Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis
Abstract
1. Introduction
2. Results
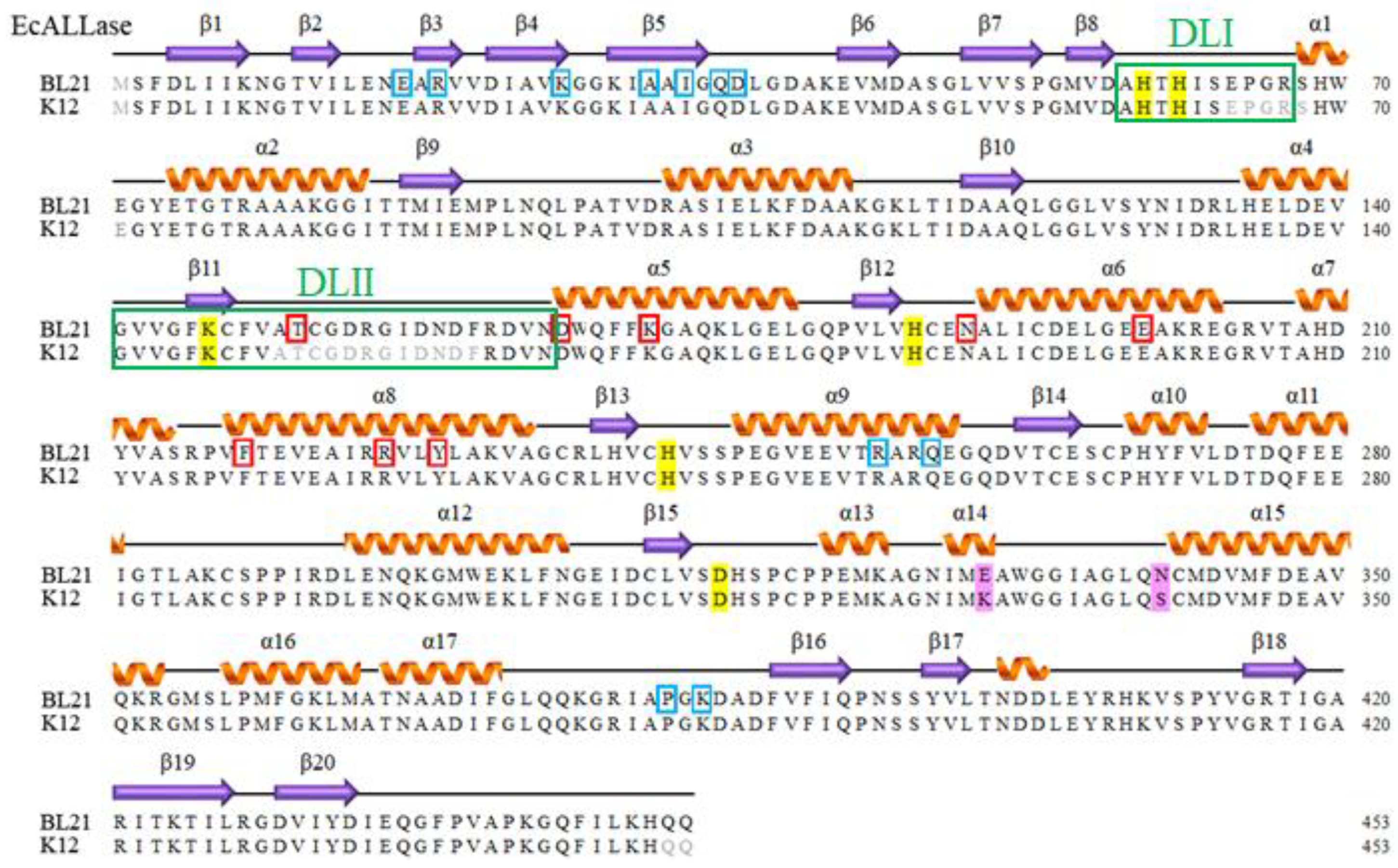
2.1. Sequence Analysis of EcALLase-BL21
2.2. Crystallization and Data Collection of EcALLase-BL21
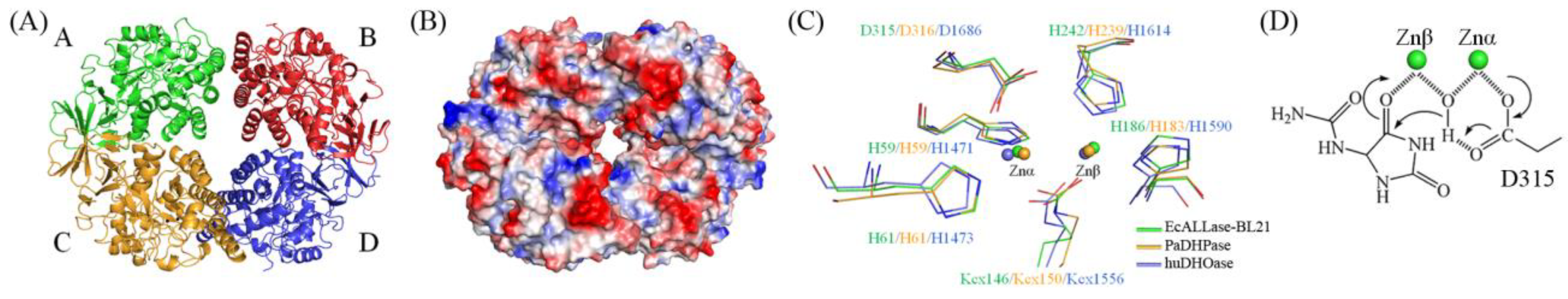
2.3. Overall Structure of EcALLase-BL21
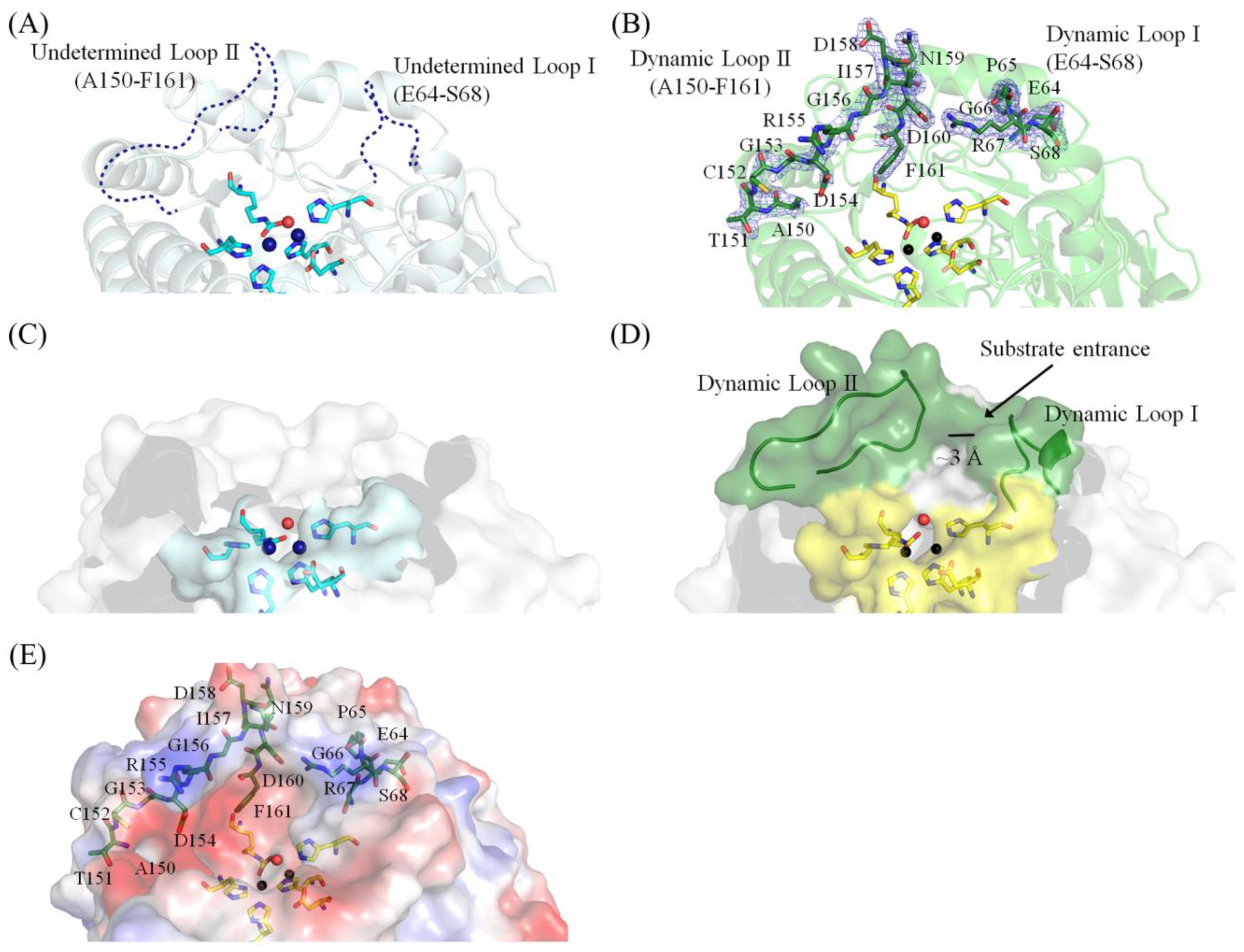
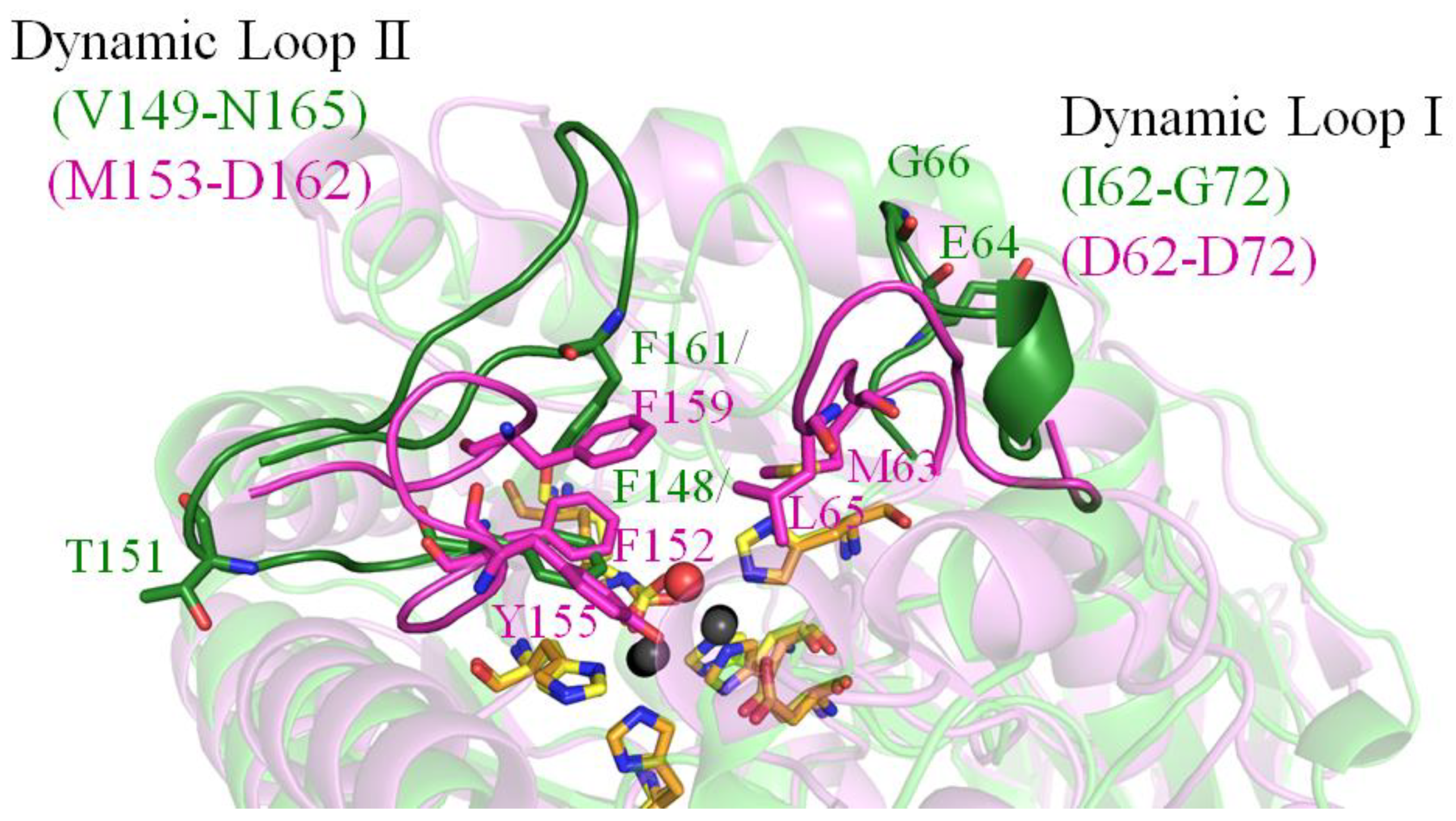
2.4. Dynamic Loops of EcALLase-BL21
2.5. Molecular Docking
2.6. Analysis of Substrate Binding Pockets of HYDase and ALLase
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Crystallization Experiments
4.3. X-Ray Diffraction Data and Structure Determination
4.4. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Werner, A.K.; Witte, C.P. The biochemistry of nitrogen mobilization: Purine ring catabolism. Trends Plant Sci. 2011, 16, 381–387. [Google Scholar] [CrossRef]
- Hayashi, S.; Fujiwara, S.; Noguchi, T. Evolution of urate-degrading enzymes in animal peroxisomes. Cell Biochem. Biophys. 2000, 32, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.Y.; Huang, Y.H.; Huang, C.Y. Chemical rescue of the post-translationally carboxylated lysine mutant of allantoinase and dihydroorotase by metal ions and short-chain carboxylic acids. Amino Acids 2013, 44, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Mulrooney, S.B.; Hausinger, R.P. Metal ion dependence of recombinant Escherichia coli allantoinase. J. Bacteriol. 2003, 185, 126–134. [Google Scholar] [CrossRef]
- Ramazzina, I.; Cendron, L.; Folli, C.; Berni, R.; Monteverdi, D.; Zanotti, G.; Percudani, R. Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases. J. Biol. Chem. 2008, 283, 23295–23304. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.Q.; Kong, C.H.; Wang, P.; Meiners, S.J. Root exudate signals in plant-plant interactions. Plant Cell Environ. 2021, 44, 1044–1058. [Google Scholar] [CrossRef]
