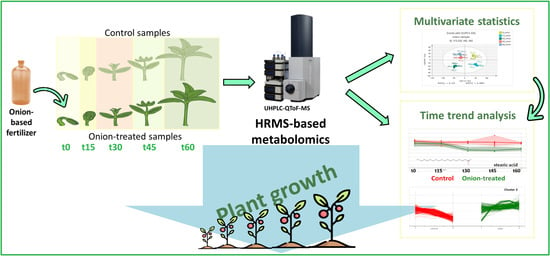
Longitudinal Plant Health Monitoring via High-Resolution Mass Spectrometry Screening Workflows: Application to a Fertilizer Mediated Tomato Growth Experiment
Abstract
:1. Introduction
2. Results
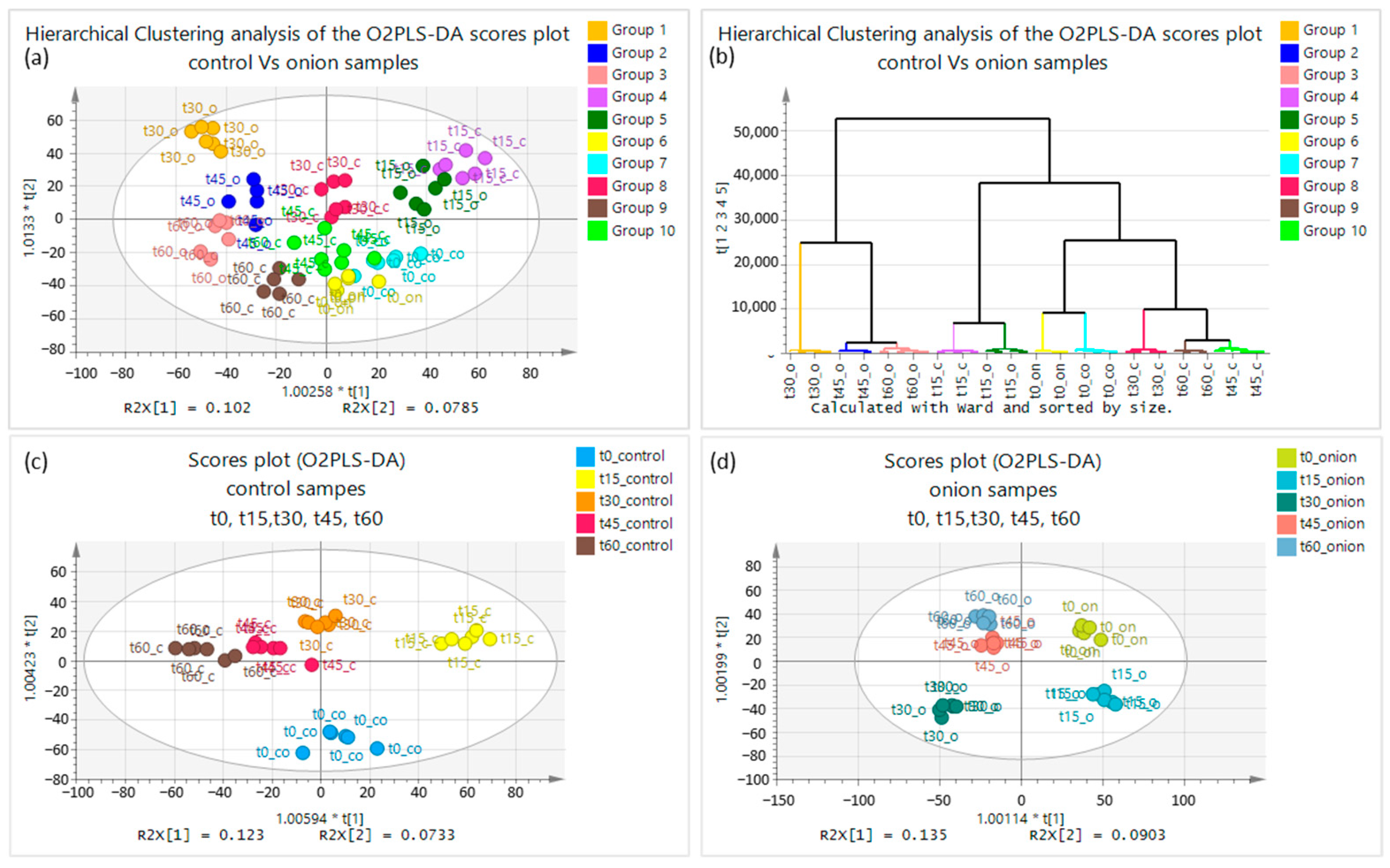
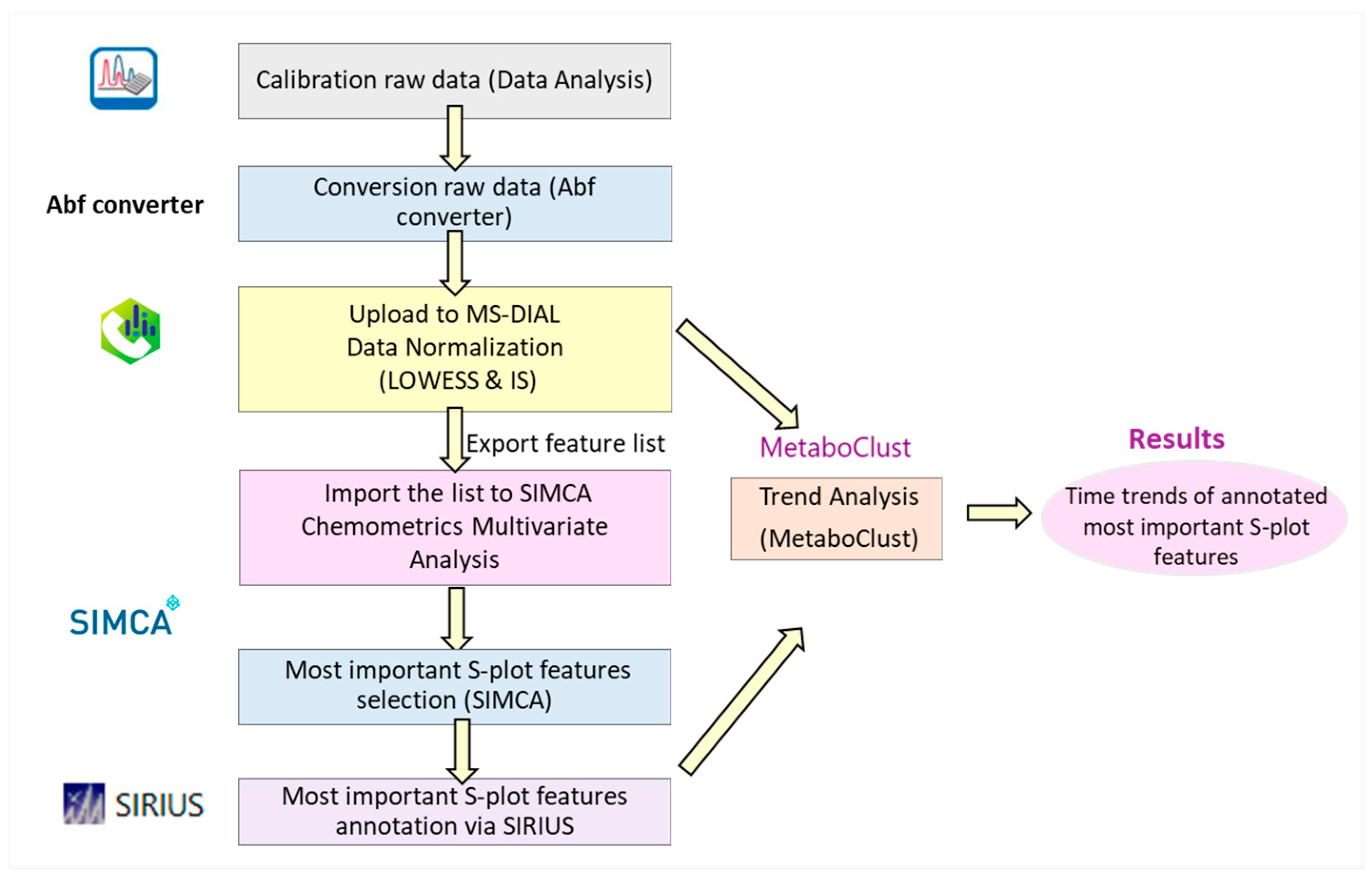
2.1. Chemometric Results
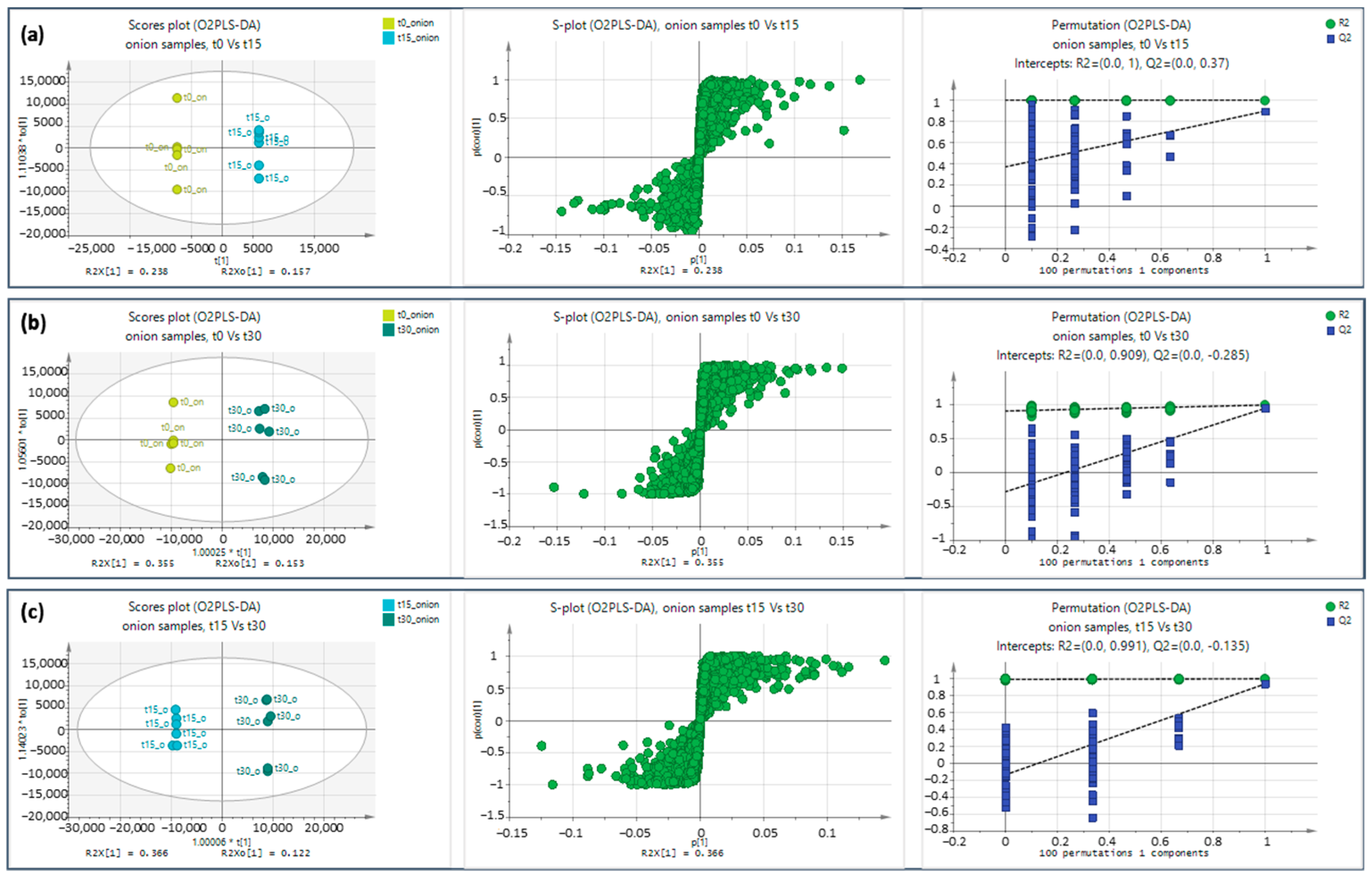
2.1.1. Multivariate Analysis of Data Obtained by +ESI
2.1.2. Multivariate Analysis of Data Obtained by ESI−
2.2. Metabolites Annotation in Both Ionization Modes
3. Discussion
3.1. Optimization of Sample Preparation
3.2. Chemometric Strategy
3.3. Role of the Identified Compounds in Plant Health and Their Trends
4. Materials and Methods
4.1. Leaf Harvesting and Time Point Definition
4.2. Reagents and Materials
4.3. Sample Preparation for HRMS Analysis
