Ginsentide-like Coffeetides Isolated from Coffee Waste Are Cell-Penetrating and Metal-Binding Microproteins
Abstract
:1. Introduction
2. Results
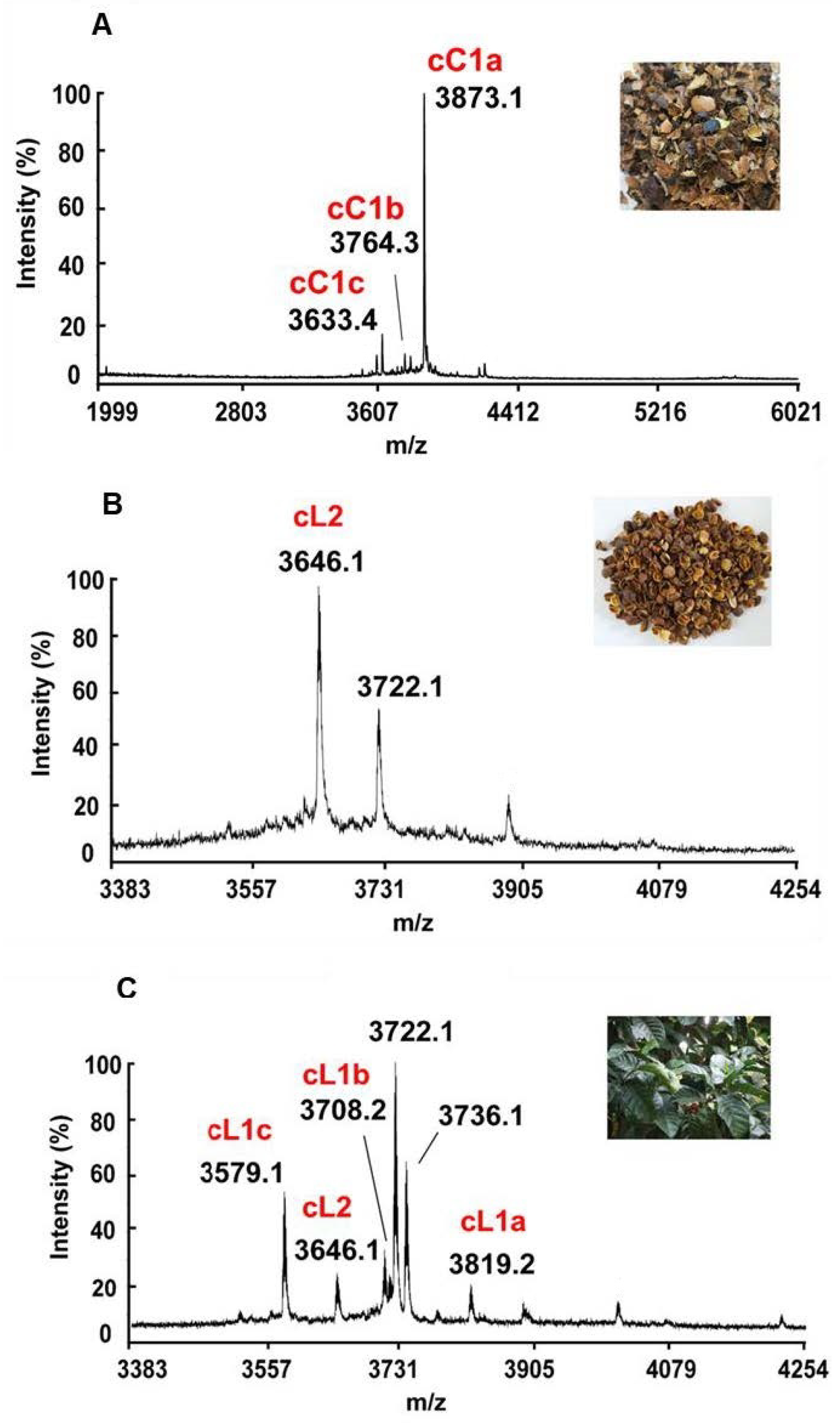
2.1. Mass-Spectrometry Screening, Isolation, and Purification of Cysteine-Rich Peptides in Aqueous Extracts of C. canephora and C. liberica
2.2. Sequencing, Database Search, and Transcriptomic Analysis of Coffeetides
2.3. Disulfide Mapping of Coffeetide cC1a
2.4. Chemical Synthesis and Oxidative Folding of Coffeetide cC1a
2.5. Solution NMR Structure of cC1a
2.6. Coffeetide cC1a Is Resistant to Heat, Acid, Proteolytic, and Human Serum-Mediated Degradation
2.7. Coffeetide cC1a Is Non-Cytotoxic
2.8. Coffeetide cC1a Is Cell-Penetrating
2.9. Coffeetide cC1a Is Metal Binding
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Plant Materials
4.3. Extraction and Screening of Coffea canephora and Coffea liberica
4.4. Isolation and Purification of Coffeetides
4.5. Data Mining and Bioinformatics Analysis
4.6. Sequence Determination of Coffeetides
4.7. Disulfide Mapping
4.8. NMR Solution Structure
4.9. Chemical Synthesis of Coffeetide
4.10. Oxidative Folding of Coffeetide cC1a
4.11. Stability Assays
4.12. Cell Culture
4.13. Toxicity Assay
4.14. Cell-Penetrating Assay
4.15. Isothermal Titration Calorimetry (ITC) Assay
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hu, G.L.; Wang, X.; Zhang, L.; Qiu, M.H. The sources and mechanisms of bioactive ingredients in coffee. Food Funct. 2019, 10, 3113–3126. [Google Scholar] [CrossRef]
- Matosinhos, R.C.; Bezerra, J.P.; Barros, C.H.; Fernandes Pereira Ferreira Bernardes, A.C.; Coelho, G.B.; Carolina de Paula Michel Araújo, M.; Dian de Oliveira Aguiar Soares, R.; Sachs, D.; Saúde-Guimarães, D.A. Coffea arabica extracts and their chemical constituents in a murine model of gouty arthritis: How they modulate pain and inflammation. J. Ethnopharmacol. 2022, 284, 114778. [Google Scholar] [CrossRef]
- Veltri, C.; Grundmann, O. Current perspectives on the impact of Kratom use. Subst. Abus. Rehabil. 2019, 10, 23–31. [Google Scholar] [CrossRef]
- ALAsmari, K.M.; Zeid, I.M.A.; Al-Attar, A.M. Medicinal properties of Arabica coffee (Coffea arabica) oil: An Overview. Adv. Life Sci. 2020, 8, 20–29. [Google Scholar]
- Bisht, S.; Sisodia, S. Coffea arabica: A wonder gift to medical science. J. Nat. Pharm. 2010, 1, 58–65. [Google Scholar] [CrossRef]
- Chen, X. A review on coffee leaves: Phytochemicals, bioactivities and applications. Crit. Rev. Food Sci. Nutr. 2019, 59, 1008–1025. [Google Scholar] [CrossRef]
- Gökcen, B.B.; Şanlier, N. Coffee consumption and disease correlations. Crit. Rev. Food Sci. Nutr. 2019, 59, 336–348. [Google Scholar] [CrossRef]
- Ali, A.; Zahid, H.F.; Cottrell, J.J.; Dunshea, F.R. A Comparative Study for Nutritional and Phytochemical Profiling of Coffea arabica (C. arabica) from Different Origins and Their Antioxidant Potential and Molecular Docking. Molecules 2022, 27, 5126. [Google Scholar] [CrossRef]
