Distribution, Typical Structure and Self-Assembly Properties of Collagen from Fish Skin and Bone
Abstract
:1. Introduction
2. Results
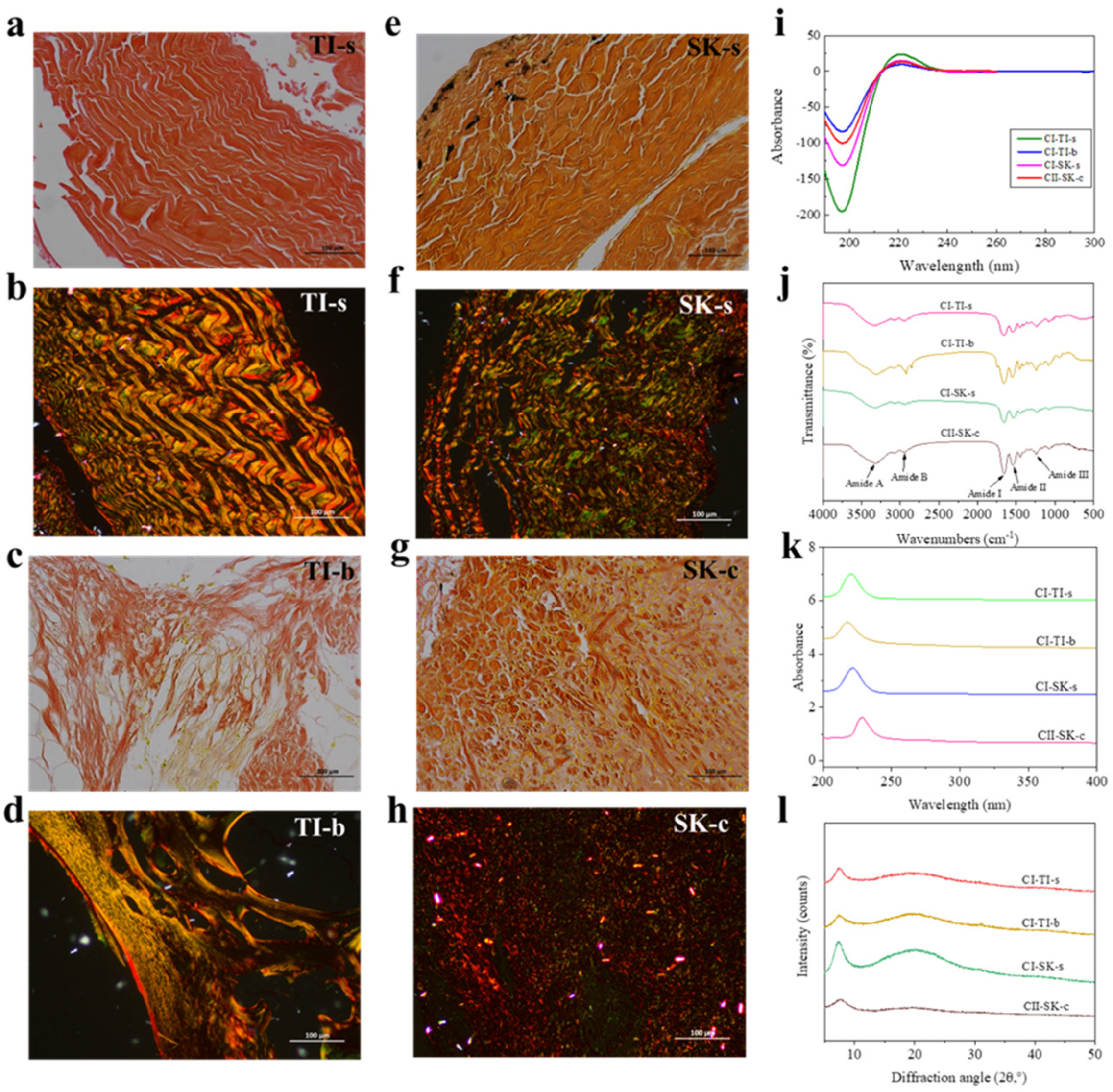
2.1. Distribution of Collagen Fibers in TI-s, TI-b, SK-s, and SK-c
2.2. Structural Characteristics of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
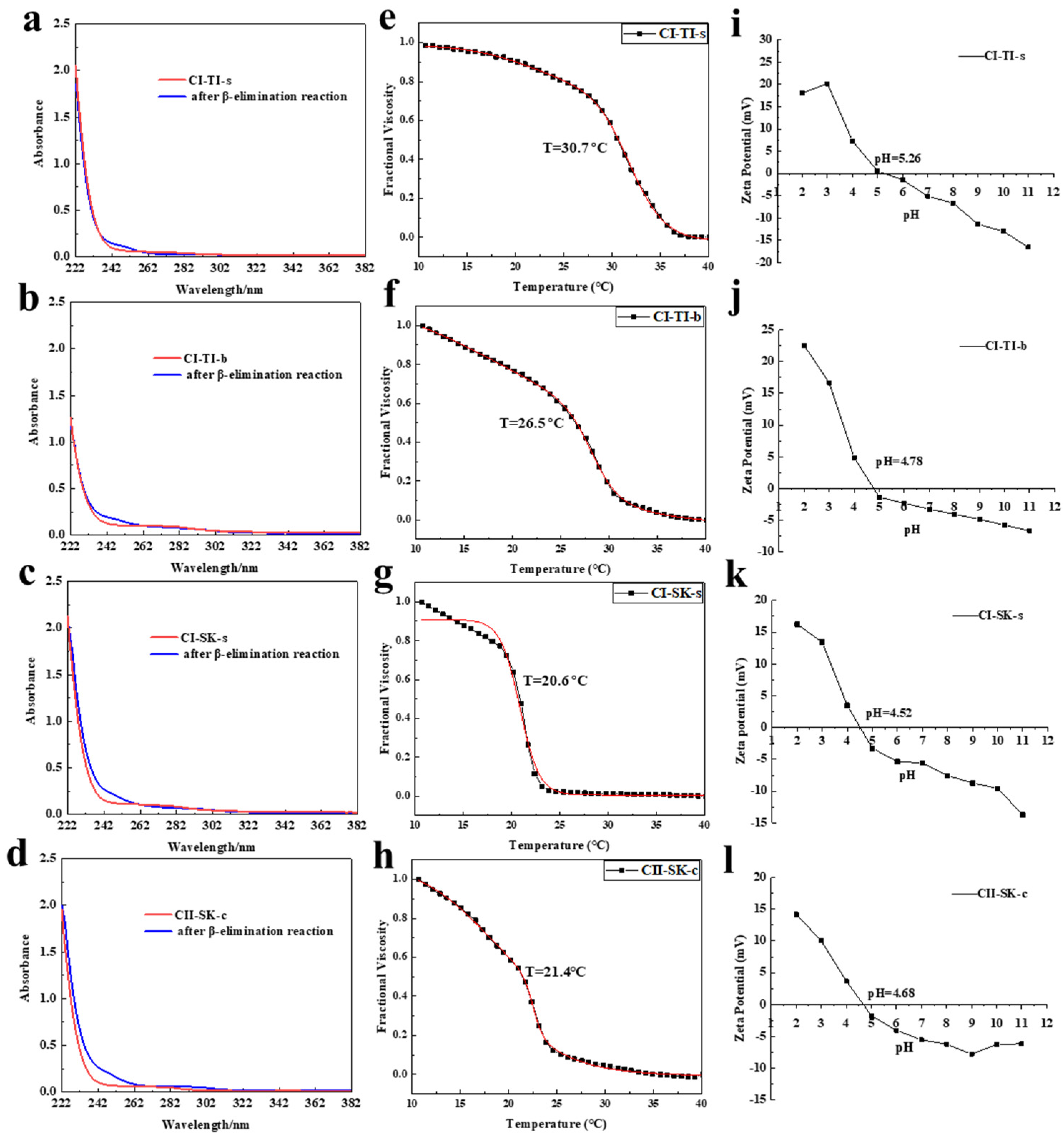
2.2.1. Analysis of the UV Spectra of CI-TI-s, CI-TI-b, CI-SK-s, and CI-SK-c
2.2.2. FTIR Analysis of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