4.4. Instrumental Analysis
4.5. Mass Spectrometry Data Analysis
4.5.1. Screening Methodology
4.5.2. Chemometrics Methodology
4.5.3. Time Series
4.5.4. Metabolite Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Adugna, G. A review on impact of compost on soil properties, water use and crop productivity. Acad. Res. J. Agric. Sci. Res. 2016, 4, 93–104. [Google Scholar] [CrossRef]
- Mohammad, H.; Golabi, M.J.D.; Iyekar, C. Use of Composted Organic Wastes As Alternative to Synthetic Fertilizers for Enhancing Crop Productivity and Agricultural Sustainability on the Tropical Island of Guam. In Proceedings of the 13th International Soil Conservation Organisation Conference, Brisbane, Australia, 4–8 July 2004; Conserving Soil and Water for Society: Sharing Solutions. Volume 234. [Google Scholar]
- Panara, A.; Gikas, E.; Thomaidis, N.S. From By-Products to Fertilizer: Chemical Characterization Using UPLC-QToF-MS via Suspect and Non-Target Screening Strategies. Molecules 2022, 27, 3498. [Google Scholar] [CrossRef]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; Sonah, H.; et al. Advances in Omics Approaches for Abiotic Stress Tolerance in Tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef]
- Kazmi, R.H.; Willems, L.A.J.; Joosen, R.V.L.; Khan, N.; Ligterink, W.; Hilhorst, H.W.M. Metabolomic analysis of tomato seed germination. Metabolomics 2017, 13, 145. [Google Scholar] [CrossRef]
- Pentimone, I.; Colagiero, M.; Rosso, L.C.; Ciancio, A. Omics applications: Towards a sustainable protection of tomato. Appl. Microbiol. Biotechnol. 2020, 104, 4185–4195. [Google Scholar] [CrossRef] [PubMed]
- Afifah, E.N.; Murti, R.H.; Nuringtyas, T.R. Metabolomics Approach for the Analysis of Resistance of Four Tomato Genotypes (Solanum lycopersicum L.) to Root-Knot Nematodes (Meloidogyne Incognita). Open Life Sci. 2019, 14, 141–149. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261.e12. [Google Scholar] [CrossRef]
- De Vos, R.C.; Hall, R.D.; Moing, A. Metabolomics of a Model Fruit: Tomato; Blackwell: Oxford, UK, 2011; Volume 43, pp. 109–156. [Google Scholar]
- Knapp, S.; Peralta, I. The Tomato (Solanum lycopersicum L., Solanaceae) and Its Botanical Relatives; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Intelligence, M. Tomato Market Size & Share Analysis—Growth Trends & Forecasts (2023–2028). Available online: https://www.mordorintelligence.com/industry-reports/tomato-market (accessed on 22 June 2023).
