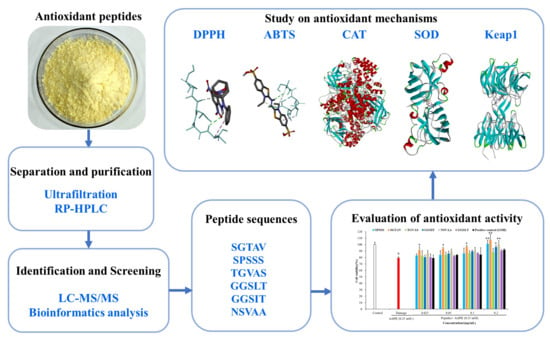
Novel Antioxidant Peptides from Pearl Shell Meat Hydrolysate and Their Antioxidant Activity Mechanism
Abstract
1. Introduction
2. Results and Discussion
2.1. Purification and Antioxidant Activities of Antioxidant Peptides
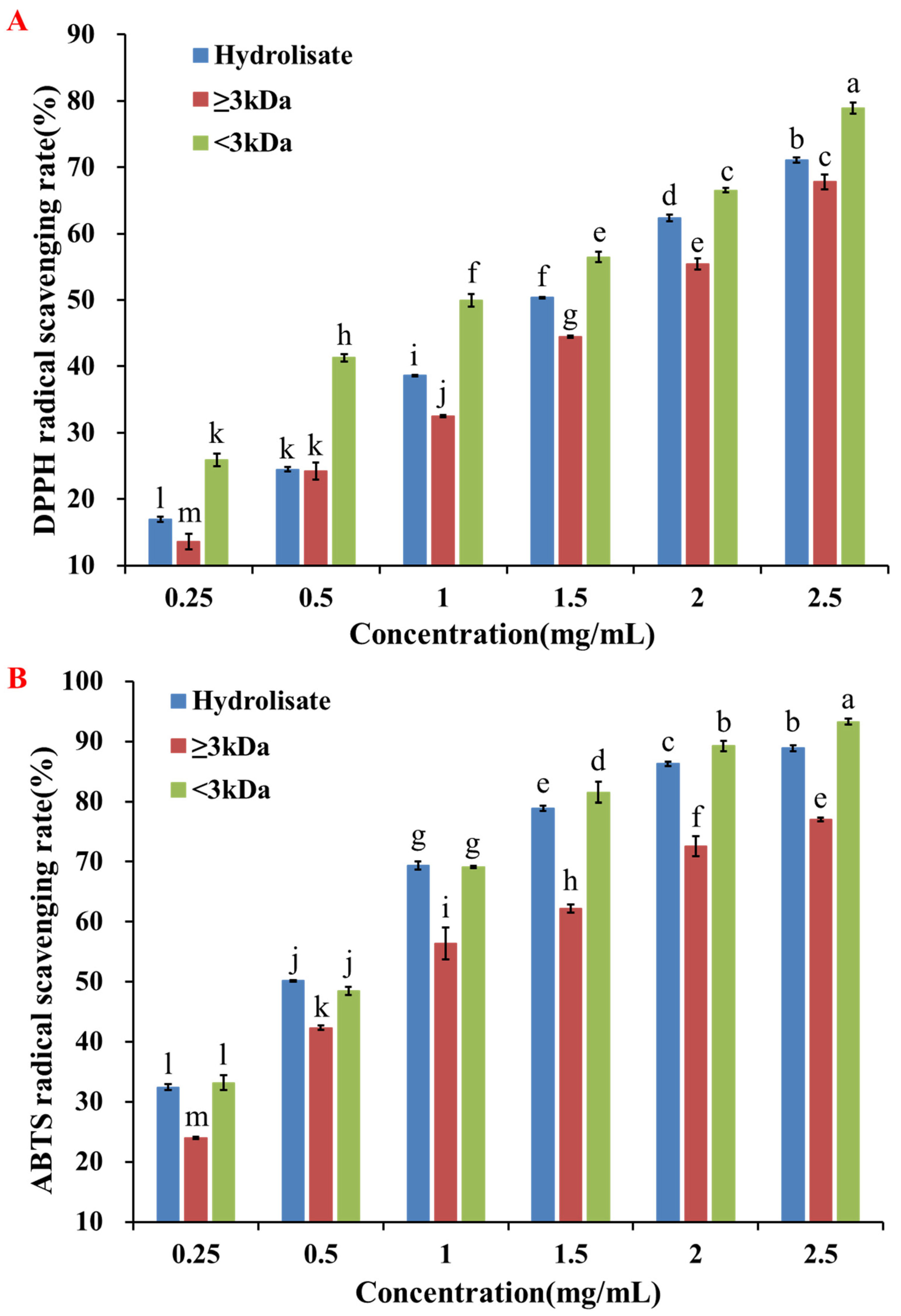
2.1.1. Separation of the Pearl Shell Meat Hydrolysate by Ultrafiltration
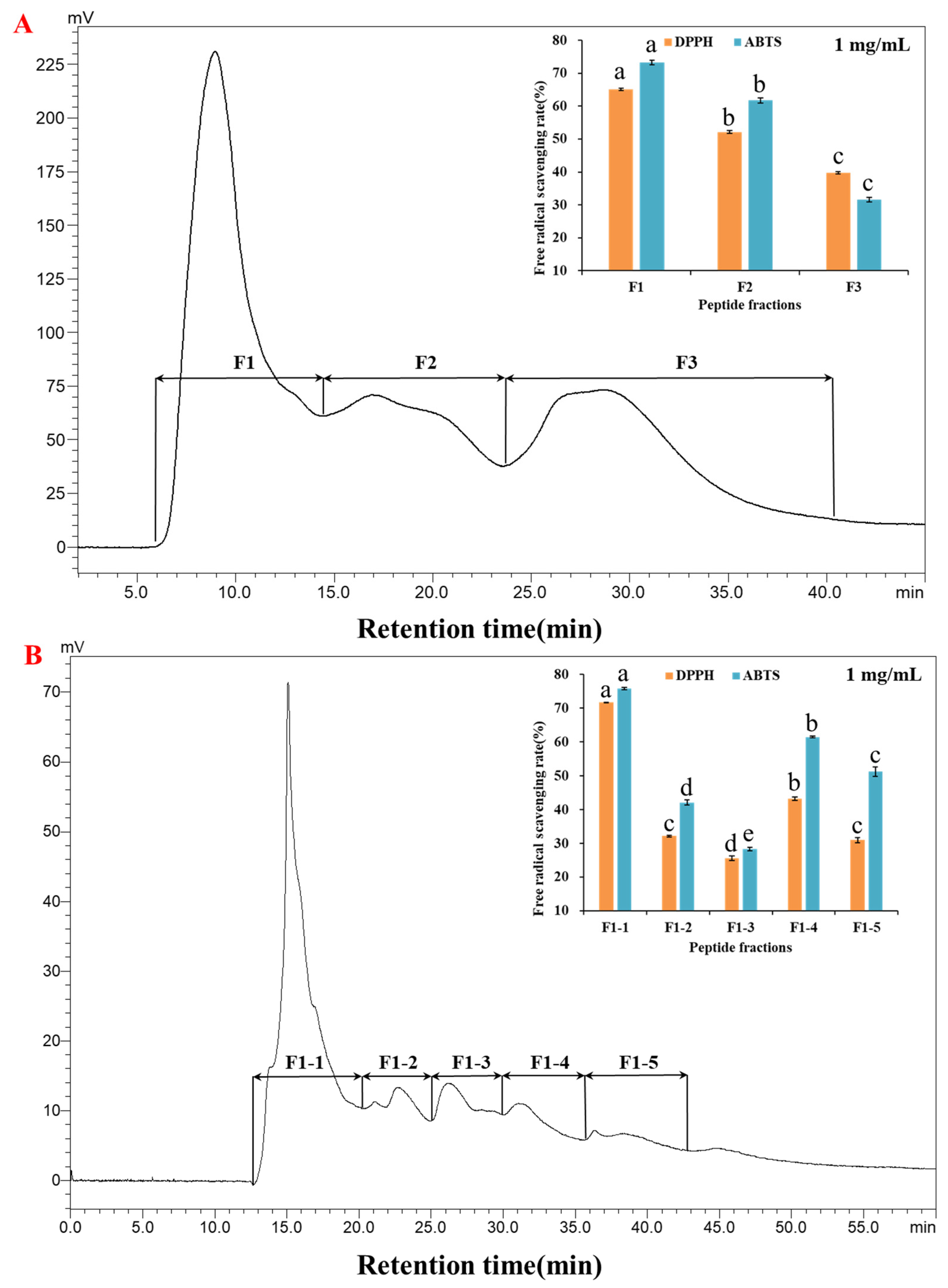
2.1.2. Purification of the Fraction <3 kDa by RP-HPLC
2.2. Identification of Fraction F1-1 and Bioinformatics Analysis
2.3. Cellular Antioxidant Activity of Antioxidant Peptides
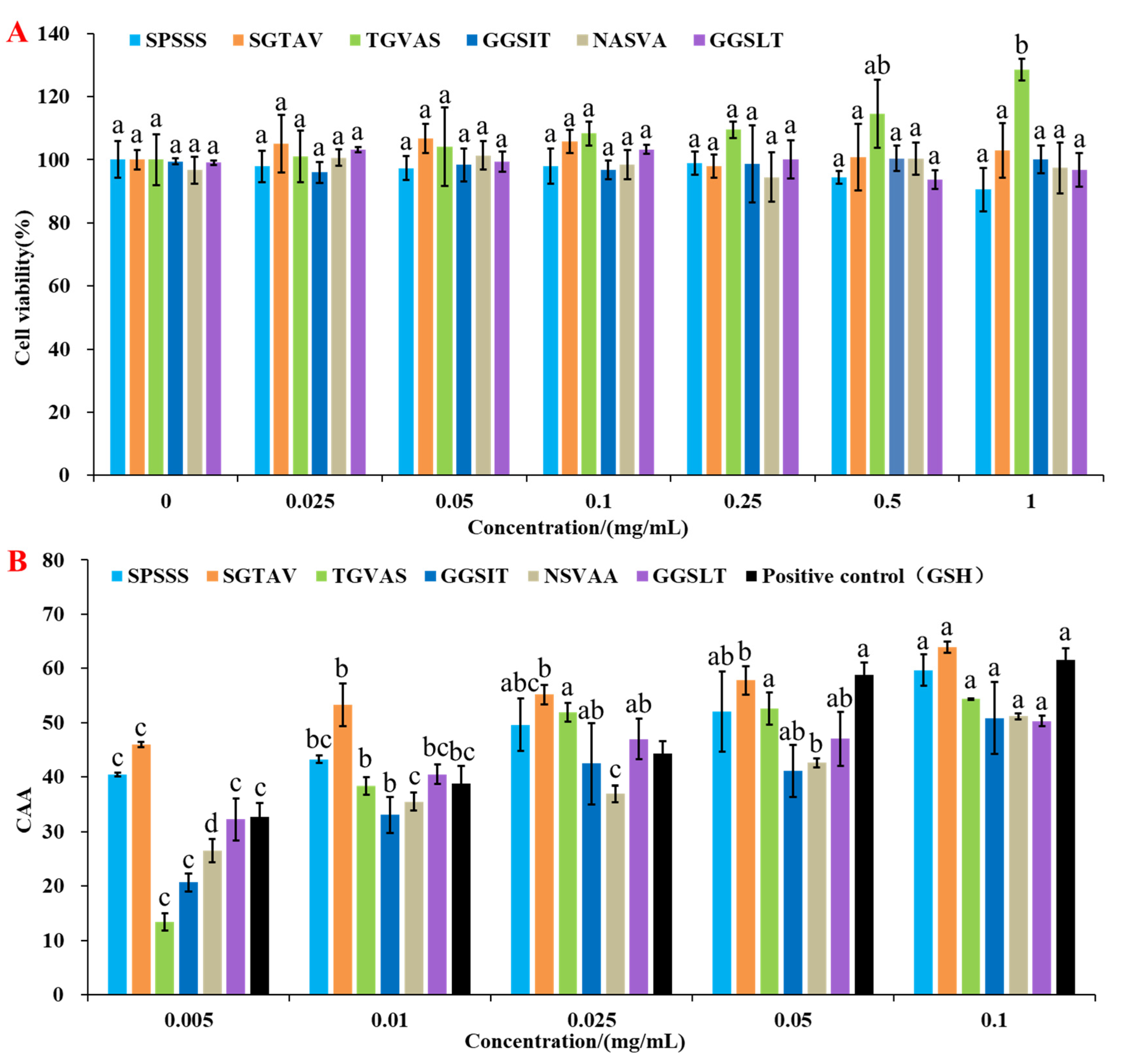
2.3.1. Cell Viability of HepG2 Cells
2.3.2. Cellular Antioxidant Activity
2.4. Cytoprotective Effect of Antioxidant Peptides against AAPH-Induced Cell Damage
2.4.1. Effect of Cell Viability Treated by AAPH
2.4.2. Cytoprotective Effect of Antioxidant Peptides
2.4.3. Effect of Antioxidant Peptides on CAT and SOD
2.5. Molecular Docking Simulation of Antioxidant Mechanisms
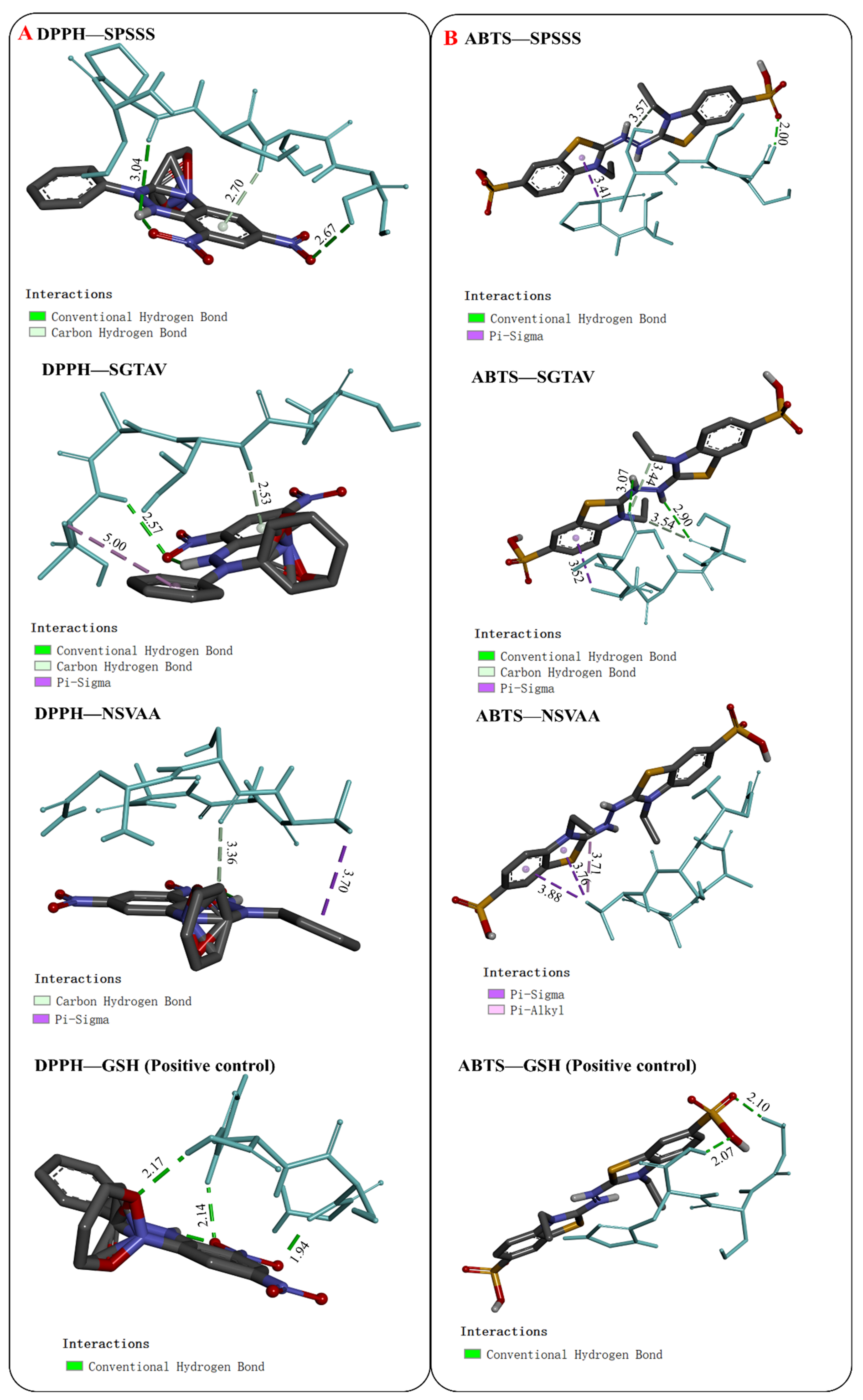
2.5.1. Interaction between Antioxidant Peptides and Free Radicals
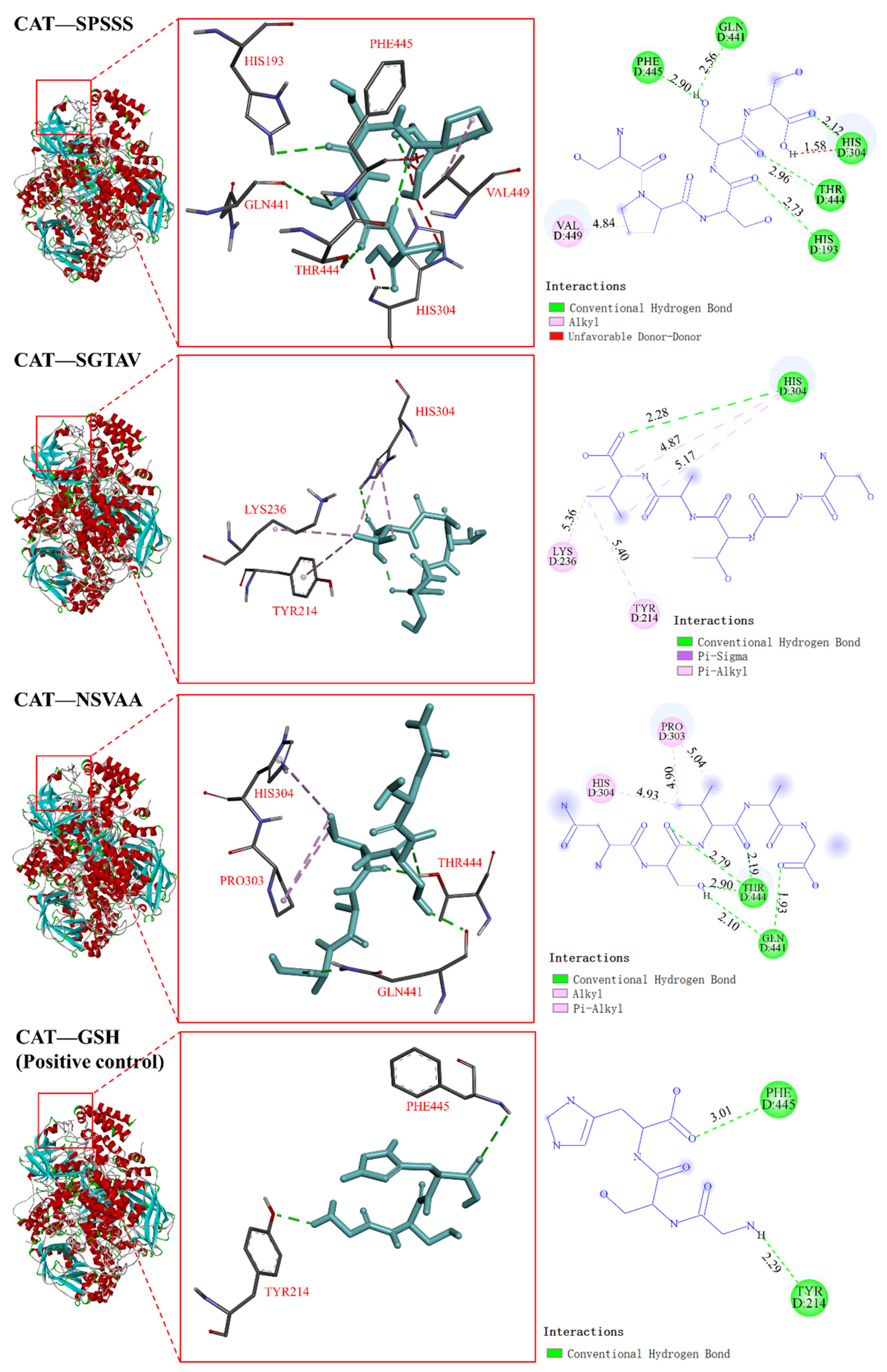
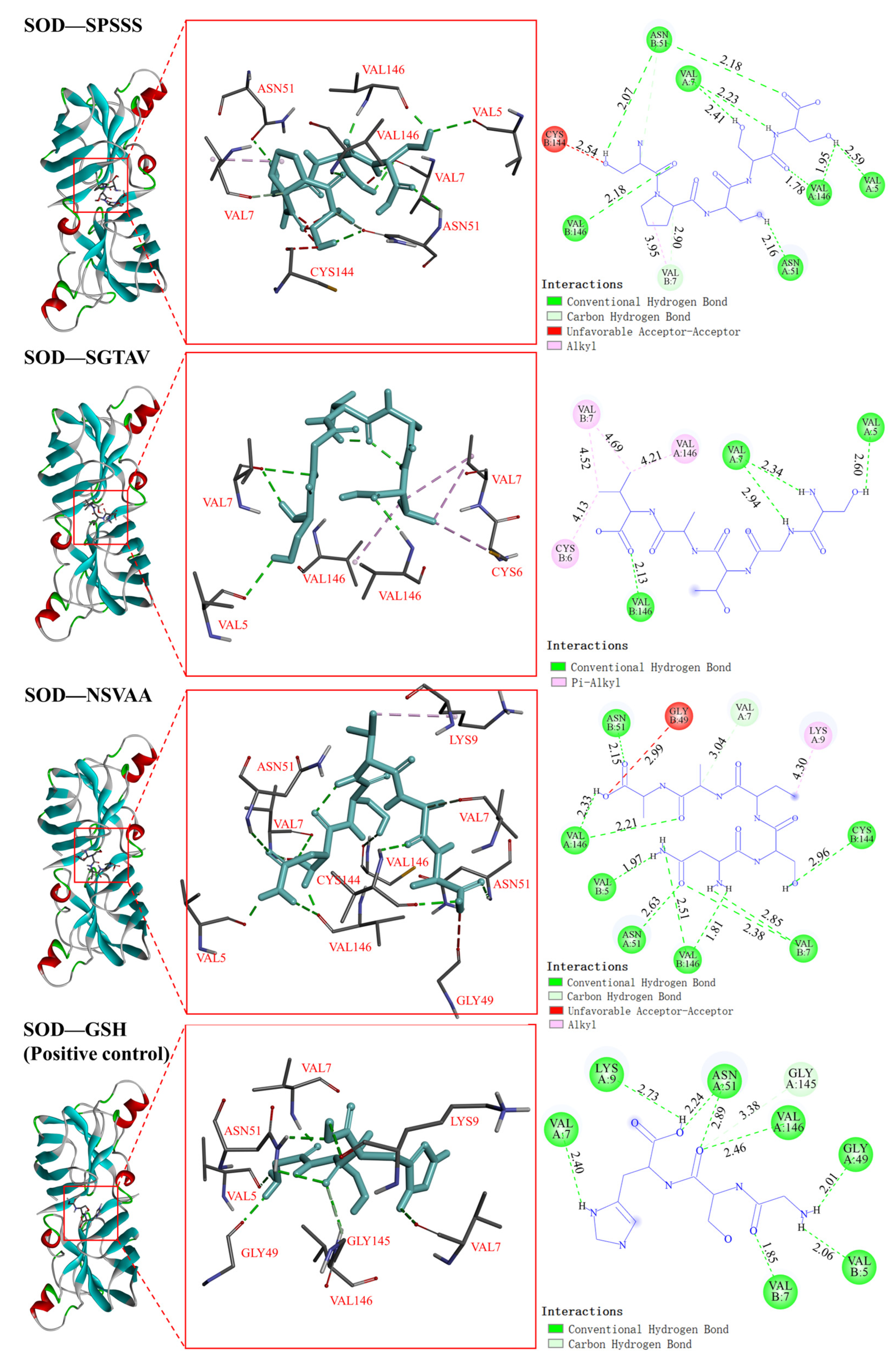
2.5.2. Interaction between Antioxidant Peptides and Antioxidant Enzymes
2.5.3. Interaction between Antioxidant Peptides and Antioxidation-Related Protein (Keap1)
3. Materials and Methods
3.1. Materials
3.2. Preparation of Pearl Shell Meat Hydrolysate
3.3. Separation and Purification of Pearl Shell Meat Hydrolysate
3.3.1. Separation of Pearl Shell Meat Hydrolysate by Ultrafiltration
3.3.2. Purification of the Highly Antioxidant Fraction by RP-HPLC
3.4. Identification of the Antioxidant Peptides Identification by Proteomics
3.5. Screening Potential Antioxidant Peptides by Bioinformatics Analysis
3.6. Synthesis of Antioxidant Peptides
3.7. Evaluation of Antioxidant Capacity In Vitro
3.7.1. DPPH Radical Scavenging Ability Assay
3.7.2. ABTS Radical Scavenging Ability Assay
3.8. Evaluation on Cellular Antioxidant Activity of Antioxidant Peptides
3.8.1. Cell Culture