- Ding, M.; Bhupathiraju, S.N.; Satija, A.; van Dam, R.M.; Hu, F.B. Long-term coffee consumption and risk of cardiovascular disease: A systematic review and a dose-response meta-analysis of prospective cohort studies. Circulation 2014, 129, 643–659. [Google Scholar] [CrossRef]
- Chieng, D.; Kistler, P.M. Coffee and tea on cardiovascular disease (CVD) prevention. Trends Cardiovasc. Med. 2022, 32, 399–405. [Google Scholar] [CrossRef]
- James, J.E. Critical review of dietary caffeine and blood pressure: A relationship that should be taken more seriously. Psychosom. Med. 2004, 66, 63–71. [Google Scholar] [CrossRef]
- Geraets, L.; Moonen, H.J.; Wouters, E.F.; Bast, A.; Hageman, G.J. Caffeine metabolites are inhibitors of the nuclear enzyme poly (ADP-ribose) polymerase-1 at physiological concentrations. Biochem. Pharmacol. 2006, 72, 902–910. [Google Scholar] [CrossRef]
- Gonzalez de Mejia, E.; Ramirez-Mares, M.V. Impact of caffeine and coffee on our health. Trends Endocrinol. Metab. TEM 2014, 25, 489–492. [Google Scholar] [CrossRef]
- Bonita, J.S.; Mandarano, M.; Shuta, D.; Vinson, J. Coffee and cardiovascular disease: In vitro, cellular, animal, and human studies. Pharmacol. Res. 2007, 55, 187–198. [Google Scholar] [CrossRef]
- Socała, K.; Szopa, A.; Serefko, A.; Poleszak, E.; Wlaź, P. Neuroprotective Effects of Coffee Bioactive Compounds: A Review. Int. J. Mol. Sci. 2020, 22, 107. [Google Scholar] [CrossRef]
- Mansour, A.; Mohajeri-Tehrani, M.R.; Samadi, M.; Qorbani, M.; Merat, S.; Adibi, H.; Poustchi, H.; Hekmatdoost, A. Effects of supplementation with main coffee components including caffeine and/or chlorogenic acid on hepatic, metabolic, and inflammatory indices in patients with non-alcoholic fatty liver disease and type 2 diabetes: A randomized, double-blind, placebo-controlled, clinical trial. Nutr. J. 2021, 20, 35. [Google Scholar] [CrossRef]
- Muchtaridi, M.; Lestari, D.; Khairul Ikram, N.K.; Gazzali, A.M.; Hariono, M.; Wahab, H.A. Decaffeination and Neuraminidase Inhibitory Activity of Arabica Green Coffee (Coffea arabica) Beans: Chlorogenic Acid as a Potential Bioactive Compound. Molecules 2021, 26, 3402. [Google Scholar] [CrossRef]
- Stefanello, N.; Spanevello, R.M.; Passamonti, S.; Porciúncula, L.; Bonan, C.D.; Olabiyi, A.A.; Teixeira da Rocha, J.B.; Assmann, C.E.; Morsch, V.M.; Schetinger, M.R.C. Coffee, caffeine, chlorogenic acid, and the purinergic system. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 123, 298–313. [Google Scholar] [CrossRef]
- Murthy, P.S.; Madhava Naidu, M. Sustainable management of coffee industry by-products and value addition—A review. Resour. Conserv. Recycl. 2012, 66, 45–58. [Google Scholar] [CrossRef]