2.2.3. CD Spectra Analysis of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
2.2.4. X-ray Diffraction of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
2.3. The O-Glycopeptide Bond of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
2.4. Td and Zeta Potential of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
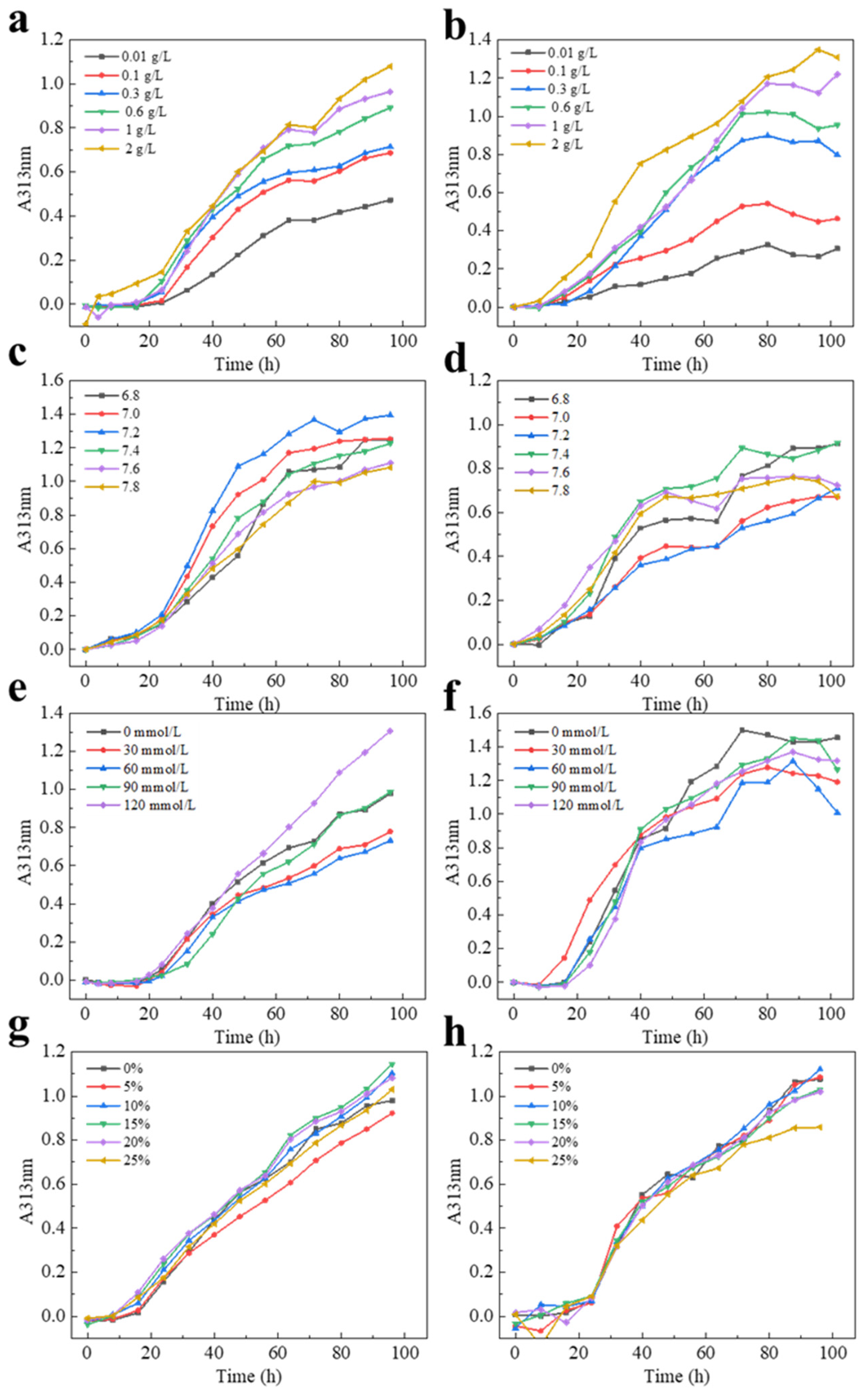
2.5. The Self-Assembly Properties of CI-TI-s, CI-TI-b, CI-SK-s, and CII-SK-c
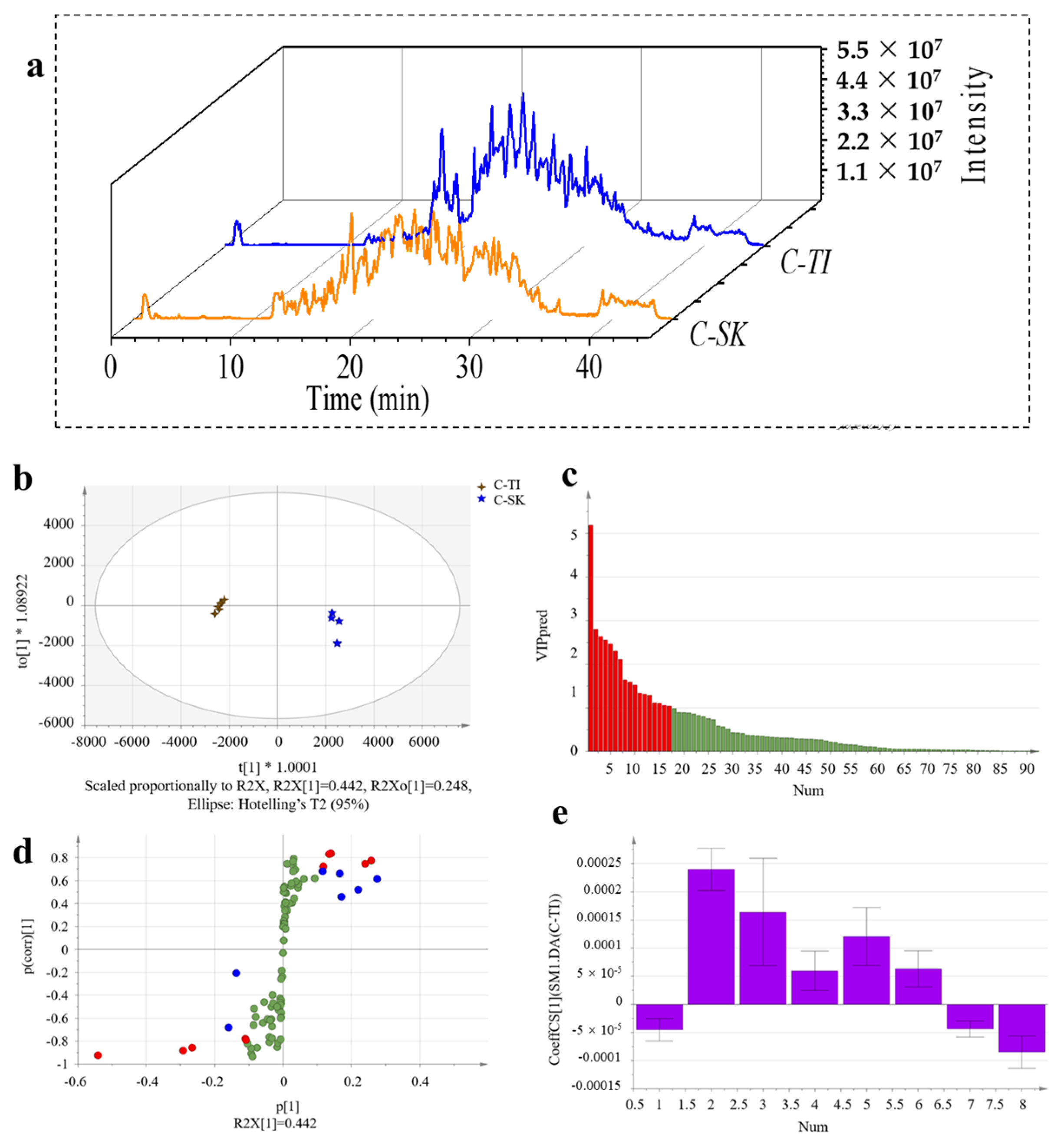
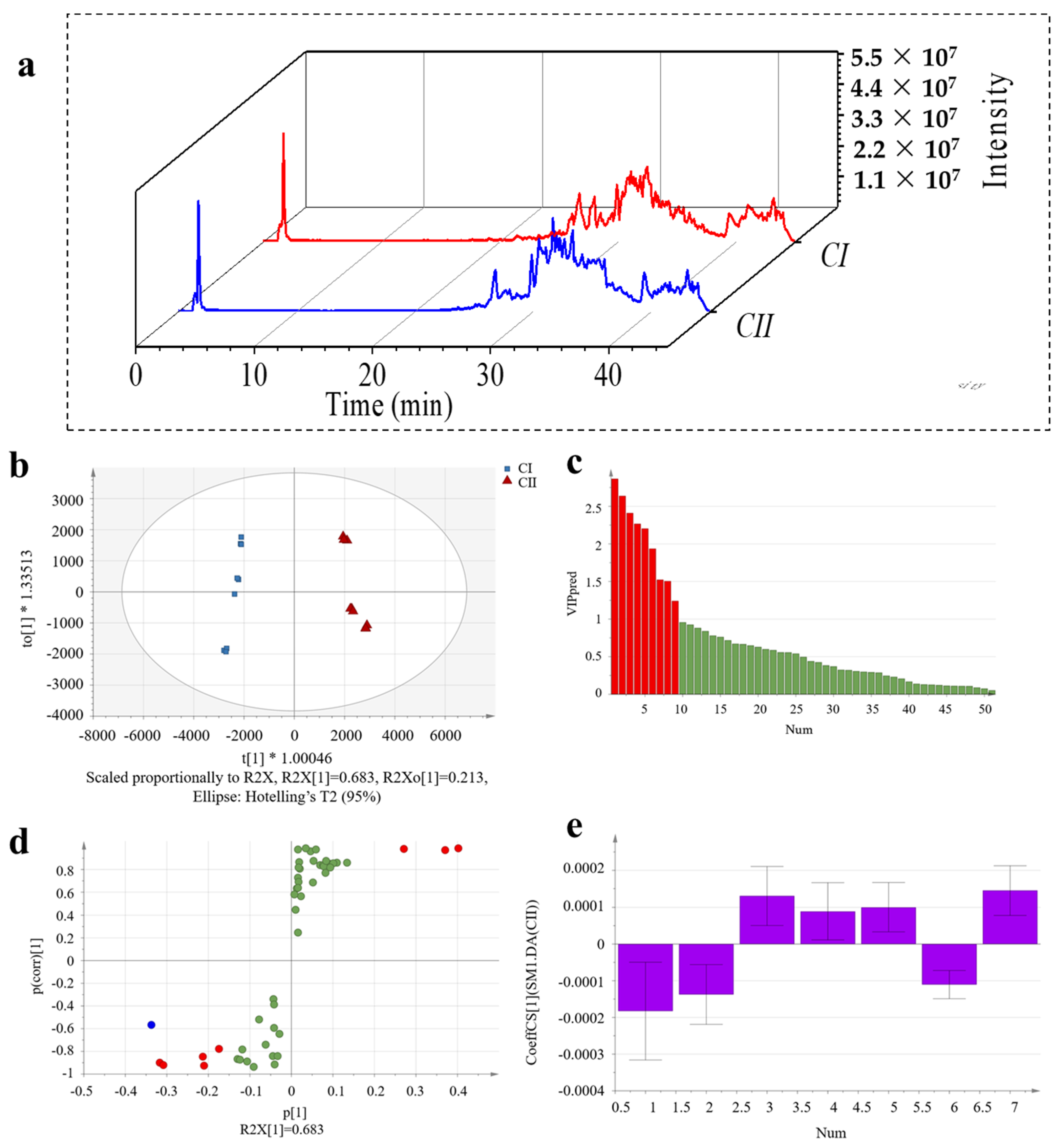
2.6. Discovery of Unique Peptides for C-TI and C-SK
