer Chemotherapy. Oncogene 2003, 22, 7524–7536. [Google Scholar] [CrossRef]
- Chen, Y.; Bicker, E.; Wu, J.Y.; Xie, M.Y.; Linder, W. Simultaneous Determination of 16 Nucleosides and Nucleobases by Hydrophilic Interaction Chromatography and Its Application to the Quality Evaluation of Ganoderma. J. Agric. Food Chem. 2012, 60, 14243–14252. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside Analogues: Mechanisms of Drug Resistance and Reversal Strategies. Leukemia 2001, 15, 875–890. [Google Scholar] [CrossRef]
- Luo, M.; Groaz, E.; Snoeck, R.; Andrei, G.; Herdewijn, P. Amidate Prodrugs of O-2-Alkylated Pyrimidine Acyclic Nucleosides Display Potent Anti-Herpesvirus Activity. ACS Med. Chem. Lett. 2020, 11, 1410–1415. [Google Scholar] [CrossRef]
- Pastor-Anglada, M.; Molina-Arcas, M.; Casado, F.J.; Bellosillo, B.; Colomer, D.; Gil, J. Nucleoside Transporters in Chronic Lymphocytic Leukaemia. Leukemia 2004, 18, 385–393. [Google Scholar] [CrossRef]
- Martinez-Chapa, S.O.; Madou, M.J. Nanotechnology for COVID-19: Therapeutics and Vaccine Research. ACS nano 2020, 14, 7760–7782. [Google Scholar]
- Racine, S.; de Nanteuil, F.; Serrano, E.; Waser, J. Synthesis of (carbo) nucleoside analogues by [3+2] annulation of aminocyclopropanes. Angew. Chem. Int. Ed. 2014, 53, 8484–8487. [Google Scholar] [CrossRef] [PubMed]
- Meanwell, M.; Silverman, S.M.; Lehmann, J.; Adluri, B.; Britton, R. A Short de Novo Synthesis of Nucleoside Analogues. Science 2020, 369, 725–730. [Google Scholar] [CrossRef]
- Kaminskas, E. Approval Summary: Azacitidine for Treatment of Myelodysplastic Syndrome Subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 3604. [Google Scholar] [CrossRef]
- Aribi, A.; Borthakur, G.; Ravandi, F.; Shan, J.; Davisson, J.; Cortes, J.; Kantarjian, H. Activity of Decitabine, a Hypomethylating Agent, in Chronic Myelomonocytic Leukemia. Cancer 2010, 109, 713–717. [Google Scholar] [CrossRef]
- Koellmann, C.; Wiechert, S.M.; Jones, P.G.; Pietschmann, T.; Werz, D.B. Synthesis of 4′/5′-Spirocyclopropanated Uridine and D-Xylouridine Derivatives and Their Activity against the Human Respiratory Syncytial Virus. Org. Lett. 2019, 21, 6966–6971. [Google Scholar] [CrossRef]
- Kollmann, C.; Sake, S.M.; Jones, P.G.; Pietschmann, T.; Werz, D.B. Protecting-Group-Mediated Diastereoselective Synthesis of C4′-Methylated Uridine Analogs and Their Activity against the Human Respiratory Syncytial Virus. J. Org. Chem. 2020, 85, 4267–4278. [Google Scholar] [CrossRef]
- Elion, G.B. Acyclovir: Discovery, Mechanism of Action, and Selectivity. J. Med. Virol. 1993, 41, 2–6. [Google Scholar] [CrossRef]
- Elion, G.B. The Quest for a Cure. Annu. Rev. Pharmacol. Toxicol. 2003, 33, 1. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Schols, D.; Meier, C. Anti-HIV-Active Nucleoside Triphosphate Prodrugs. J. Med. Chem. 2020, 63, 6003–6027. [Google Scholar] [CrossRef]
- Yamamoto, S.; Wang, M.F.; Adjei, A.A.; Ameho, C.K. Role of Nucleosides and Nucleotides in the Immune System, Gut Reparation after Injury, and Brain Function. Nutrition 1997, 13, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the Microbiota, Immune and Nervous Systems in Health and Disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef]
- Niu, G.; Li, Z.; Huang, P.; Tan, H. Engineering Nucleoside Antibiotics toward the Development of Novel Antimicrobial Agents. J. Antibiot. 2019, 72, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.A.; Ablenas, C.J.; Desrochers, G.F.; Powdrill, M.H.; Pezacki, J.P. A Bifunctional Nucleoside Probe for the Inhibition of the Human Immunodeficiency Virus-Type 1 Reverse Transcriptase. Bioconjug. Chem. 2020, 31, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Borbone, N.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Nucleoside Analogues and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses. Molecules 2021, 26, 986. [Google Scholar] [CrossRef] [PubMed]