- Lu, S.; Jia, Z.; Meng, X.; Chen, Y.; Wang, S.; Fu, C.; Yang, L.; Zhou, R.; Wang, B.; Cao, Y. Combined Metabolomic and Transcriptomic Analysis Reveals Allantoin Enhances Drought Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 14172. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Wang, Y.; Du, F.; Zhang, Y.; Yin, M.; Zhao, X.; Xu, J.; Yang, Y.; Wang, W.; et al. Transcriptome and Metabolome Analyses Reveal Complex Molecular Mechanisms Involved in the Salt Tolerance of Rice Induced by Exogenous Allantoin. Antioxidants 2022, 11, 2045. [Google Scholar] [CrossRef]
- Voß, L.; Heinemann, K.J.; Herde, M.; Medina-Escobar, N.; Witte, C.P. Enzymes and cellular interplay required for flux of fixed nitrogen to ureides in bean nodules. Nat. Commun. 2022, 13, 5331. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Y.; Zhang, J.; Hong, T. Allantoin Inhibits Compound 48/80-Induced Pseudoallergic Reactions In Vitro and In Vivo. Molecules 2022, 27, 3473. [Google Scholar] [CrossRef]
- Seo, C.S.; Lee, M.Y. Simultaneous Determination of Fourteen Marker Compounds in the Traditional Herbal Prescription, Geumgwesingihwan, Using Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry. Molecules 2022, 27, 3890. [Google Scholar] [CrossRef]
- Aguilar-Guadarrama, A.B.; Yáñez-Ibarra, G.; Cancino-Marentes, M.E.; González-Ibarra, P.; Ortiz-Andrade, R.; Sánchez-Recillas, A.; Rodríguez-Carpena, J.G.; Aguirre-Vidal, Y.; Medina-Diaz, I.M.; Ávila-Villarreal, G. Chromatographic Techniques and Pharmacological Analysis as a Quality Control Strategy for Serjania triquetra a Traditional Medicinal Plant. Pharmaceuticals 2022, 15, 1289. [Google Scholar] [CrossRef]
- Thornfeldt, C. Cosmeceuticals containing herbs: Fact, fiction, and future. Dermatol. Surg. 2005, 31, 873–880. [Google Scholar] [CrossRef]
- Peng, W.F.; Huang, C.Y. Allantoinase and dihydroorotase binding and inhibition by flavonols and the substrates of cyclic amidohydrolases. Biochimie 2014, 101, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y. Structure, catalytic mechanism, posttranslational lysine carbamylation, and inhibition of dihydropyrimidinases. Adv. Protein Chem. Struct. Biol. 2020, 122, 63–96. [Google Scholar]
- Gerlt, J.A.; Babbitt, P.C. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001, 70, 209–246. [Google Scholar] [CrossRef]