3.8.2. Cell Viability Assay
3.8.3. Cellular Antioxidant Activity Assay
3.9. Evaluation on Cytoprotective Effects of Antioxidant Peptides against AAPH-Induced Cell Damage
3.9.1. AAPH-Induced Cell Damage
3.9.2. Cytoprotective Effect of Antioxidant Peptides
3.9.3. Protective Effect of Antioxidant Peptides on CAT and SOD
3.10. Study on the Activity Mechanism of Antioxidant Peptides
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zhang, J.; Li, M.; Zhang, G.; Tian, Y.; Kong, F.; Xiong, S.; Zhao, S.; Jia, D.; Manyande, A.; Du, H. Identification of novel antioxidant peptides from snakehead (Channa argus) soup generated during gastrointestinal digestion and insights into the anti-oxidation mechanisms. Food Chem. 2021, 337, 127921. [Google Scholar] [CrossRef]
- Ahn, C.-B.; Kim, J.-G.; Je, J.-Y. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014, 147, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Feng, Y.; Duan, Y.; Ma, H.; Zhang, H. Purification and identification of novel antioxidant peptides from watermelon seed protein hydrolysates and their cytoprotective effects on H2O2-induced oxidative stress. Food Chem. 2020, 327, 127059. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhong, Y.; Duan, Y.; Chen, Q.; Li, F. Antioxidant mechanism of tea polyphenols and its impact on health benefits. Anim. Nutr. 2020, 6, 115–123. [Google Scholar] [CrossRef]
- Zhong, Q.; Wei, B.; Wang, S.; Ke, S.; Chen, J.; Zhang, H.; Wang, H. The antioxidant activity of polysaccharides derived from marine organisms: An overview. Mar. Drugs 2019, 17, 674. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Nikoo, M.; Benjakul, S. Potential application of seafood-derived peptides as bifunctional ingredients, antioxidant–cryoprotectant: A review. J. Funct. Foods 2015, 19, 753–764. [Google Scholar] [CrossRef]
- Jayaprakash, R.; Perera, C.O. Partial Purification and Characterization of Bioactive Peptides from Cooked New Zealand Green-Lipped Mussel (Perna canaliculus) Protein Hydrolyzates. Foods 2020, 9, 879. [Google Scholar] [CrossRef]