- Moussa, Z.; El-Sharief, M.A.M.S.; Abbas, S.Y. New Imidazolidineiminothione Derivatives: Synthesis, Spectral Characterization and Evaluation of Antitumor, Antiviral, Antibacterial. and Antifungal Activities. Eur. J. Med. Chem. Chim. Ther. 2016, 122, 419–428. [Google Scholar] [CrossRef]
- Serpi, M.; Ferrari, V.; Pertusati, F. Nucleoside Derived Antibiotics to Fight Microbial Drug Resistance: New Utilities for an Established Class of Drugs? J. Med. Chem. 2016, 59, 10343–10382. [Google Scholar] [CrossRef]
- Choi, J.H.; Matsuzaki, N.; Wu, J.; Kotajima, M.; Kawagishi, H. Ribosides and Ribotide of a Fairy Chemical, Imidazole-4-Carboxamide, as Its Metabolites in Rice. Org. Lett. 2019, 21, 7841–7845. [Google Scholar] [CrossRef] [PubMed]
- Slavickova, M.; Pohl, R.; Hocek, M. Additions of Thiols to 7-Vinyl-7-Deazaadenine Nucleosides and Nucleotides. Synthesis of Hydrophobic Derivatives of 2′-Deoxyadenosine, dATP and DNA. J. Org. Chem. 2016, 81, 11115–11125. [Google Scholar] [CrossRef] [PubMed]
- Galbis, J.A.; de García-Martín, M.G.; De Paz, M.V.; Galbis, E. Synthetic Polymers from Sugar-Based Monomers. Chem. Rev. 2015, 116, 1600–1636. [Google Scholar] [CrossRef]
- Chiu, C.P.; Liu, S.C.; Tang, C.H.; Chan, Y.; El-Shazly, M.; Lee, C.L.; Du, Y.C.; Wu, T.Y.; Chang, F.R.; Wu, Y.C. Anti-Inflammatory Cerebrosides from Cultivated Cordyceps Militaris. J. Agric. Food Chem. 2016, 64, 1540–1548. [Google Scholar] [CrossRef]
- Lu, M.-Y.; Chen, C.-C.; Kuo, C.-F.; Lin, T.-W.; Lee, L.-Y. N6-(2-Hydroxyethyl) adenosine in the Medicinal Mushroom Cordyceps Cicadae Attenuates Lipopolysaccharide-Stimulated Pro-Inflammatory Responses by Suppressing TLR4-Mediated NF-KB Signaling Pathways. J. Nat. Prod. 2015, 78, 2452–2460. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Chang, T.; Chang, C.Y.; Chen, C.J. Crystal Structure of Nucleoside Diphosphate Kinase Required for Coleoptile Elongation in Rice (Oryza sativa L.). J. Struct. Biol. 2005, 150, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Torres-Torronteras, J.; Cabrera-Pérez, R.; Vila-Julià, F.; Viscomi, C.F.; Martí, R. Long-Term Sustained Effect of Liver-Targeted Adeno-Associated Virus Gene Therapy for Mitochondrial Neurogastrointestinal Encephalomyopathy. Hum. Gene Ther. 2018, 29, 708–718. [Google Scholar] [CrossRef]
- Jaenisch, R. Germ Line Integration and Mendelian Transmission of the Exogenous Moloney Leukemia Virus. Proc. Natl. Acad. Sci. USA 1976, 73, 1260–1264. [Google Scholar] [CrossRef]
- Radu, C.G.; Shu, C.J.; Nair-Gill, E.; Shelly, S.M.; Barrio, J.R.; Satyamurthy, N.; Phelps, M.E.; Witte, O.N. Molecular Imaging of Lymphoid Organs and Immune Activation by Positron Emission Tomography with a New [18F]-Labeled 2′-deoxycytidine Analog. Infect. Dis. Aquac. 2008, 14, 3–68. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Belda, M.; Fernández-Caballero, I.; Campillo, N.; Arroyo-Manzanares, N.; Hernández-Córdoba, M.; Vias, P. Hydrophilic Interaction Liquid Chromatography Coupled to Quadrupole-Time-of-Flight Mass Spectrometry for Determination of Nuclear and Cytoplasmatic Contents of Nucleotides, Nucleosides and Their Nucleobases in Food Yeasts. Talanta Open. 2021, 4, 100064. [Google Scholar] [CrossRef]
- Babu, K.N.; Pallavi, P.N. Isolation, Identification and Mass Multiplication of Trichodermaan Important Bio-Control Agent. Int. J. Pharm. Life Sci. 2013, 4, 2320–2323. [Google Scholar]
- Correa, Y.; Cabanillas, B.; Jullian, V.; Lvarez, D.; Castillo, D. Identification and Characterization of Compounds from Chrysosporium Multifidum, a Fungus with Moderate Antimicrobial Activity Isolated from Hermetia Illucens Gut Microbiota. PLoS ONE 2019, 14, e0218837. [Google Scholar] [CrossRef]
- Mcintosh, J.A.; Benkovics, T.; Silverman, S.M.; Huffman, M.A.; Fier, P.S. Engineered Ribosyl-1-Kinase Enables Concise Synthesis of Molnupiravir, an Antiviral for COVID-19. ACS Cent. Sci. 2021, 12, 1980–1985. [Google Scholar] [CrossRef] [PubMed]