- Pagett, M.; Teng, K.S.; Sullivan, G.; Zhang, W. Reusing Waste Coffee Grounds as Electrode Materials: Recent Advances and Future Opportunities. Glob. Chall. 2023, 7, 2200093. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef]
- Edwards, C.; Cohen, M.; Bloom, S. Peptides as drugs. QJM 1999, 92, 1–4. [Google Scholar] [CrossRef]
- Dietrich, U.; Dürr, R.; Koch, J. Peptides as drugs: From screening to application. Curr. Pharm. Biotechnol. 2013, 14, 501–512. [Google Scholar] [CrossRef]
- Tam, J.P.; Nguyen, G.K.T.; Loo, S.; Wang, S.; Yang, D.; Kam, A. Ginsentides: Cysteine and Glycine-rich Peptides from the Ginseng Family with Unusual Disulfide Connectivity. Sci. Rep. 2018, 8, 16201. [Google Scholar] [CrossRef]
- Kam, A.; Loo, S.; Fan, J.S.; Sze, S.K.; Yang, D.; Tam, J.P. Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation. J. Biol. Chem. 2019, 294, 19604–19615. [Google Scholar] [CrossRef]
- Loo, S.; Kam, A.; Xiao, T.; Nguyen, G.K.; Liu, C.F.; Tam, J.P. Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa. Sci. Rep. 2016, 6, 39401. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.; Kam, A.; Tam, J.P. Hyperstable EGF-like bleogen derived from cactus accelerates corneal healing in rats. Front. Pharmacol. 2022, 13, 942168. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.; Kam, A.; Li, B.B.; Feng, N.; Wang, X.; Tam, J.P. Discovery of hyperstable noncanonical plant-derived epidermal growth factor receptor agonist and analogs. J. Med. Chem. 2021, 64, 7746–7759. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.; Tay, S.V.; Kam, A.; Lee, W.; Tam, J.P. Hololectin Interdomain Linker Determines Asparaginyl Endopeptidase-Mediated Maturation of Antifungal Hevein-Like Peptides in Oats. Front. Plant Sci. 2022, 13, 899740. [Google Scholar] [CrossRef]
- Wong, C.T.T.; Taichi, M.; Nishio, H.; Nishiuchi, Y.; Tam, J.P. Optimal Oxidative Folding of the Novel Antimicrobial Cyclotide from Hedyotis biflora Requires High Alcohol Concentrations. Biochemistry 2011, 50, 7275–7283. [Google Scholar] [CrossRef] [PubMed]
- Archer, B. The proteins of Hevea brasiliensis latex. 4. Isolation and characterization of crystalline hevein. Biochem. J. 1960, 75, 236. [Google Scholar] [CrossRef]
- Rodríguez-Romero, A.; Ravichandran, K.G.; Soriano-García, M. Crystal structure of hevein at 2.8 Å resolution. FEBS Lett. 1991, 291, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Van Parijs, J.; Broekaert, W.F.; Goldstein, I.J.; Peumans, W.J. Hevein: An antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta 1991, 183, 258–264. [Google Scholar] [CrossRef]
- Andersen, N.H.; Cao, B.; Rodriguez-Romero, A.; Arreguin, B. Hevein: NMR assignment and assessment of solution-state folding for the agglutinin-toxin motif. Biochemistry 1993, 32, 1407–1422. [Google Scholar] [CrossRef]