- Abreu, A.C.; Fernandez, I. NMR Metabolomics Applied on the Discrimination of Variables Influencing Tomato (Solanum lycopersicum). Molecules 2020, 25, 3738. [Google Scholar] [CrossRef]
- Mazzei, P.; Vinale, F.; Woo, S.L.; Pascale, A.; Lorito, M.; Piccolo, A. Metabolomics by Proton High-Resolution Magic-Angle-Spinning Nuclear Magnetic Resonance of Tomato Plants Treated with Two Secondary Metabolites Isolated from Trichoderma. J. Agric. Food Chem. 2016, 64, 3538–3545. [Google Scholar] [CrossRef] [PubMed]
- Mun, H.I.; Kwon, M.C.; Lee, N.R.; Son, S.Y.; Song, D.H.; Lee, C.H. Comparing Metabolites and Functional Properties of Various Tomatoes Using Mass Spectrometry-Based Metabolomics Approach. Front. Nutr. 2021, 8, 659646. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; de Groot, J.; van Beek, T.A.; Vervoort, J.; de Vos, C.H. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef]
- de Oliveira, A.N.; Bolognini, S.R.F.; Navarro, L.C.; Delafiori, J.; Sales, G.M.; de Oliveira, D.N.; Catharino, R.R. Tomato classification using mass spectrometry-machine learning technique: A food safety-enhancing platform. Food Chem. 2023, 398, 133870. [Google Scholar] [CrossRef]
- Messaili, S.; Qu, Y.; Fougere, L.; Colas, C.; Desneux, N.; Lavoir, A.V.; Destandau, E.; Michel, T. Untargeted metabolomic and molecular network approaches to reveal tomato root secondary metabolites. Phytochem. Anal. PCA 2021, 32, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Yuan, M.; Tang, L.; Shen, Y.; Yu, Q.; Li, S. Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 2022, 817, 152878. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, W.; Zhang, Y.; Guo, Y.; Duan, Z.; Tang, Z.; Abozeid, A. Metabolomics Analysis Reveals Potential Mechanisms in Bupleurum L. (Apiaceae) Induced by Three Levels of Nitrogen Fertilization. Agronomy 2021, 11, 2291. [Google Scholar] [CrossRef]
- Bodein, A.; Scott-Boyer, M.P.; Perin, O.; Le Cao, K.A.; Droit, A. timeOmics: An R package for longitudinal multi-omics data integration. Bioinformatics 2022, 38, 577–579. [Google Scholar] [CrossRef]
- Rusilowicz, M.J.; Dickinson, M.; Charlton, A.J.; O’Keefe, S.; Wilson, J. MetaboClust: Using interactive time-series cluster analysis to relate metabolomic data with perturbed pathways. PLoS ONE 2018, 13, e0205968. [Google Scholar] [CrossRef]
- Bertinetto, C.; Engel, J.; Jansen, J. ANOVA simultaneous component analysis: A tutorial review. Anal. Chim. Acta X 2020, 6, 100061. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Reifycs Abf Converter. Available online: https://www.reifycs.com/AbfConverter (accessed on 12 July 2023).
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- MS Dial. Available online: http://prime.psc.riken.jp/compms/msdial/main.html (accessed on 12 July 2023).