- Je, J.-Y.; Qian, Z.-J.; Byun, H.-G.; Kim, S.-K. Purification and characterization of an antioxidant peptide obtained from tuna backbone protein by enzymatic hydrolysis. Process. Biochem. 2007, 42, 840–846. [Google Scholar] [CrossRef]
- Shang, W.-H.; Tang, Y.; Su, S.-Y.; Han, J.-R.; Yan, J.-N.; Wu, H.-T.; Zhu, B.-W. In silico assessment and structural characterization of antioxidant peptides from major yolk protein of sea urchin Strongylocentrotus nudus. Food Funct. 2018, 9, 6435–6443. [Google Scholar] [CrossRef]
- Wu, J.; Huo, J.; Huang, M.; Zhao, M.; Luo, X.; Sun, B. Structural Characterization of a Tetrapeptide from Sesame Flavor-Type Baijiu and Its Preventive Effects against AAPH-Induced Oxidative Stress in HepG2 Cells. J. Agric. Food Chem. 2017, 65, 10495–10504. [Google Scholar] [CrossRef]
- Ren, D.; Xue, G.; Zheng, H.; Yang, W.; Cao, W.; Lin, H.; Gao, J.; Qin, X.; Zhang, C. Thermal stabilization effects of κ-Carrageenan on water-soluble protein extracted from Pinctada martensii meat. J. Food Meas. Charact. 2022, 16, 4985–4995. [Google Scholar] [CrossRef]
- You, L.; Li, Y.; Zhao, H.; Regenstein, J.; Zhao, M.; Ren, J. Purification and Characterization of an Antioxidant Protein from Pearl Oyster (Pinctada fucata martensii). J. Aquat. Food Prod. Technol. 2015, 24, 661–671. [Google Scholar] [CrossRef]
- Yang, F.; Qin, X.; Zhang, T.; Lin, H.; Zhang, C. Evaluation of small molecular polypeptides from the mantle of Pinctada martensii on promoting skin wound healing in mice. Molecules 2019, 24, 4231. [Google Scholar] [CrossRef]
- Tian, Y.; Jiang, P.; Liu, X.; Wei, L.; Bai, Y.; Liu, X.; Li, S. Production and identification of peptides with activity promoting osteoblast proliferation from meat dregs of Pinctada martensii. J. Food Biochem. 2021, 45, e13890. [Google Scholar] [CrossRef]
- Huang, P.; Chen, B.; Shen, J.; Huang, W.; Xia, Z.; Li, Y.; Wang, X.; Yu, Y.; Cao, Y.; Miao, J. Preparation Process Optimization for the Antioxidant Peptide from Pinctada martensii Meat and Its Tyrosinase Inhibitory Activity. Mod. Food Sci. Technol. 2022, 38, 52–61. [Google Scholar] [CrossRef]