- Gidrol, X.; Chrestin, H.; Tan, H.-L.; Kush, A. Hevein, a lectin-like protein from Hevea brasiliensis (rubber tree) is involved in the coagulation of latex. J. Biol. Chem. 1994, 269, 9278–9283. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceutical 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Loo, S.; Tay, S.V.; Kam, A.; Tang, F.; Fan, J.-S.; Yang, D.; Tam, J.P. Anti-fungal Hevein-like peptides biosynthesized from quinoa cleavable hololectins. Molecules 2021, 26, 5909. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Loo, S.; Kam, A.; Dutta, B.; Zhang, X.; Feng, N.; Sze, S.K.; Liu, C.-F.; Wang, X.; Tam, J.P. Decoding the Cure-all Effects of Ginseng. bioRxiv 2023. bioRxiv:2023.2004.2005.535784. [Google Scholar] [CrossRef]
- Dutta, B.; Loo, S.; Kam, A.; Sze, S.K.; Tam, J.P. Ginsentide TP1 Protects Hypoxia-Induced Dysfunction and ER Stress-Linked Apoptosis. Cells 2023, 12, 1401. [Google Scholar] [CrossRef]
- Kam, A.; Loo, S.; Dutta, B.; Sze, S.K.; Tam, J.P. Plant-derived mitochondria-targeting cysteine-rich peptide modulates cellular bioenergetics. J. Biol. Chem. 2019, 294, 4000–4011. [Google Scholar] [CrossRef]
- Padmapriya, R.; Tharian, J.A.; Thirunalasundari, T. Coffee waste management—An overview. Int. J. Curr. Sci 2013, 9, 83–91. [Google Scholar]
- Lee, J.-C.; Son, Y.-O.; Pratheeshkumar, P.; Shi, X. Oxidative stress and metal carcinogenesis. Free Radic. Biol. Med. 2012, 53, 742–757. [Google Scholar] [CrossRef]
- Jomova, K.; Baros, S.; Valko, M. Redox active metal-induced oxidative stress in biological systems. Transit. Met. Chem. 2012, 37, 127–134. [Google Scholar] [CrossRef]
- Jomova, K.; Valko, M. Importance of iron chelation in free radical-induced oxidative stress and human disease. Curr. Pharm. Des. 2011, 17, 3460–3473. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef]
- Farah, A. Coffee: Production, Quality and Chemistry. In Coffee by-Products; Castillo, M., Fernández-Gómez, B., Martínez Sáez, N., Iriondo-DeHond, A., Mesa, M.D., Eds.; Royal Society of Chemistry: London, UK, 2019. [Google Scholar]
- Lee, H.; Broekaert, W.; Raikhel, N.; Lee, H. Co-and post-translational processing of the hevein preproprotein of latex of the rubber tree (Hevea brasiliensis). J. Biol. Chem. 1991, 266, 15944–15948. [Google Scholar] [CrossRef]
- Henriques, S.T.; Craik, D.J. Cyclotides as templates in drug design. Drug Discov. Today 2010, 15, 57–64. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Craik, D.J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 2004, 43, 5965–5975. [Google Scholar] [CrossRef]
- Pelegrini, P.B.; Quirino, B.F.; Franco, O.L. Plant cyclotides: An unusual class of defense compounds. Peptides 2007, 28, 1475–1481. [Google Scholar] [CrossRef]
- Wang, C.K.; Craik, D.J. Cyclic peptide oral bioavailability: Lessons from the past. Pept. Sci. 2016, 106, 901–909. [Google Scholar] [CrossRef]
- Wong, C.T.; Rowlands, D.K.; Wong, C.H.; Lo, T.W.; Nguyen, G.K.; Li, H.Y.; Tam, J.P. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew. Chem. Int. Ed. 2012, 51, 5620–5624. [Google Scholar] [CrossRef]