- Rice, A.J.; Pesavento, R.P.; Ren, J.; Youn, I.; Kwon, Y.; Ellepola, K.; Che, C.T.; Johnson, M.E.; Lee, H. Identification of Small Molecule Inhibitors against Staphylococcus aureus Dihydroorotase via HTS. Int. J. Mol. Sci. 2021, 22, 9984. [Google Scholar] [CrossRef]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Structural Analysis of Saccharomyces cerevisiae Dihydroorotase Reveals Molecular Insights into the Tetramerization Mechanism. Molecules 2021, 26, 7249. [Google Scholar] [CrossRef]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Complexed Crystal Structure of Saccharomyces cerevisiae Dihydroorotase with Inhibitor 5-Fluoroorotate Reveals a New Binding Mode. Bioinorg. Chem. Appl. 2021, 2021, 2572844. [Google Scholar] [CrossRef]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Structural basis for the interaction modes of dihydroorotase with the anticancer drugs 5-fluorouracil and 5-aminouracil. Biochem. Biophys. Res. Commun. 2021, 551, 33–37. [Google Scholar] [CrossRef]
- Lipowska, J.; Miks, C.D.; Kwon, K.; Shuvalova, L.; Zheng, H.; Lewinski, K.; Cooper, D.R.; Shabalin, I.G.; Minor, W. Pyrimidine biosynthesis in pathogens—Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae. Int. J. Biol. Macromol. 2019, 136, 1176–1187. [Google Scholar] [CrossRef]
- Del Cano-Ochoa, F.; Moreno-Morcillo, M.; Ramon-Maiques, S. CAD, A Multienzymatic Protein at the Head of de Novo Pyrimidine Biosynthesis. Subcell Biochem. 2019, 93, 505–538. [Google Scholar]
- Rice, A.J.; Lei, H.; Santarsiero, B.D.; Lee, H.; Johnson, M.E. Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase). Bioorg. Med. Chem. 2016, 24, 4536–4543. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.S.; Luo, R.H.; Yang, Y.C.; Huang, C.Y. Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil. Bioinorg. Chem. Appl. 2022, 2022, 1817745. [Google Scholar] [CrossRef] [PubMed]