- Wang, L.; Ma, M.; Yu, Z.; Du, S.-K. Preparation and identification of antioxidant peptides from cottonseed proteins. Food Chem. 2021, 352, 129399. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Sandoval-Sicairos, E.S.; Milán-Noris, A.K.; Luna-Vital, D.A.; Milán-Carrillo, J.; Montoya-Rodríguez, A. Anti-inflammatory and antioxidant effects of peptides released from germinated amaranth during in vitro simulated gastrointestinal digestion. Food Chem. 2021, 343, 128394. [Google Scholar] [CrossRef]
- Xia, Z.; Miao, J.; Chen, B.; Guo, J.; Ou, Y.; Liang, X.; Yin, Y.; Tong, X.; Cao, Y. Purification, identification, and antioxidative mechanism of three novel selenium-enriched oyster antioxidant peptides. Food Res. Int. 2022, 157, 111359. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Liu, R.H. Cellular Antioxidant Activity (CAA) Assay for Assessing Antioxidants, Foods, and Dietary Supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef]
- Liang, R.; Zhang, Z.; Lin, S. Effects of pulsed electric field on intracellular antioxidant activity and antioxidant enzyme regulating capacities of pine nut (Pinus koraiensis) peptide QDHCH in HepG2 cells. Food Chem. 2017, 237, 793–802. [Google Scholar] [CrossRef]
- Wang, L.; Ding, L.; Yu, Z.; Zhang, T.; Ma, S.; Liu, J. Intracellular ROS scavenging and antioxidant enzyme regulating capacities of corn gluten meal-derived antioxidant peptides in HepG2 cells. Food Res. Int. 2016, 90, 33–41. [Google Scholar] [CrossRef]
- Yarnpakdee, S.; Benjakul, S.; Kristinsson, H.G.; Bakken, H.E. Preventive effect of Nile tilapia hydrolysate against oxidative damage of HepG2 cells and DNA mediated by H2O2 and AAPH. J. Food Sci. Technol. 2015, 52, 6194–6205. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, S.; Liang, J.; Tang, M.; Wang, S. Protective effects of crimson snapper scales peptides against oxidative stress on Drosophila melanogaster and the action mechanism. Food Chem. Toxicol. 2021, 148, 111965. [Google Scholar] [CrossRef]