- Westarp, S.; Kaspar, F.; Neubauer, P.; Kurreck, A. Industrial Potential of the Enzymatic Synthesis of Nucleoside Analogues: Existing Challenges and Perspectives. Curr. Opin. Biotechnol. 2022, 78, 102829. [Google Scholar] [CrossRef]
- Cheng, X.; Ma, L. Enzymatic Synthesis of Fluorinated Compounds. Appl. Microbiol. Biotechnol. 2021, 105, 8033–8058. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.C.; Miller, G.J. Advances in Biocatalytic and Chemoenzymatic Synthesis of Nucleoside Analogues. Expert Opin. Drug Discov. 2022, 17, 355–364. [Google Scholar] [CrossRef]
- Nie, S.; Li, W.; Yu, B. Total Synthesis of Nucleoside Antibiotic A201A. J. Am. Chem. Soc. 2014, 136, 4157–4160. [Google Scholar] [CrossRef] [PubMed]
- Trost, B.M.; Xu, S.; Sharif, E.U. New Catalytic Asymmetric Formation of Oxygen Heterocycles Bearing Nucleoside Bases at the Anomeric Carbon. J. Am. Chem. Soc. 2019, 141, 10199–10204. [Google Scholar] [CrossRef]
- Xu, J.; Green, N.J.; Gibard, C.; Krishnamurthy, R.; Sutherland, J.D. Prebiotic Phosphorylation of 2-Thiouridine Provides Either Nucleotides or DNA Building Blocks via Photoreduction. Nat. Chem. 2019, 11, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Duchek, J.; Adams, D.R.; Hudlicky, T. Chemoenzymatic Synthesis of Inositols, Conduritols, and Cyclitol Analogues. Chem. Rev. 2011, 111, 4223–4258. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.J. Unifying the Synthesis of Nucleoside Analogues. Science 2020, 369, 623. [Google Scholar] [CrossRef]
- Li, Q.; Groaz, E.; Persoons, L.; Daelemans, D.; Herdewijn, P. Synthesis and Antitumor Activity of C-7-Alkynylated and Arylated Pyrrolotriazine C-Ribonucleosides. ACS Med. Chem. Lett. 2020, 11, 1605–1610. [Google Scholar] [CrossRef]
- Fujino, H.; Fukuda, T.; Nagatomo, M.; Inoue, M. Convergent Total Synthesis of Hikizimycin Enabled by Intermolecular Radical Addition to Aldehyde. J. Am. Chem. Soc. 2020, 142, 13227–13234. [Google Scholar] [CrossRef]
- Das, P.; Almond, D.W.; Tumbelty, L.N.; Austin, B.E.; Moura-Letts, G. From Heterocycles to Carbacycles: Synthesis of Carbocyclic Nucleoside Analogues from Enals and Hydroxylamines. Org. Lett. 2020, 22, 5491–5495. [Google Scholar] [CrossRef]
- Xie, M.S.; Wang, Y.; Li, J.P.; Du, C.; Zhang, Y.Y.; Hao, E.J.; Zhang, Y.M.; Qu, G.R.; Guo, H.M. A Straightforward Entry to Chiral Carbocyclic Nucleoside Analogues via the Enantioselective [3+2] Cycloaddition of α-Nucleobase Substituted Acrylates. Chem. Commun. 2015, 51, 12451–12454. [Google Scholar] [CrossRef]
- Downey, A.M.; Pohl, R.; Roithová, J.; Hocek, M. Synthesis of Nucleosides through Direct Glycosylation of Nucleobases with 5-O-Monoprotected or 5-Modified Ribose: Improved Protocol, Scope, and Mechanism. Chem. Eur. J. 2017, 23, 3910–3917. [Google Scholar] [CrossRef]
- Downey, A.M.; Hocek, M. Strategies toward Protecting Group-Free Glycosylation through Selective Activation of the Anomeric Center. Beilstein J. Org. Chem. 2017, 13, 1239–1279. [Google Scholar] [CrossRef]
- Merino, P. Chemical Synthesis of Nucleoside Analogues; John Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Romeo, G.; Chiacchio, U.; Corsaro, A.; Merino, P. Chemical Synthesis of Heterocyclic-Sugar Nucleoside Analogues. Chem. Rev. 2010, 110, 3337–3370. [Google Scholar] [CrossRef]
- De Benedetti, E.C.; Rivero, C.W.; Britos, C.N.; Lozano, M.E.; Trelles, J.A. Biotransformation of 2,6-Diaminopurine Nucleosides by Immobilized Geobacillus Stearothermophilus. Biotechnol. Prog. 2012, 28, 1251–1256. [Google Scholar] [CrossRef]
- Hellendahl, K.F.; Kamel, S.; Wetterwald, A.; Neubauer, P.; Wagner, A. Human Deoxycytidine Kinase Is a Valuable Biocatalyst for the Synthesis of Nucleotide Analogues. Catalysts 2019, 9, 997. [Google Scholar] [CrossRef]