- Basbous, J.; Aze, A.; Chaloin, L.; Lebdy, R.; Hodroj, D.; Ribeyre, C.; Larroque, M.; Shepard, C.; Kim, B.; Pruvost, A.; et al. Dihydropyrimidinase protects from DNA replication stress caused by cytotoxic metabolites. Nucleic Acids Res. 2020, 48, 1886–1904. [Google Scholar] [CrossRef]
- Huang, Y.H.; Ning, Z.J.; Huang, C.Y. Crystal structure of dihydropyrimidinase in complex with anticancer drug 5-fluorouracil. Biochem. Biophys. Res. Commun. 2019, 519, 160–165. [Google Scholar] [CrossRef]
- Cheng, J.H.; Huang, Y.H.; Lin, J.J.; Huang, C.Y. Crystal structures of monometallic dihydropyrimidinase and the human dihydroorotase domain K1556A mutant reveal no lysine carbamylation within the active site. Biochem. Biophys. Res. Commun. 2018, 505, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; Huang, C.C.; Huang, Y.H.; Huang, C.Y. Structural Basis for pH-Dependent Oligomerization of Dihydropyrimidinase from Pseudomonas aeruginosa PAO1. Bioinorg. Chem. Appl. 2018, 2018, 9564391. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, C.T.; Huang, Y.H.; Huang, C.Y. Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1: Insights into the molecular basis of formation of a dimer. Biochem. Biophys. Res. Commun. 2016, 478, 1449–1455. [Google Scholar] [CrossRef]
- Huang, C.Y. Inhibition of a putative dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by flavonoids and substrates of cyclic amidohydrolases. PLoS ONE 2015, 10, e0127634. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Chen, M.C.; Hsu, C.C.; Chan, S.I.; Yang, Y.S.; Chen, C.J. Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: Metal and structural requirements for post-translational modification and function. J. Biol. Chem. 2013, 288, 30645–30658. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Hsu, C.C.; Chen, M.C.; Yang, Y.S. Effect of metal binding and posttranslational lysine carboxylation on the activity of recombinant hydantoinase. J. Biol. Inorg. Chem. 2009, 14, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Cheon, Y.H.; Kim, H.S.; Han, K.H.; Abendroth, J.; Niefind, K.; Schomburg, D.; Wang, J.; Kim, Y. Crystal structure of D-hydantoinase from Bacillus stearothermophilus: Insight into the stereochemistry of enantioselectivity. Biochemistry 2002, 41, 9410–9417. [Google Scholar] [CrossRef]