- Verdine, G.L. Drugging the undruggable using stapled peptides. In Proceedings of the Abstracts of Papers of the American Chemical Society, American Chemical Society, Washington, WA, USA; 2010. [Google Scholar]
- Kostova, I.; Balkansky, S. Metal complexes of biologically active ligands as potential antioxidants. Curr. Med. Chem. 2013, 20, 4508–4539. [Google Scholar] [CrossRef]
- Horniblow, R.D.; Henesy, D.; Iqbal, T.H.; Tselepis, C. Modulation of iron transport, metabolism and reactive oxygen status by quercetin–iron complexes in vitro. Mol. Nutr. Food Res. 2017, 61, 1600692. [Google Scholar] [CrossRef] [PubMed]
- Jungwirth, U.; Kowol, C.R.; Keppler, B.K.; Hartinger, C.G.; Berger, W.; Heffeter, P. Anticancer activity of metal complexes: Involvement of redox processes. Antioxid. Redox Signal. 2011, 15, 1085–1127. [Google Scholar] [CrossRef] [PubMed]
- Koskenkorva-Frank, T.S.; Weiss, G.; Koppenol, W.H.; Burckhardt, S. The complex interplay of iron metabolism, reactive oxygen species, and reactive nitrogen species: Insights into the potential of various iron therapies to induce oxidative and nitrosative stress. Free Radic. Biol. Med. 2013, 65, 1174–1194. [Google Scholar] [CrossRef]
- Bouché, M.; Hognon, C.; Grandemange, S.; Monari, A.; Gros, P.C. Recent advances in iron-complexes as drug candidates for cancer therapy: Reactivity, mechanism of action and metabolites. Dalton Trans. 2020, 49, 11451–11466. [Google Scholar] [CrossRef] [PubMed]
- Chunkao, S.; Youravong, W.; Yupanqui, C.T.; Alashi, A.M.; Aluko, R.E. Structure and function of mung bean protein-derived iron-binding antioxidant peptides. Foods 2020, 9, 1406. [Google Scholar] [CrossRef] [PubMed]
- Blat, D.; Weiner, L.; Youdim, M.B.; Fridkin, M. A novel iron-chelating derivative of the neuroprotective peptide NAPVSIPQ shows superior antioxidant and antineurodegenerative capabilities. J. Med. Chem. 2008, 51, 126–134. [Google Scholar] [CrossRef]
- Cuajungco, M.P.; Fagét, K.Y.; Huang, X.; Tanzi, R.E.; Bush, A.I. Metal Chelation as a Potential Therapy for Alzheimer’s Disease. Ann. N. Y. Acad. Sci. 2000, 920, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shushan, S.; Miller, Y. Neuropeptides: Roles and Activities as Metal Chelators in Neurodegenerative Diseases. J. Phys. Chem. B 2021, 125, 2796–2811. [Google Scholar] [CrossRef]
- Amit, T.; Avramovich-Tirosh, Y.; Youdim, M.B.H.; Mandel, S. Targeting multiple Alzheimer’s disease etiologies with multimodal neuroprotective and neurorestorative iron chelators. FASEB J. 2008, 22, 1296–1305. [Google Scholar] [CrossRef]
- Budimir, A. Metal ions, Alzheimer’s disease and chelation therapy. Acta Pharm. 2011, 61, 1–14. [Google Scholar] [CrossRef]
- Weinreb, O.; Mandel, S.; Bar-Am, O.; Amit, T. Iron-chelating backbone coupled with monoamine oxidase inhibitory moiety as novel pluripotential therapeutic agents for Alzheimer’s disease: A tribute to Moussa Youdim. J. Neural Transm. 2011, 118, 479–492. [Google Scholar] [CrossRef]