- Yokozeki, K.; Tsuji, T. A Novel Enzymatic Method for the Production of Purine-2′-Deoxyribonucleosides. J. Mol. Catal. B Enzym. 2000, 10, 207–213. [Google Scholar] [CrossRef]
- Yao, Y.; Xiong, J.; Chen, Y.; Tang, J.; Ying, H. Enhanced Adenosine Triphosphate Production by Saccharomyces Cerevisiae Using an Efficient Energy Regeneration System. Korean J. Chem. Eng. 2011, 28, 178–183. [Google Scholar] [CrossRef]
- Ben’itez-Mateos, A.I.; Klein, C.; Padrosa, D.R.; Paradisi, F. A Novel Thymidine Phosphorylase to Synthesize (Halogenated) Anticancer and Antiviral Nucleoside Drugs in Continuous Flow. Catal. Sci. Technol. 2022, 12, 6231–6238. [Google Scholar] [CrossRef]
- Cattaneo, G.; Rabuffetti, M.; Speranza, G.; Kupfer, T.; Peters, B.; Massolini, G.; Ubiali, D.; Calleri, E. Synthesis of Adenine Nucleosides by Transglycosylation Using Two Sequential Nucleoside Phosphorylase-Based Bioreactors with on-Line Reaction Monitoring by Using HPLC. ChemCatChem 2017, 9, 4614–4620. [Google Scholar] [CrossRef]
- Rivero, C.W.; De Benedetti, E.C.; Lozano, M.E.; Trelles, J.A. Bioproduction of Ribavirin by Green Microbial Biotransformation. Process Biochem. 2015, 50, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, M.; Nidetzky, B. Biocatalytic Cascade Transformations for the Synthesis of C-Nucleosides and N-Nucleoside Analogues. Curr. Opin. Biotechnol. 2023, 79, 102873. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, L.; Huang, H.; Linhardt, R.J. Chemoenzymatic Synthesis of Glycosaminoglycans. Acc. Chem. Res. 2019, 53, 335–346. [Google Scholar] [CrossRef]
- Cohen, S.; Jordheim, L.P.; Megherbi, M.; Dumontet, C.; Guitton, J. Liquid Chromatographic Methods for the Determination of Endogenous Nucleotides and Nucleotide Analogues Used in Cancer Therapy: A Review. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010, 878, 1912. [Google Scholar] [CrossRef]
- Vinogradov, S.V.; Zeman, A.D.; Batrakova, E.V.; Kabanov, A. V Polyplex Nanogel Formulations for Drug Delivery of Cytotoxic Nucleoside Analogues-ScienceDirect. J. Control. Release 2005, 107, 143–157. [Google Scholar] [CrossRef]
- Ikeda, R.; Nishimura, M.; Sun, Y.; Wada, M.; Nakashima, K. Simple HPLC-UV Determination of Nucleosides and Its Application to the Authentication of Cordyceps and Its Allies. Biomed. Chromatogr. 2010, 22, 630–636. [Google Scholar] [CrossRef]
- Sun, H.; Zheng, L.; Greenberg, M.M. Independent Generation of Reactive Intermediates Leads to an Alternative Mechanism for Strand Damage Induced by Hole Transfer in Poly (dA--T) Sequences. J. Am. Chem. Soc. 2018, 140, 11308–11316. [Google Scholar] [CrossRef]
- Davison, E.K.; Petrone, D.A.; Meanwell, M.; Nodwell, M.B.; Silverman, S.M.; Campeau, L.-C.; Britton, R. Practical and Concise Synthesis of Nucleoside Analogues. Nat. Protoc. 2022, 17, 2008–2024. [Google Scholar] [CrossRef]
- Mohammadi, S.; Alfaro, A.C.; Baroutian, S.; Seyfoddin, A. Extraction of Bioactive Compounds from Black-footed Abalone (Haliotis Iris) Using Subcritical Water Extraction. J. Chem. Technol. Biotechnol. 2022, 97, 3511–3519. [Google Scholar] [CrossRef]
- Holzer, S.; Rzechorzek, N.J.; Short, I.R.; Jenkyn-Bedford, M.; Kilkenny, M.L. Structural Basis for Inhibition of Human Primase by Arabinofuranosyl Nucleoside Analogues Fludarabine and Vidarabine. ACS Chem. Biol. 2019, 14, 1904–1912. [Google Scholar] [CrossRef]
- Wang, L.; Sun, D.W. Rapid Cooling of Porous and Moisture Foods by Using Vacuum Cooling Technology. Trends Food Sci. Technol. 2001, 12, 174–184. [Google Scholar] [CrossRef]
- Glasgow, S.M. Fermentation and Biochemical Engineering Handbook. In Crystallization; William Andrew: Norwich, NY, USA, 1996; pp. 535–557. [Google Scholar]
- Accelerating Crystallization of Open Organic Materials by Poly(ionic Liquid)s. Angew. Chem. Int. Ed. 2020, 59, 22109–22116. [CrossRef] [PubMed]