- Soong, C.L.; Ogawa, J.; Honda, M.; Shimizu, S. Cyclic-imide-hydrolyzing activity of D-hydantoinase from Blastobacter sp. strain A17p-4. Appl. Environ. Microbiol. 1999, 65, 1459–1462. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Yang, Y.S. A novel cold-adapted imidase from fish Oreochromis niloticus that catalyzes hydrolysis of maleimide. Biochem. Biophys. Res. Commun. 2003, 312, 467–472. [Google Scholar] [CrossRef]
- Huang, C.Y.; Yang, Y.S. The role of metal on imide hydrolysis: Metal content and pH profiles of metal ion-replaced mammalian imidase. Biochem. Biophys. Res. Commun. 2002, 297, 1027–1032. [Google Scholar] [CrossRef]
- Ogawa, J.; Soong, C.L.; Honda, M.; Shimizu, S. Imidase, a dihydropyrimidinase-like enzyme involved in the metabolism of cyclic imides. Eur. J. Biochem 1997, 243, 322–327. [Google Scholar] [CrossRef]
- Lin, E.S.; Huang, C.Y. Cytotoxic Activities and the Allantoinase Inhibitory Effect of the Leaf Extract of the Carnivorous Pitcher Plant Nepenthes miranda. Plants 2022, 11, 2265. [Google Scholar] [CrossRef]
- Guan, H.H.; Huang, Y.H.; Lin, E.S.; Chen, C.J.; Huang, C.Y. Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis. Int. J. Mol. Sci. 2021, 22, 6861. [Google Scholar] [CrossRef]
- Huang, Y.H.; Lien, Y.; Chen, J.H.; Lin, E.S.; Huang, C.Y. Identification and characterization of dihydropyrimidinase inhibited by plumbagin isolated from Nepenthes miranda extract. Biochimie 2020, 171–172, 124–135. [Google Scholar] [CrossRef]
- Kim, K.; Kim, M.I.; Chung, J.; Ahn, J.H.; Rhee, S. Crystal structure of metal-dependent allantoinase from Escherichia coli. J. Mol. Biol. 2009, 387, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Del Cano-Ochoa, F.; Grande-Garcia, A.; Reverte-Lopez, M.; D’Abramo, M.; Ramon-Maiques, S. Characterization of the catalytic flexible loop in the dihydroorotase domain of the human multi-enzymatic protein CAD. J. Biol. Chem. 2018, 293, 18903–18913. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.H.; Park, H.S.; Kim, H.S. Evolutionary relationship and application of a superfamily of cyclic amidohydrolase enzymes. Chem. Rec. 2005, 5, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Schoemaker, H.E.; Mink, D.; Wubbolts, M.G. Dispelling the myths--biocatalysis in industrial synthesis. Science 2003, 299, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.H.; Couloux, A.; Lee, S.W.; Yoon, S.H.; Cattolico, L.; et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef]
- Ho, Y.Y.; Hsieh, H.C.; Huang, C.Y. Biochemical characterization of allantoinase from Escherichia coli BL21. Protein J. 2011, 30, 384–394. [Google Scholar] [CrossRef]
- Gojkovic, Z.; Rislund, L.; Andersen, B.; Sandrini, M.P.; Cook, P.F.; Schnackerz, K.D.; Piskur, J. Dihydropyrimidine amidohydrolases and dihydroorotases share the same origin and several enzymatic properties. Nucleic Acids Res. 2003, 31, 1683–1692. [Google Scholar] [CrossRef]
- Kim, G.J.; Lee, D.E.; Kim, H.S. Functional expression and characterization of the two cyclic amidohydrolase enzymes, allantoinase and a novel phenylhydantoinase, from Escherichia coli. J. Bacteriol. 2000, 182, 7021–7028. [Google Scholar] [CrossRef]
- Clemente-Jimenez, J.M.; Martinez-Rodriguez, S.; Rodriguez-Vico, F.; Heras-Vazquez, F.J. Optically pure alpha-amino acids production by the “Hydantoinase Process”. Recent Pat. Biotechnol. 2008, 2, 35–46. [Google Scholar]
- Thoden, J.B.; Phillips, G.N., Jr.; Neal, T.M.; Raushel, F.M.; Holden, H.M. Molecular structure of dihydroorotase: A paradigm for catalysis through the use of a binuclear metal center. Biochemistry 2001, 40, 6989–6997. [Google Scholar] [CrossRef]
- Lohkamp, B.; Andersen, B.; Piskur, J.; Dobritzsch, D. The crystal structures of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J. Biol. Chem. 2006, 281, 13762–13776. [Google Scholar] [CrossRef] [PubMed]
- Cheon, Y.H.; Park, H.S.; Kim, J.H.; Kim, Y.; Kim, H.S. Manipulation of the active site loops of D-hydantoinase, a (beta/alpha)8-barrel protein, for modulation of the substrate specificity. Biochemistry 2004, 43, 7413–7420. [Google Scholar] [CrossRef] [PubMed]