- Hatai, B.; Banerjee, S.K. Molecular docking interaction between superoxide dismutase (receptor) and phytochemicals (ligand) from Heliotropium indicum Linn for detection of potential phytoconstituents: New drug design for releasing oxidative stress condition/inflammation of osteoarthritis patients. J. Pharmacogn. Phytochem. 2019, 2, 1700–1706. [Google Scholar]
- Ma, B.-X.; Meng, X.-S.; Tong, J.; Ge, L.-L.; Zhou, G.; Wang, Y.-W. Protective effects of Coptis chinensis inflorescence extract and linarin against carbon tetrachloride-induced damage in HepG2 cells through the MAPK/Keap1-Nrf2 pathway. Food Funct. 2018, 9, 2353–2361. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Study on the structure-activity relationship of watermelon seed antioxidant peptides by using molecular simulations. Food Chem. 2021, 364, 130432. [Google Scholar] [CrossRef] [PubMed]
- Koohshekan, B.; Divsalar, A.; Saiedifar, M.; Saboury, A.; Ghalandari, B.; Gholamian, A.; Seyedarabi, A. Protective effects of aspirin on the function of bovine liver catalase: A spectroscopy and molecular docking study. J. Mol. Liq. 2016, 218, 8–15. [Google Scholar] [CrossRef]
- Hashemi, S.A.; Bathaie, S.Z.; Mohagheghi, M.A. Interaction of saffron carotenoids with catalase: In vitro, in vivo and molecular docking studies. J. Biomol. Struct. Dyn. 2020, 38, 3916–3926. [Google Scholar] [CrossRef] [PubMed]
- Buendia, I.; Michalska, P.; Navarro, E.; Gameiro, I.; Egea, J.; León, R. Nrf2–ARE pathway: An emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol. Ther. 2016, 157, 84–104. [Google Scholar] [CrossRef]
- Xiong, K.; Tan, L.; Wang, A.; Yang, L. Progress on Anti-oxidation Mechanisms and Antioxidants of the Keap1-Nrf2/ARE Signaling Pathway. Bachelor Vet. Med. 2021, 42, 89–94. [Google Scholar] [CrossRef]
- Yang, M.; Jiang, Z.-H.; Li, C.-G.; Zhu, Y.-J.; Li, Z.; Tang, Y.-Z.; Ni, C.-L. Apigenin prevents metabolic syndrome in high-fructose diet-fed mice by Keap1-Nrf2 pathway. Biomed. Pharmacother. 2018, 105, 1283–1290. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides exhibit antioxidant activity through the Keap1-Nrf2 signaling pathway. J. Funct. Foods 2020, 64, 103696. [Google Scholar] [CrossRef]