- Fleming, A.M.; Alenko, A.; Kitt, J.P.; Orendt, A.M.; Burrows, C.J. Structural Elucidation of Bisulfite Adducts to Pseudouridine That Result in Deletion Signatures during Reverse Transcription of RNA. J. Am. Chem. Soc. 2019, 141, 16450–16460. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Korshin, G.; Wang, D.; Cai, Z. Characterization of Dissolved Organic Matter Using High-Performance Liquid Chromatography (HPLC)-Size Exclusion Chromatography (SEC) with a Multiple Wavelength Absorbance Detector. Chemosphere 2012, 87, 879–885. [Google Scholar] [CrossRef]
- Okuda, T.; Naoi, D.; Tenmoku, M.; Tanaka, S.; He, K.; Ma, Y.; Yang, F.; Lei, Y.; Jia, Y.; Zhang, D. Polycyclic Aromatic Hydrocarbons (PAHs) in the Aerosol in Beijing, China, Measured by Aminopropylsilane Chemically-Bonded Stationary-Phase Column Chromatography and HPLC/fluorescence Detection. Chemosphere 2006, 65, 427–435. [Google Scholar] [CrossRef]
- Broeckhoven, K.; Desmet, G. Advances and Innovations in Liquid Chromatography Stationary Phase Supports. Anal. Chem. 2020, 93, 257–272. [Google Scholar] [CrossRef]
- Sun, P.; Binter, E.A.; Liang, Z.; Brown, M.A.; Schlossman, M.L. Antagonistic Role of Aqueous Complexation in the Solvent Extraction and Separation of Rare Earth Ions. ACS Cent. Sci. 2021, 7, 1908–1918. [Google Scholar] [CrossRef]
- Li, X.B.; Wei, C.; Deng, Z.g.; Li, C.X.; Fan, G.; Rong, H.; Zhang, F. Extraction and Separation of Indium and Copper from Zinc Residue Leach Liquor by Solvent Extraction. Sep. Purif. Technol. 2015, 156, 348–355. [Google Scholar] [CrossRef]
- Banerjee, D.; Simon, C.M.; Plonka, A.M.; Motkuri, R.K.; Liu, J.; Chen, X.; Smit, B.; Parise, J.B.; Haranczyk, M.; Thallapally, P.K. Metal-Organic Framework with Optimally Selective Xenon Adsorption and Separation. Nat. Commun. 2016, 7, 11831. [Google Scholar] [CrossRef]
- Mohammed, N.; Lian, H.; Islam, M.S.; Strong, M.K.; Tam, K.C. Selective Adsorption and Separation of Organic Dyes Using Functionalized Cellulose Nanocrystals. Chem. Eng. J. 2021, 417, 129237. [Google Scholar] [CrossRef]
- Mohamed, M.H.; Elsaidi, S.K.; Pham, T.; Forrest, K.A.; Schaef, H.T.; Hogan, A.; Wojtas, L.; Xu, W.; Space, B.; Zaworotko, M.J. Hybrid Ultra-Microporous Materials for Selective Xenon Adsorption and Separation. Angew. Chem. 2016, 128, 8425–8429. [Google Scholar] [CrossRef]
- Yang, P.; Wen, Q.; Wu, J.; Zhuang, W.; Zhang, Y.; Ying, H. Determination of Solubility of cAMPNa in Water + (Ethanol, Methanol, and Acetone) within 293.15–313.15 K. Ind. Eng. Chem. Res. 2014, 53, 10803–10809. [Google Scholar] [CrossRef]
- Abou-Elkhair, R.A.I.; Moustafa, A.H.; Haikal, A.Z.; Ibraheem, A.M. Synthesis and Biological Evaluation of 2-Oxonicotinonitriles and 2-Oxonicotinonitrile Based Nucleoside Analogues. Eur. J. Med. Chem. 2014, 74, 388–397. [Google Scholar] [CrossRef]
- Wozniak, L.A.; Okruszek, A. The Stereospecific Synthesis of P-Chiral Biophosphates and Their Analogues by the Stec Reaction. Chem. Soc. Rev. 2003, 32, 158–169. [Google Scholar] [CrossRef]
- Weimann, G.; Khorana, H.G. Studies on Polynucleotides. XIII.1 Stepwise Synthesis of Deoxyribo-Oligonucleotides. An Alternative General Approach and the Synthesis of Thymidine Di-, Tri- and Tetranucleotides Bearing 3”-Phosphomonoester End Groups2. J. Am. Chem. Soc. 1962, 84, 419–430. [Google Scholar] [CrossRef]
- Suárez-Marina, I.; Abul-Haija, Y.M.; Turk-Macleod, R.; Gromski, P.S.; Cooper, G.J.T.; Olivé, A.O.; Colón-Santos, S.; Cronin, L. Integrated Synthesis of Nucleotide and Nucleosides Influenced by Amino Acids. Commun. Chem. 2019, 2, 28. [Google Scholar] [CrossRef]
- Traube, F.R.; Schiffers, S.; Iwan, K.; Kellner, S.; Spada, F.; Müller, M.; Carell, T. Isotope-Dilution Mass Spectrometry for Exact Quantification of Noncanonical DNA Nucleosides. Nat. Protoc. 2018, 14, 283–312. [Google Scholar] [CrossRef]