- Nyhan, W.L. Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 2005, 86, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.; Knorr, S.; Hamacher, K.; Schmidt, B. DOCKTITE-a highly versatile step-by-step workflow for covalent docking and virtual screening in the molecular operating environment. J. Chem. Inf. Model. 2015, 55, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Vogels, G.D.; Van der Drift, C. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 1976, 40, 403–468. [Google Scholar] [CrossRef]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar]
- Chou, H.C.; Lee, C.Z.; Ma, L.C.; Fang, C.T.; Chang, S.C.; Wang, J.T. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 2004, 72, 3783–3792. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Grande-Garcia, A.; Lallous, N.; Diaz-Tejada, C.; Ramon-Maiques, S. Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD. Structure 2014, 22, 185–198. [Google Scholar] [CrossRef]
- Lee, M.; Chan, C.W.; Graham, S.C.; Christopherson, R.I.; Guss, J.M.; Maher, M.J. Structures of ligand-free and inhibitor complexes of dihydroorotase from Escherichia coli: Implications for loop movement in inhibitor design. J. Mol. Biol. 2007, 370, 812–825. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Adams, P.D.; Read, R.J.; McCoy, A.J.; Moriarty, N.W.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Zwart, P.H.; Hung, L.W. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 582–601. [Google Scholar] [CrossRef]
- Lebedev, A.A.; Young, P.; Isupov, M.N.; Moroz, O.V.; Vagin, A.A.; Murshudov, G.N. JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 61–69. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]


| Data Collection | |
|---|---|
| Crystal | EcALLase-BL21 |
| Wavelength (Å) | 1 |
| Resolution (Å) | 30–2.07 |
| Space group | C121 |
| Cell dimension (Å) | a = 203.16 α = 90° |
| b = 77.14 β = 100.8° | |
| c = 144.85 γ = 90° | |
| Completeness (%) | 99.02 (97.21) * |
| <I/σI> | 7.39 (2.14) |
| CC1/2 | 0.992 (0.855) |