- Bailly, C. The steroidal alkaloids alpha-tomatine and tomatidine: Panorama of their mode of action and pharmacological properties. Steroids 2021, 176, 108933. [Google Scholar] [CrossRef]
- Nakayasu, M.; Ohno, K.; Takamatsu, K.; Aoki, Y.; Yamazaki, S.; Takase, H.; Shoji, T.; Yazaki, K.; Sugiyama, A. Tomato roots secrete tomatine to modulate the bacterial assemblage of the rhizosphere. Plant Physiol. 2021, 186, 270–284. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Sawada, Y.; Yamada, Y.; Suzuki, M.; Hirai, M.Y.; Sakurai, T.; Saito, K. Combination of liquid chromatography-Fourier transform ion cyclotron resonance-mass spectrometry with 13C-labeling for chemical assignment of sulfur-containing metabolites in onion bulbs. Anal. Chem. 2013, 85, 1310–1315. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R.; et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The role of quercetin in plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Gorni, P.H.; de Lima, G.R.; Pereira, L.M.d.O.; Spera, K.D.; Lapaz, A.d.M.; Pacheco, A.C. Increasing plant performance, fruit production and nutritional value of tomato through foliar applied rutin. Sci. Hortic. 2022, 294, 110755. [Google Scholar] [CrossRef]
- Parvin, K.; Hasanuzzaman, M.; Bhuyan, M.; Mohsin, S.M.; Fujita, A.M. Quercetin Mediated Salt Tolerance in Tomato through the Enhancement of Plant Antioxidant Defense and Glyoxalase Systems. Plants 2019, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; O’Connell, A.P. Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J. Exp. Bot. 2002, 53, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues de Queiroz, A.; Hines, C.; Brown, J.; Sahay, S.; Vijayan, J.; Stone, J.M.; Bickford, N.; Wuellner, M.; Glowacka, K.; Buan, N.R.; et al. The effects of exogenously applied antioxidants on plant growth and resilience. Phytochem. Rev. 2023, 22, 407–447. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Christie, W.W. The Lipid Web, Glycosyldiacylglycerols. Available online: https://lipidmaps.org/resources/lipidweb/lipidweb_html/lipids/complex/mg-dgdg/index.htm (accessed on 20 July 2023).