- Pertusati, F.; McGuigan, C. Diastereoselective Synthesis of P-Chirogenic Phosphoramidate Prodrugs of Nucleoside Analogues (ProTides) via Copper Catalysed Reaction. Chem. Commun. 2015, 51, 8070–8073. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H.X.; Li, Y.; Lam, C.W.K.; Wang, C.Y.; Zhang, W.J.; Wong, V.K.W.; Pang, S.S.; Yao, M.C.; Zhang, W. Method for Quantification of Ribonucleotides and Deoxyribonucleotides in Human Cells Using (Trimethylsilyl) Diazomethane Derivatization Followed by Liquid Chromatography--Tandem Mass Spectrometry. Anal. Chem. 2018, 91, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Otieno, D.A.; Jondiko, I.J.; Mcdowell, P.G.; Kezdy, F.J. Quantitative Analysis of the Pyrethrins by HPLC. J. Chromatogr. Sci. 1982, 20, 12. [Google Scholar] [CrossRef]
- Monosik, R.; Magdolen, P.; Stredansky, M.; Sturdik, E. Monitoring of Monosaccharides, Oligosaccharides, Ethanol and Glycerol during Wort Fermentation by Biosensors, HPLC and Spectrophotometry. Food Chem. 2013, 138, 220–226. [Google Scholar] [CrossRef]
- Chang, H.; Bajaj, I.; Motagamwala, A.H.; Somasundaram, A.; Huber, G.W.; Maravelias, C.T.; Dumesic, J.A. Sustainable Production of 5-Hydroxymethyl Furfural from Glucose for Process Integration with High Fructose Corn Syrup Infrastructure. Green Chem. 2021, 23, 3277–3288. [Google Scholar] [CrossRef]
- Yun, J.; Wu, H.; Liu, J.; Shen, S.; Zhang, S.; Xu, L.; Yao, K.; Yao, S.J. Strategy of Combining Prefiltration and Chromatography Using Composite Cryogels for Large-Scale Separation of Biotransformation Compounds from Crude High-Cell-Density Broth. Ind. Eng. Chem. Res. 2015, 54, 2564–2572. [Google Scholar] [CrossRef]
- Li, X.; Dumbre, S.G.; Lescrinier, E.; Groaz, E.; Herdewijn, P. Synthesis and Conformation of Pentopyranoside Nucleoside Phosphonates. J. Org. Chem. 2019, 84, 6589–6603. [Google Scholar] [CrossRef]
- Pathak, V.; Pathak, A.K.; Reynolds, R.C. Synthesis of Aza-Acyclic Nucleoside Libraries of Purine, Pyrimidine and 1,2,4-Triazole. ACS Comb. Sci. 2019, 21, 183–191. [Google Scholar] [CrossRef]
- Bracegirdle, J.; Gordon, D.P.; Harvey, J.E.; Keyzers, R.A. Kinase-Inhibitory Nucleoside Derivatives from the Pacific Bryozoan Nelliella Nelliiformis. J. Nat. Prod. 2020, 83, 547–551. [Google Scholar] [CrossRef]
- Jin, Y.; Fan, J.T.; Gu, X.L.; Zhang, L.Y.; Han, J.; Du, S.H.; Zhang, A.X. Neuroprotective Activity of Cerebrosides from Typhonium Giganteum by Regulating Caspase-3 and Bax/Bcl-2 Signaling Pathways in PC12 Cells. J. Nat. Prod. 2017, 80, 1734–1741. [Google Scholar] [CrossRef]
- Zheng, H.; Lin, H.; Sui, J.; Yin, J.; Wang, B.; Pavase, T.R.; Cao, L. Preparation of a Boronate-Functionalized Affinity Silica Hybrid Monolith Column for the Specific Capture of Nucleosides. ChemistrySelect 2019, 4, 623–628. [Google Scholar] [CrossRef]
- Li, S.P.; Li, P.; Lai, C.M.; Gong, Y.X.; Kan, K.K.W.; Dong, T.T.X.; Tsim, K.W.K.; Wang, Y.T. Simultaneous Determination of Ergosterol, Nucleosides and Their Bases from Natural and Cultured Cordyceps by Pressurised Liquid Extraction and High-Performance Liquid Chromatography. J. Chromatogr. A 2004, 1036, 239–243. [Google Scholar] [CrossRef]
- Yang, F.Q.; Li, S.P. Effects of Sample Preparation Methods on the Quantification of Nucleosides in Natural and Cultured Cordyceps. J. Pharm. Biomed. Anal. 2008, 48, 231–235. [Google Scholar] [CrossRef]
- Rahman, M.M.; Abd El-Aty, A.M.; Kim, S.W.; Na, T.W.; Shin, H.C.; Hong, S.M.; Shim, J.H. A Simple Extraction Method for the Detection and Quantification of Polyoxin D, a Nucleoside Antibiotic, in Butterbur Using UPLC-MS/MS. Food Chem. 2017, 221, 683–688. [Google Scholar] [CrossRef]
- Matějícek, P.; Cígler, P.; Olejniczak, A.B.; Andrysiak, A.; Wojtczak, B.; Procházka, K.; Lesnikowski, Z.J. Aggregation Behavior of NucleosideBoron Cluster Conjugates in Aqueous Solutions. Langmuir 2008, 24, 2625–2630. [Google Scholar] [CrossRef]
- Kuś, P.M.; Rola, R. LC-QqQ-MS/MS Methodology for Determination of Purine and Pyrimidine Derivatives in Unifloral Honeys and Application of Chemometrics for Their Classification. Food Chem. 2021, 348, 129076. [Google Scholar] [CrossRef]