- Gao, L.; Laude, K.; Cai, H. Mitochondrial pathophysiology, reactive oxygen species, and cardiovascular diseases. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 137–155. [Google Scholar] [CrossRef]
- Patten, D.A.; Germain, M.; Kelly, M.A.; Slack, R.S. Reactive oxygen species: Stuck in the middle of neurodegeneration. J. Alzheimer’s Dis. 2010, 20, S357–S367. [Google Scholar] [CrossRef]
- Brieger, K.; Schiavone, S.; Miller, F.J., Jr.; Krause, K.H. Reactive oxygen species: From health to disease. Swiss. Med. Wkly. 2012, 142, w13659. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Jeener, J.; Meier, B.; Bachmann, P.; Ernst, R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979, 71, 4546–4553. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Wüthrich, K.; Billeter, M.; Braun, W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J. Mol. Biol. 1983, 169, 949–961. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2014, 31, 1325–1327. [Google Scholar] [CrossRef]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 1998, 54, 905–921. [Google Scholar]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Cryst. 2002, 40, 82–92. [Google Scholar]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]










| Peptide | Species | Amino Acid Sequence | Mass (Da) 1 | Charge 2 | PI | Approach 3 |
|---|---|---|---|---|---|---|
| Loops | 1 2 3 4 5 6 | |||||
| cC1a | C. canephora | ZEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3873.1 | −3 | 4.00 | T, P |
| cC1b | C. canephora | -EGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3764.3 | −3 | 4.00 | T, P |
| cC1c | C. canephora | --GECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3633.4 | −2 | 4.14 | T, P |
| cC2 | C. canephora | QEGECSPFGKPCRYNPWGCCDSCVCVAT-PADE-GRCLGNC | 4145.6 | −2 | 4.51 | T |
| cC3 | C. canephora | QEGECSPLGKPCKYNPWGCCGSCLCIVDQP-THEGTCVGNC | 4206.7 | −2 | 4.83 | T |
| cC4 | C. canephora | QEGECSPLGKPCRYNPWGCCGSCLCIVDQP-THEGTCVGNC | 4234.7 | −2 | 4.83 | T |
| cC5 | C. canephora | QEGECSALGKPCRYNPSGCCGLCVCVIPDPTDE-GSCIGIC | 4067.7 | −3 | 4.18 | T |
| cC6 | C. canephora | QEVECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3931.6 | −3 | 4.00 | T |
| cC7 | C. canephora | QEPSCIPVLGSCVGNPWGCCPGCMCIRQ-LTDR---CHGYC | 3976.6 | 0 | 6.69 | T |
| cL1a | C. liberica | ZEGECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3819.2 | −2 | 5.36 | T, P |
| cL1b | C. liberica | -EGECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3708.2 | −2 | 5.36 | T, P |
| cL1c | C. liberica | --GECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3579.1 | −1 | 6.01 | T, P |
| cL2 | C. liberica | --GECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3649.2 | −2 | 4.14 | T, P |
| cL3 | C. liberica | GKDTCIGLLESCKDDPWGCCFGCVCLWP--GDL---CRGSC | 3834.4 | −2 | 4.36 | T |