- Hortensteiner, S.; Wuthrich, K.L.; Matile, P.; Ongania, K.H.; Krautler, B. The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide a macrocycle by a monooxygenase. J. Biol. Chem. 1998, 273, 15335–15339. [Google Scholar] [CrossRef]
- Kundu, A.; Vadassery, J. Chlorogenic acid-mediated chemical defence of plants against insect herbivores. Plant Biol. 2019, 21, 185–189. [Google Scholar] [CrossRef]
- Volpi, E.S.N.; Mazzafera, P.; Cesarino, I. Should I stay or should I go: Are chlorogenic acids mobilized towards lignin biosynthesis? Phytochemistry 2019, 166, 112063. [Google Scholar] [CrossRef]
- Yang, J.; Guo, J.; Yuan, J. In vitro antioxidant properties of rutin. LWT—Food Sci. Technol. 2008, 41, 1060–1066. [Google Scholar] [CrossRef]
- Arya, A.; Al-Obaidi, M.M.; Shahid, N.; Bin Noordin, M.I.; Looi, C.Y.; Wong, W.F.; Khaing, S.L.; Mustafa, M.R. Synergistic effect of quercetin and quinic acid by alleviating structural degeneration in the liver, kidney and pancreas tissues of STZ-induced diabetic rats: A mechanistic study. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2014, 71, 183–196. [Google Scholar] [CrossRef]
- Muthamil, S.; Balasubramaniam, B.; Balamurugan, K.; Pandian, S.K. Synergistic Effect of Quinic Acid Derived From Syzygium cumini and Undecanoic Acid Against Candida spp. Biofilm and Virulence. Front. Microbiol. 2018, 9, 2835. [Google Scholar] [CrossRef] [PubMed]
- Menelaou, M.; Mateescu, C.; Salifoglou, A. D-(-)-Quinic acid: An efficient physiological metal ion ligand. J. Agroaliment. Process. Technol. 2011, 17, 344–347. [Google Scholar]
- Marsh, K.B.; Boldingh, H.L.; Shilton, R.S.; Laing, W.A. Changes in quinic acid metabolism during fruit development in three kiwifruit species. Funct. Plant Biol. 2009, 36, 463–470. [Google Scholar] [CrossRef]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. PCA 2010, 21, 4–13. [Google Scholar] [CrossRef]
- Duhrkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Bocker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Duhrkop, K.; Nothias, L.F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Duhrkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Bocker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed]
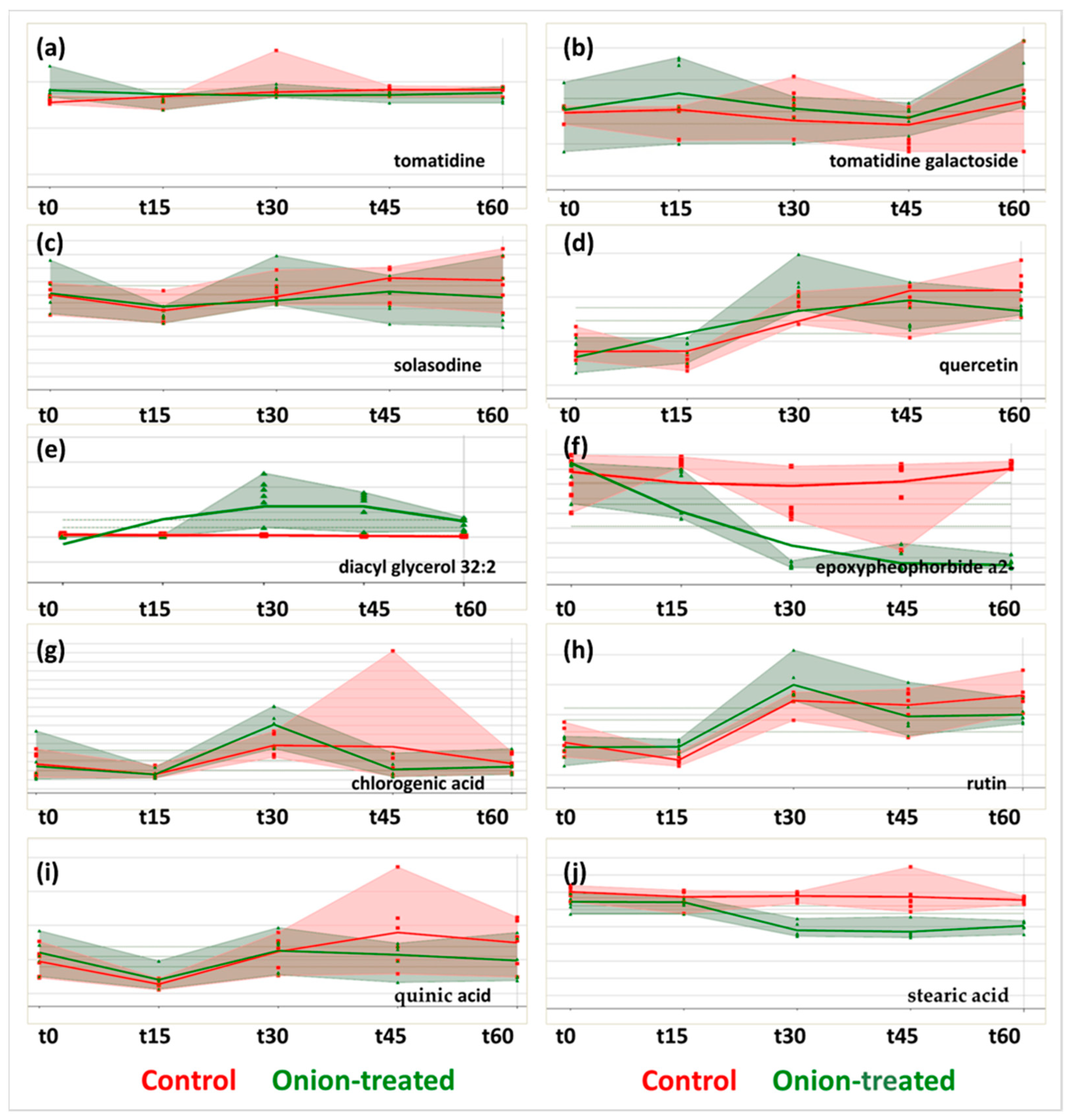
| Compound Name | Chemical Formula | Exp. tR (min) | Exp. m/z Values | Theor. m/z | ESI Mode | CSI Finger ID Score | Similarity | Level a | Cluster k |