- Jin, H.; Lao, Y.M.; Zhou, J.; Zhang, H.J.; Cai, Z.H. A Rapid UHPLC-HILIC Method for Algal Guanosine 5′-Diphosphate 3′-Diphosphate (ppGpp) and the Potential Separation Mechanism. J. Chromatogr. B 2018, 1096, 143–153. [Google Scholar] [CrossRef]
- Datta, K.K.R.; Vinu, A.; Mandal, S.; Al-Deyab, S.; Hill, J.P.; Ariga, K. Base-Selective Adsorption of Nucleosides to Pore-Engineered Nanocarbon, Carbon Nanocage. J. Nanosci. Nanotechnol. 2011, 11, 3959–3964. [Google Scholar] [CrossRef]
- Burcu, O.; Ceren, T.; Aktas, U.; Tuncal, A.; Vurmaz, D.; Deniz, A. Boronate Affinity Nanoparticles for Nucleoside Separation. Artif. Cells Nanomed. Biotechnol. 2016, 44, 322–327. [Google Scholar]
- Zhao, S.; Zou, Y.; Wang, Y.; Zhang, H.; Liu, X. Organized Cryogel Composites with 3D Hierarchical Porosity as an Extraction Adsorbent for Nucleosides. J. Sep. Sci. 2019, 42, 2140–2147. [Google Scholar] [CrossRef]
- Cheng, T.; Liu, X.; Huihui, L.; Wang, S.; Zhang, H.; Zhu, B. Boronic Acid-Functionalized Attapulgite with High Adsorption Capacity for Selective Capture of Nucleosides at Acidic pH Values. Mikrochim. Acta Int. J. Phys. Chem. Methods Anal. 2016, 183, 1779–1786. [Google Scholar]
- Jia, M.; Zhang, Z.; Li, J.; Ma, X.; Chen, L.; Yang, X. Molecular Imprinting Technology for Microorganism Analysis. TrAC Trends Anal. Chem. 2018, 106, 190–201. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular Imprinting: Perspectives and Applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar]
- Liu, J.; Wang, P.; Zhou, M.; Ma, Y.; Niu, X.; Pan, G.; Pan, J. Tailored Janus Silica Nanosheets Integrating Bispecific Artificial Receptors for Simultaneous Adsorption of 2, 6-Dichlorophenol and Pb (II). J. Mater. Chem. A 2019, 7, 16161–16175. [Google Scholar] [CrossRef]
- Wang, P.; Liu, J.; Ma, Y.; Tang, J.; Yang, K.; Liu, Z.; Pan, J. Mosaic-Inspired Magnetic Alginate Composite Sorbents Derived from Coalesce of Two Emulsion Droplets for Selective Recognition of 2′-Deoxyadenosine. Chem. Eng. J. 2020, 394, 124931. [Google Scholar] [CrossRef]
- Mouro, C.A.; Bokeloh, F.; Xu, J.; Prost, E.; Duma, L.; Merlier, F.; Bueno, S.M.A.; Haupt, K.; Bernadette, T.S.B. Dual-Oriented Solid-Phase Molecular Imprinting: Toward Selective Artificial Receptors for Recognition of Nucleotides in Water. Macromolecules 2017, 50, 7484–7490. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, P.; Song, Y.; Li, H.; Luo, J.; Pan, J. Hybrid Hydrogel Microspheres Loading Single-Hole Hollow Imprinted Particles for Fast and Selective Uptake of 2′-Deoxyadenosine. Sep. Purif. Technol. 2022, 287, 120472. [Google Scholar] [CrossRef]
- Breton, F.; Delépée, R.; Agrofoglio, L.A. Molecular Imprinting of AMP by an Ionic-Noncovalent Dual Approach. J. Sep. Sci. 2009, 32, 3285–3291. [Google Scholar] [CrossRef]
- Siyal, A.A.; Shamsuddin, M.R.; Khan, M.I.; Rabat, N.E.; Zulfiqar, M.; Man, Z.; Siame, J.; Azizli, K.A. A Review on Geopolymers as Emerging Materials for the Adsorption of Heavy Metals and Dyes. J. Environ. Manag. 2018, 224, 327–339. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; He, G.; Yu, L.; Noor, N.; Sun, Y.; Zhou, X.; Hu, J.; Parkin, I.P. Enhanced Adsorption Capacity of Ultralong Hydrogen Titanate Nanobelts for Antibiotics. J. Mater. Chem. A 2017, 5, 4352–4358. [Google Scholar] [CrossRef]
- Liao, Q.; Rong, H.; Zhao, M.; Luo, H.; Chu, Z.; Wang, R. Strong Adsorption Properties and Mechanism of Action with Regard to Tetracycline Adsorption of Double-Network Polyvinyl Alcohol-Copper Alginate Gel Beads. J. Hazard. Mater. 2022, 422, 126863. [Google Scholar] [CrossRef]
- Kaya, S.I.; Cetinkaya, A.; Ozkan, S.A. Molecularly Imprinted Polymers as Highly Selective Sorbents in Sample Preparation Techniques and Their Applications in Environmental Water Analysis. Trends Environ. Anal. Chem. 2022, 37, e00193. [Google Scholar] [CrossRef]