2.7. Discovery of Unique Peptides for Type I and Type II Collagen
3. Materials and Methods
3.1. Materials
3.2. Van Gieson and Picric Acid-Sirius Red Staining
3.3. Preparation of Collagen
3.4. Fourier Transform Infrared (FTIR) Spectroscopy Analysis
3.5. Determination of Ultraviolet-Visible Absorption Spectra
3.6. Circular Dichroism (CD) Spectra Analysis
3.7. Determination of X-ray Diffraction (XRD)
3.8. Thermal Denaturation Temperature (Td) Analysis
3.9. Zeta Potential Measurement
3.10. LC/MS Analysis of Collagen of Different Types and Sources
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Reátegui-Pinedo, N.; Salirrosas, D.; Sánchez-Tuesta, L.; Quiñones, C.; Jáuregui-Rosas, S.R.; Barraza, G.; Cabrera, A.; Ayala-Jara, C.; Martinez, R.M.; Baby, A.R.; et al. Characterization of Collagen from Three Genetic Lines (Gray, Red and F1) of Oreochromis niloticus (Tilapia) Skin in Young and Old Adults. Molecules 2022, 27, 1123. [Google Scholar] [CrossRef] [PubMed]
- Menezes, M.d.L.L.R.; Ribeiro, H.L.; Abreu, F.d.O.M.d.S.; Feitosa, J.P.d.A.; Filho, M.d.S.M.d.S. Optimization of the Collagen Extraction