- Agrawal, H.; Joshi, R.; Gupta, M. Purification, identification and characterization of two novel antioxidant peptides from finger millet (Eleusine coracana) protein hydrolysate. Food Res. Int. 2019, 120, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xie, N.; Li, B. Influence of peptide characteristics on their stability, intestinal transport, and in vitro bioavailability: A review. J. Food Biochem. 2019, 43, e12571. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.L.; Liceaga, A.M.; Yoon, K.Y. Purification, characterisation and stability of an antioxidant peptide derived from sandfish (Arctoscopus japonicus) protein hydrolysates. J. Funct. Foods 2016, 20, 433–442. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Ong, M.G.-L.; Pang, M.-J.; Wong, S.-J.; Teh, L.K.; Chai, T.-T. Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chem. 2019, 271, 614–622. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Yu, W.; Wang, Z.; Li, J.; Feng, X.; Wang, J.; Shan, A. De novo design of a pH-triggered self-assembled β-hairpin nanopeptide with the dual biological functions for antibacterial and entrapment. J. Nanobiotechnol. 2021, 19. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, J.; Dong, X.; Zhang, Y.; Zhou, X.; Huang, M.; Zhou, G. Purification and identification of antioxidant peptides from duck plasma proteins. Food Chem. 2020, 319, 126534. [Google Scholar] [CrossRef] [PubMed]
- Floegel, A.; Kim, D.-O.; Chung, S.-J.; Koo, S.I.; Chun, O.K. Comparison of ABTS/DPPH assays to measure antioxidant capacity in popular antioxidant-rich US foods. J. Food Compos. Anal. 2011, 24, 1043–1048. [Google Scholar] [CrossRef]
- Türkez, H.; Arslan, M.E.; Sönmez, E.; Tatar, A.; Geyikoğlu, F.; Açikyildiz, M.; Mardinoğlu, A. Safety Assessments of Nickel Boride Nanoparticles on the Human Pulmonary Alveolar Cells by Using Cell Viability and Gene Expression Analyses. Biol. Trace Elem. Res. 2020, 199, 2602–2611. [Google Scholar] [CrossRef]