| cL4 | C. liberica | QEGECSPAGKSCR--PVRCCDFCVCVVDYP-THVGTCRGNC | 4069.7 | −2 | 4.36 | T |
| cL5 | C. liberica | QEGECSPAGKPCR--PVRCCDFCVCVVDYP-THVGTCRGNC | 4082.7 | 0 | 6.70 | T |
| cL6 | C. liberica | QEPSCIPVLGSCVGNPWGCCPGCMCIRQ-LTDR---CHGYC | 3976.7 | 0 | 6.69 | T |
| cA1 | C. arabica | QEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3903.5 | −3 | 4.00 | T |
| cA2 | C. arabica | QEGECSPLGEACAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3863.5 | −3 | 4.00 | T |
| cA3 | C. arabica | QEPSCLPAGESCTGNPWGCCPGCICIWQ-LTER---CVGNC | 3905.6 | −2 | 4.25 | T |
| cA4 | C. arabica | QEPSCIPVGEPCAGNPGGCCDGCICIWQ-LTDR---CAGSC | 3733.5 | −3 | 3.92 | T |
| cA5 | C. arabica | QEGECSPLGKPCRYNPRGCCDFCVCVVADVTDEEGSCRGNC | 4389.7 | −3 | 4.44 | T |
| cA6 | C. arabica | RKDTCIGLLESCKDDPYGCCPGCVCLWP--GDL---CRGDC | 3884.6 | −2 | 4.46 | T |
| cA7 | C. arabica | QEGECSPAGKPCR--PVRCCDSCLCIVDYP-THVGTCRGNC | 4047.7 | 0 | 6.70 | T |
| cA8 | C. arabica | QEGECSPLGKPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3902.5 | −1 | 4.68 | T |
| cA9 | C. arabica | QEPSCIPVGEPCAGNPGGCCDGCICIWQ-LTDR---CAGSC | 3733.3 | −3 | 3.92 | T |
| cA10 | C. arabica | QEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3890.1 | −3 | 4.00 | T |
| cR1 | C. racemosa | QEPRCIPALGSCVGNPWGCCFGCMCIRQ-LTDR---CLGYC | 4043.7 | +1 | 7.70 | T |
| cR2 | C. racemosa | QEPRCIPVFGSCVGNPWGCCFGCMCIRQ-HTNR---CLGYC | 4128.7 | +2 | 8.22 | T |
| cR3 | C. racemosa | QEGECSPFGKPCRYNPWGCCGSCLCVVDHP-THEGTCVGNC | 4263.7 | −2 | 5.36 | T |
| cR4 | C. racemosa | QEGECSPFGKPCRYNPWGCCGSCVCVVDHP-THEGTCLGNC | 4263.7 | −2 | 5.36 | T |
| cR5 | C. racemosa | QEGKCSPAGKPC--DPWGCCDFCVCVVDFPGGE-GRCAGNC | 3885.5 | −2 | 4.44 | T |
| cR6 | C. racemosa | QEEKCSPAGKPCRYNPRGCCDFCVCVVDFPGGE-GSCLGNC | 4218.7 | −1 | 4.94 | T |
| cR7 | C. racemosa | GKDTCIGLLESCKDDPWGCCPGCVCLWP--GDL---CRGSC | 3780.5 | −2 | 4.36 | T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, J.P.; Huang, J.; Loo, S.; Li, Y.; Kam, A. Ginsentide-like Coffeetides Isolated from Coffee Waste Are Cell-Penetrating and Metal-Binding Microproteins. Molecules 2023, 28, 6556. https://doi.org/10.3390/molecules28186556
Tam JP, Huang J, Loo S, Li Y, Kam A. Ginsentide-like Coffeetides Isolated from Coffee Waste Are Cell-Penetrating and Metal-Binding Microproteins. Molecules. 2023; 28(18):6556. https://doi.org/10.3390/molecules28186556
Chicago/Turabian StyleTam, James P., Jiayi Huang, Shining Loo, Yimeng Li, and Antony Kam. 2023. "Ginsentide-like Coffeetides Isolated from Coffee Waste Are Cell-Penetrating and Metal-Binding Microproteins" Molecules 28, no. 18: 6556. https://doi.org/10.3390/molecules28186556
APA StyleTam, J. P., Huang, J., Loo, S., Li, Y., & Kam, A. (2023). Ginsentide-like Coffeetides Isolated from Coffee Waste Are Cell-Penetrating and Metal-Binding Microproteins. Molecules, 28(18), 6556. https://doi.org/10.3390/molecules28186556