- Ansari, S.; Karimi, M. Novel Developments and Trends of Analytical Methods for Drug Analysis in Biological and Environmental Samples by Molecularly Imprinted Polymers. TrAC Trends Anal. Chem. 2017, 89, 146–162. [Google Scholar] [CrossRef]
- Gong, X.; Tang, B.; Liu, J.J.; You, X.Y.; Gu, J.; Deng, J.Y.; Xie, W.-H. Synthesis of Adenosine-Imprinted Microspheres for the Recognition of ADP-Ribosylated Proteins. Biosens. Bioelectron. 2017, 87, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Iwanowska, A.; Yusa, S.-I.; Nowakowska, M.; Szczubiałka, K. Selective Adsorption of Modified Nucleoside Cancer Biomarkers by Hybrid Molecularly Imprinted Adsorbents. J. Sep. Sci. 2016, 39, 3072–3080. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Liu, B.; Liu, J. Incorporation of Boronic Acid into Aptamer-Based Molecularly Imprinted Hydrogels for Highly Specific Recognition of Adenosine. ACS Appl. Bio Mater. 2019, 3, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, J.; Ma, Y.; Tian, X.; Li, Y.; Niu, X.; Luo, J.; Pan, J. Sequential Assembly Enabled Surface Precise Imprinting on Janus Nanosheets for Highly Specific Separation of Adenosine 5′-Monophosphate. Chem. Eng. J. 2022, 432, 134349. [Google Scholar] [CrossRef]
- Wang, P.; Liu, J.; Chen, X.; Ma, X.; Guo, D.; Li, Z.; Pan, J. Janus Silica Nanosheets-Based MMIPs Platform for Synergetic Selective Capture and Fast Separation of 2′-Deoxyadenosine: Two Different Components Segmented on the Surface of One Object. Chem. Eng. J. 2019, 369, 793–802. [Google Scholar] [CrossRef]
- Cheng, T.; Li, H.; Ma, Y.; Liu, X.; Zhang, H. Synthesis of Boronic-Acid-Functionalized Magnetic Attapulgite for Selective Enrichment of Nucleosides. Anal. Bioanal. Chem. 2015, 407, 3525–3529. [Google Scholar] [CrossRef] [PubMed]

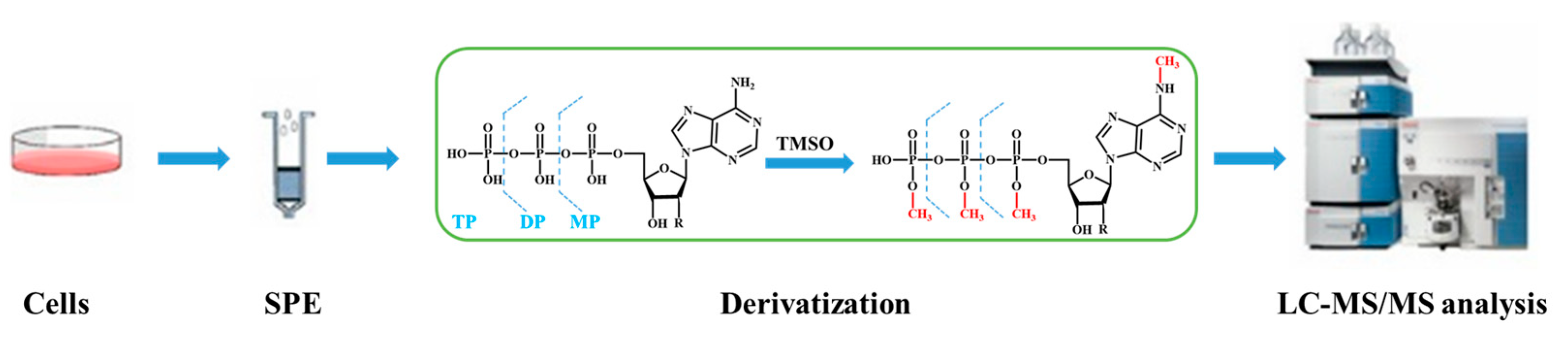
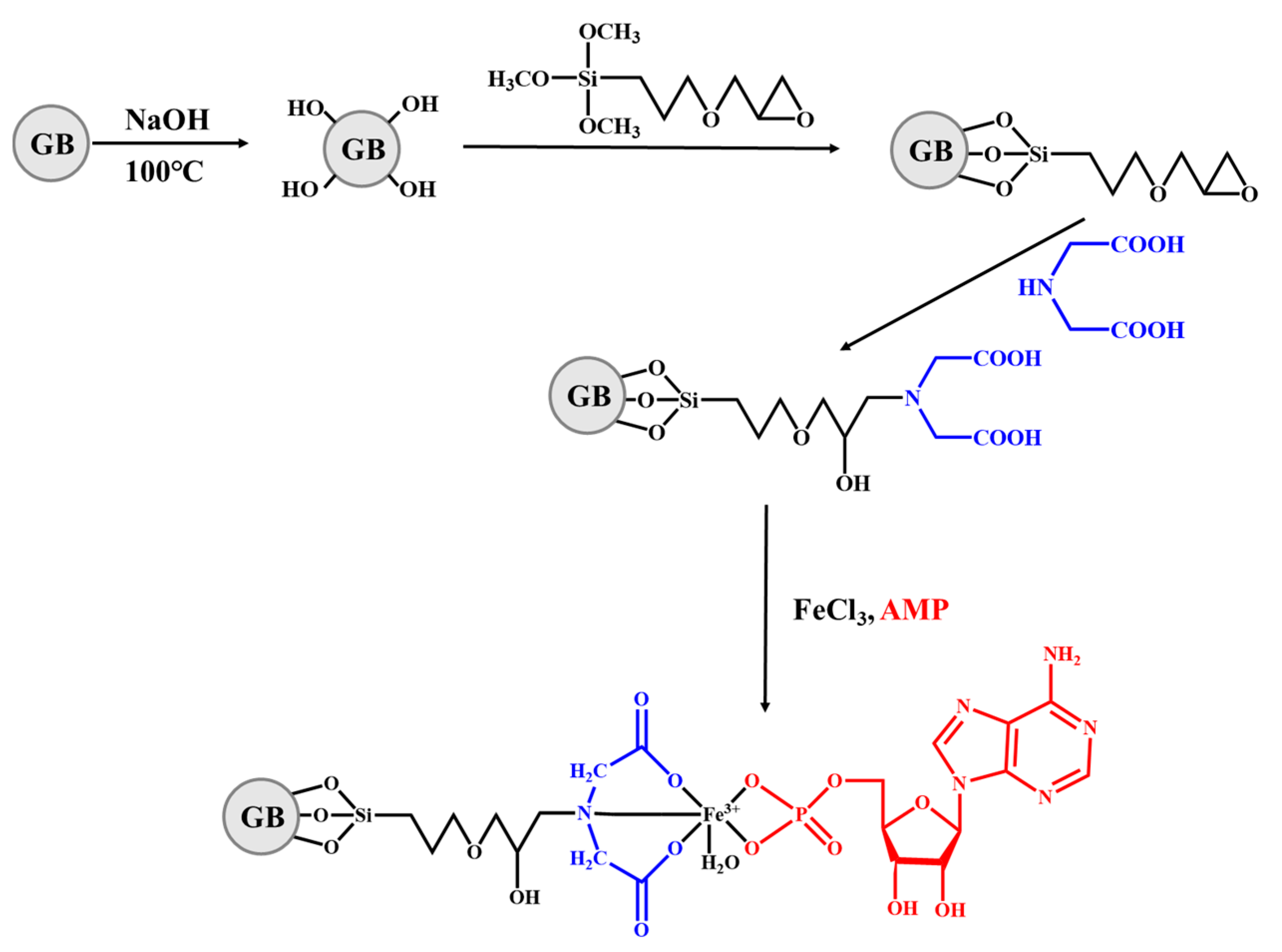
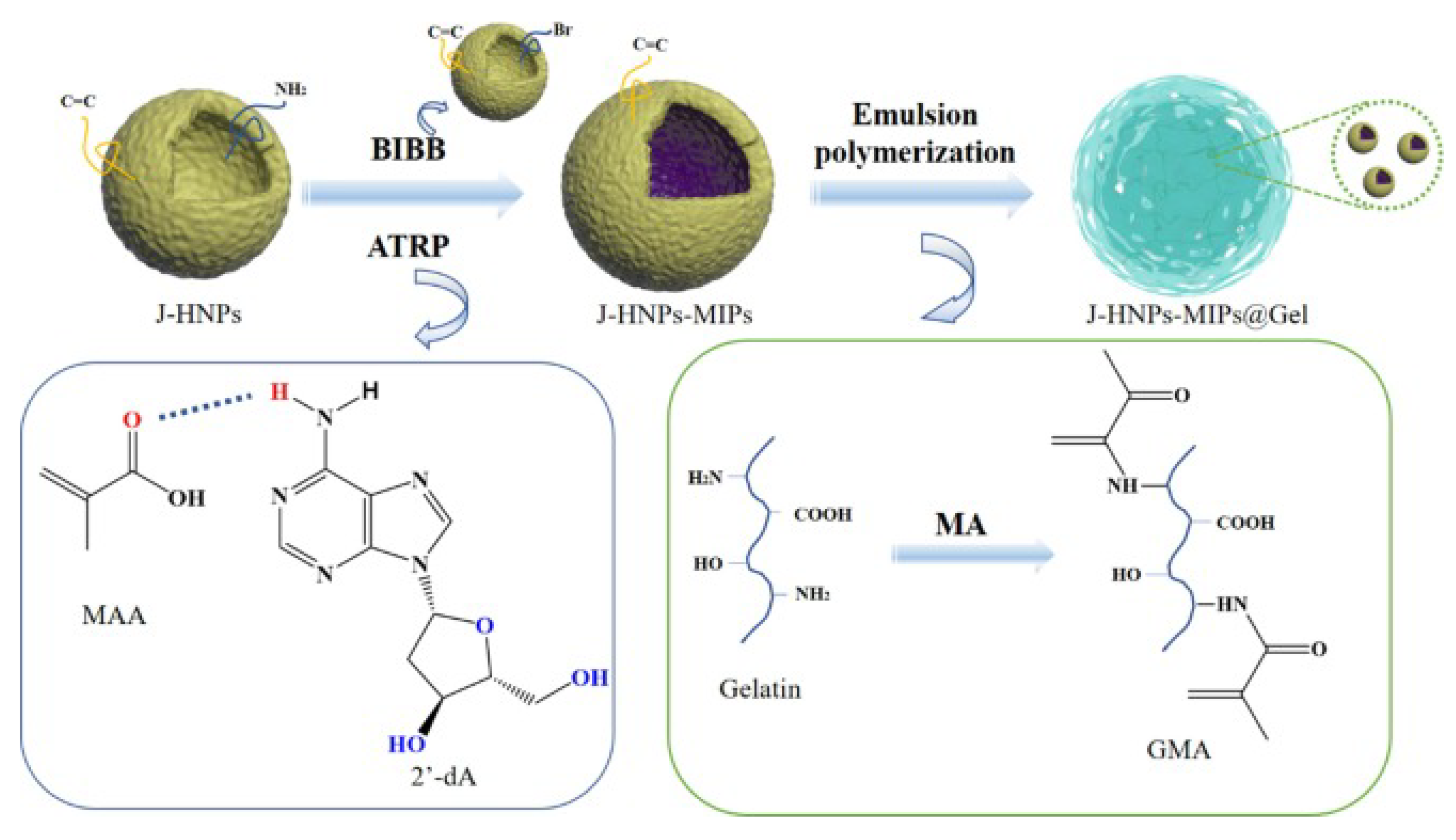


| Methods | Advantage | Disadvantage | Reference |
|---|---|---|---|
| Crystallization | Inexpensive and easy to industrialize | Toxic solvents or heavy metal salts, environmental pollution, unsuitable for low concentrations products | [82,83,84] |
| HPLC | Rapid analysis and high-sensitivity | Difficult to implement in industrial applications and high operating costs | [85,86] |
| Column chromatography | High separation efficiency and simple operation | Organic mobile phase and environmental pollution | [87,88] |
| Solvent extraction | Large processing capacity and low energy consumption | Organic extraction solvents and selectivity need to be improved | [89,90] |
| Adsorption | Simple process, inexpensive, easy to regenerate, and environmentally friendly | Selectivity needs to be improved | [91,92,93] |
| Sorbents | Capacity (μmol g−1) | Equilibrium (Min) | Imprinting Factor (IF) | Reference |
|---|---|---|---|---|
| J-SNs-MMIPs-Pickering | 73.04 | 60 | 1.499 | [124] |
| J-MIPs | 13.69 | 240 | 2.182 | [136] |
| J-SNs-MMIPs | 61.22 | 70 | 1.570 | [137] |
| MIPs shell | 0.363 | 120 | 2.011 | [133] |
| J-HNPs-MIPs@Gel | 10.31 | 40 | 1.730 | [126] |
| ATP-Fe3O4-NH2-DFFPBA | 27.17 | 9 | - | [138] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Cheng, T.; Pan, J. Nucleoside Analogs: A Review of Its Source and Separation Processes. Molecules 2023, 28, 7043. https://doi.org/10.3390/molecules28207043
Wang P, Cheng T, Pan J. Nucleoside Analogs: A Review of Its Source and Separation Processes. Molecules. 2023; 28(20):7043. https://doi.org/10.3390/molecules28207043
Chicago/Turabian StyleWang, Pan, Tao Cheng, and Jianming Pan. 2023. "Nucleoside Analogs: A Review of Its Source and Separation Processes" Molecules 28, no. 20: 7043. https://doi.org/10.3390/molecules28207043
APA StyleWang, P., Cheng, T., & Pan, J. (2023). Nucleoside Analogs: A Review of Its Source and Separation Processes. Molecules, 28(20), 7043. https://doi.org/10.3390/molecules28207043