- Kankanala, R.; Ganugapati, J.; Vutukuru, S.S.; Sai, K.S.S.R. Molecular Docking Analysis of Fullerene (C60) with Human Antioxidant Enzymes: Implications in Inhibition of Enzymes. J. Comput. Theor. Nanosci. 2013, 10, 1403–1407. [Google Scholar] [CrossRef]



| Peptide | Length | Mass (Da) | −10 lgP | pI | Toxicity | Post-digestive Fragments | Hydrophobicity (Kcal/mol) |
|---|---|---|---|---|---|---|---|
| SPSSS | 5 | 463.1914 | 15.40 | 5.38 | None | SPSSS | +9.88 |
| SGTAV | 5 | 433.2173 | 17.02 | 5.54 | None | SGTAV | +9.80 |
| TGVAS | 5 | 433.2173 | 15.71 | 5.36 | None | TGVAS | +9.80 |
| GGSIT | 5 | 433.2173 | 20.65 | 5.36 | None | GGSIT | +9.79 |
| NSVAA | 5 | 460.2281 | 16.37 | 5.38 | None | NSVAA | +9.75 |
| GGSLT | 5 | 433.2173 | 20.65 | 5.36 | None | GGSLT | +9.66 |
| ITIGG | 5 | 459.2693 | 21.13 | 5.60 | None | ITIGG | +8.21 |
| ITTSS | 5 | 507.2540 | 18.19 | 5.46 | None | ITTSS | +8.20 |
| LTIGG | 5 | 459.2693 | 21.13 | 5.60 | None | LTIGG | +8.08 |
| ITLGG | 5 | 459.2693 | 21.13 | 5.60 | None | ITLGG | +8.08 |
| LTTSS | 5 | 507.2540 | 18.19 | 5.45 | None | LTTSS | +8.07 |
| LTLGG | 5 | 459.2693 | 21.13 | 5.60 | None | LTLGG | +7.95 |
| Peptides | EC50 (mg/mL) | CAA Value (mM TE/g Peptide) | |
|---|---|---|---|
| Antioxidant peptides | SGTAV | 0.009 | 15.072 |
| SPSSS | 0.027 | 5.130 | |
| TGVAS | 0.037 | 3.704 | |
| GGSLT | 0.064 | 2.152 | |
| GGSIT | 0.073 | 1.874 | |
| NSVAA | 0.095 | 1.443 | |
| Positive control | GSH | 0.030 | 4.593 |
| Peptides | Receptors | Binding Energy (kcal/mol) | Hydrogen Bonds | Hydrophobic Interaction | |||
|---|---|---|---|---|---|---|---|
| Quantity | Binding Sites | Quantity | Binding Sites | ||||
| Antioxidant peptides | SPSSS | DPPH | −3.5 | 3 | DPPH | 0 | DPPH |
| SGTAV | −3.3 | 2 | DPPH | 1 | DPPH | ||
| NSVAA | −2.8 | 1 | DPPH | 1 | DPPH | ||
| Positive control | GSH | −2.8 | 3 | DPPH | 0 | — | |
| Antioxidant peptides | SPSSS | ABTS | −2.6 | 2 | ABTS | 1 | ABTS |
| SGTAV | −2.3 | 4 | ABTS | 1 | ABTS | ||
| NSVAA | −2.5 | 0 | — | 3 | ABTS | ||