| Redundancy | 3.5 (3.2) |
| Refinement | |
| Resolution (Å) | 29.92–2.07 |
| No. reflections | 132,804 |
| Rwork/Rfree | 0.208/0.248 |
| No. atoms | |
| Protein | 14,759 |
| Ligands | 110 |
| Water | 926 |
| R.m.s deviation | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.10 |
| Ramachandran Plot | |
| In preferred regions | 96.89% |
| In allowed regions | 2.66% |
| Outliers | 0.45% |
| PDB entry | 8HFD |
| Compound | S Score | Residue | Interaction | Receptor–Ligand Distance (Å) | E (kcal/mol) |
|---|---|---|---|---|---|
| 8-HQSA | −5.6100 | Asn94 | H-donor | 2.82 | −7.4 |
| Plumbagin | −5.2950 | Asp315 | H-donor | 2.94 | −3.1 |
| Asp315 | H-donor | 3.34 | −0.7 | ||
| Lupenone | −5.7230 | No important residue | |||
| Palmitic acid | −5.4744 | Ser317 | H-donor | 3.13 | −0.7 |
| Zn-β | Metal | 1.98 | −2.8 | ||
| Stigmast-5-en-3-ol | −6.4260 | Asp315 | H-donor | 3.05 | −0.6 |
| Zn-α | Metal | 2.43 | −0.9 | ||
| Trp332 | H-pi | 4.50 | −1.9 | ||
| Neophytadiene | −4.6771 | No important residue | |||
| Citraconic anhydride | −1.3467 | Gln168 | pi-H | 4.32 | −0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-H.; Yang, P.-C.; Lin, E.-S.; Ho, Y.-Y.; Peng, W.-F.; Lu, H.-P.; Huang, C.-C.; Huang, C.-Y. Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis. Molecules 2023, 28, 827. https://doi.org/10.3390/molecules28020827
Huang Y-H, Yang P-C, Lin E-S, Ho Y-Y, Peng W-F, Lu H-P, Huang C-C, Huang C-Y. Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis. Molecules. 2023; 28(2):827. https://doi.org/10.3390/molecules28020827
Chicago/Turabian StyleHuang, Yen-Hua, Po-Chun Yang, En-Shyh Lin, Ya-Yeh Ho, Wei-Feng Peng, Hsin-Pin Lu, Chien-Chih Huang, and Cheng-Yang Huang. 2023. "Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis" Molecules 28, no. 2: 827. https://doi.org/10.3390/molecules28020827
APA StyleHuang, Y.-H., Yang, P.-C., Lin, E.-S., Ho, Y.-Y., Peng, W.-F., Lu, H.-P., Huang, C.-C., & Huang, C.-Y. (2023). Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis. Molecules, 28(2), 827. https://doi.org/10.3390/molecules28020827