|---|---|---|---|---|---|---|---|---|---|
| tomatidine | C27H45NO2 | 8.0 | 416.3548 | 416.3523 | +ESI | −110.14 | 59.8 | 2a | 3 |
| tomatidine galactoside | C33H55NO7 | 7.3 | 578.4055 | 578.4051 | +ESI | −94.64 | 67.8 | 2a | 6 |
| solasodine | C27H43NO2 | 7.7 | 414.3384 | 414.3367 | +ESI | −150.3 | 54.1 | 2a | 2 |
| quercetin | C15H10O7 | 6.8 | 303.0506 | 303.0499 | +ESI | −31.2 | 84.3 | 1 | 2 |
| diacyl glycerol 32:2 | C35H66O4 | 15.6 | 551.5031 | 551.5034 | +ESI | Lipid map | 3 | 3 | |
| epoxypheophorbide a(2−) | C35H34N4O6 | 14.0 | 607.2529 | 607.2551 | +ESI | TomatoCyc | 3 | 7 | |
| quinic acid | C7H12O6 | 1.3 | 191.0567 | 191.0561 | −ESI | −67.6 | 47.7 | 1 | 5 |
| stearic acid | C18H36O2 | 14.5 | 283.2654 | 283.2643 | −ESI | −11.3 | 100 | 2a | 3 |
| chlorogenic acid | C16H18O9 | 2.9 | 353.0873 | 353.0878 | −ESI | −8.1 | 100 | 1 | 1 |
| rutin | C27H30O16 | 5.7 | 609.1459 | 609.1461 | −ESI | −57.98 | 89.2 | 1 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panara, A.; Gikas, E.; Koupa, A.; Thomaidis, N.S. Longitudinal Plant Health Monitoring via High-Resolution Mass Spectrometry Screening Workflows: Application to a Fertilizer Mediated Tomato Growth Experiment. Molecules 2023, 28, 6771. https://doi.org/10.3390/molecules28196771
Panara A, Gikas E, Koupa A, Thomaidis NS. Longitudinal Plant Health Monitoring via High-Resolution Mass Spectrometry Screening Workflows: Application to a Fertilizer Mediated Tomato Growth Experiment. Molecules. 2023; 28(19):6771. https://doi.org/10.3390/molecules28196771
Chicago/Turabian StylePanara, Anthi, Evagelos Gikas, Anastasia Koupa, and Nikolaos S. Thomaidis. 2023. "Longitudinal Plant Health Monitoring via High-Resolution Mass Spectrometry Screening Workflows: Application to a Fertilizer Mediated Tomato Growth Experiment" Molecules 28, no. 19: 6771. https://doi.org/10.3390/molecules28196771
APA StylePanara, A., Gikas, E., Koupa, A., & Thomaidis, N. S. (2023). Longitudinal Plant Health Monitoring via High-Resolution Mass Spectrometry Screening Workflows: Application to a Fertilizer Mediated Tomato Growth Experiment. Molecules, 28(19), 6771. https://doi.org/10.3390/molecules28196771