| Positive control | GSH | −2.2 | 2 | ABTS | 0 | — | |
| Antioxidant peptides | SPSSS | CAT | −6.2 | 5 | HIS193, GLN441, HIS304, THR444, PHE445 | 1 | VAL449 |
| SGTAV | −5.4 | 1 | HIS304 | 4 | LYS236, TYR214, HIS304 | ||
| NSVAA | −5.5 | 5 | GLN441, THR444 | 3 | PRO303, HIS304 | ||
| Positive control | GSH | −4.9 | 2 | PHE445, TYR214 | 0 | — | |
| Antioxidant peptides | SPSSS | SOD | −5.2 | 11 | VAL5, VAL146, VAL7, ASN51 | 1 | VAL7 |
| SGTAV | −4.4 | 4 | VAL5, VAL7, VAL146 | 4 | VAL7, CYS6, VAL164 | ||
| NSVAA | −5.1 | 11 | CYS144, VAL7, VAL146, ASN51, VAL5 | 1 | LYS9 | ||
| Positive control | GSH | −6.7 | 9 | VAL7, VAL5, GLY49, VAL146, GLY145, ASN51, LYS9 | 0 | — | |
| Antioxidant peptides | SPSSS | Keap1 | −7.5 | 5 | SER363, ASN382, ASN387, SER555 | 0 | — |
| SGTAV | −8.0 | 7 | TYR334, ASN382, SER383, ASN414, ARG415, SER555, TYR572 | 2 | TYR334 | ||
| NSVAA | −7.6 | 8 | SER363, ASN387, ASN414, ARG415, SER508, SER555, GLY603 | 3 | PHE577, TYR572 | ||
| Positive control | GSH | −6.4 | 5 | ASN387, SER383, SER602, TYR334 | 0 | — | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.; Miao, J.; Li, J.; Li, Y.; Wang, X.; Yu, Y.; Cao, Y. Novel Antioxidant Peptides from Pearl Shell Meat Hydrolysate and Their Antioxidant Activity Mechanism. Molecules 2023, 28, 864. https://doi.org/10.3390/molecules28020864
Huang P, Miao J, Li J, Li Y, Wang X, Yu Y, Cao Y. Novel Antioxidant Peptides from Pearl Shell Meat Hydrolysate and Their Antioxidant Activity Mechanism. Molecules. 2023; 28(2):864. https://doi.org/10.3390/molecules28020864
Chicago/Turabian StyleHuang, Pantian, Jianyin Miao, Jialing Li, Yingkun Li, Xianghua Wang, Yan Yu, and Yong Cao. 2023. "Novel Antioxidant Peptides from Pearl Shell Meat Hydrolysate and Their Antioxidant Activity Mechanism" Molecules 28, no. 2: 864. https://doi.org/10.3390/molecules28020864
APA StyleHuang, P., Miao, J., Li, J., Li, Y., Wang, X., Yu, Y., & Cao, Y. (2023). Novel Antioxidant Peptides from Pearl Shell Meat Hydrolysate and Their Antioxidant Activity Mechanism. Molecules, 28(2), 864. https://doi.org/10.3390/molecules28020864