from Nile Tilapia Skin (Oreochromis niloticus) and its Hydrogel with Hyaluronic Acid. Colloids Surf. B Biointerfaces 2020, 189, 110852. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liang, Y.; Cui, Y.; Li, F.; Sun, Y.; Yang, J.; Song, H.; Bao, Z.; Nian, R. Development of Tilapia Collagen and Chitosan Composite Hydrogels for Nanobody Delivery. Colloids Surf. B Biointerfaces 2020, 195, 111261. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, J.; Wang, Y.; Sun, X.; Li, B.; Poungchawanwong, S.; Hou, H. Structural Feature and Self-assembly Properties of Type II Collagens from the Cartilages of Skate and Sturgeon. Food Chem. 2020, 331, 127340. [Google Scholar] [CrossRef] [PubMed]
- Ge, B.; Wang, H.; Li, J.; Liu, H.; Yin, Y.; Zhang, N.; Qin, S. Comprehensive Assessment of Nile Tilapia Skin (Oreochromis niloticus) Collagen Hydrogels for Wound Dressings. Marine Drugs 2020, 18, 178. [Google Scholar] [CrossRef] [PubMed]
- Naomi, R.; Ridzuan, P.M.; Bahari, H. Current Insights into Collagen Type I. Polymers 2021, 13, 2642. [Google Scholar] [CrossRef]
- Tamer, T.M.; Kenawy, E.R.; Agwa, M.M.; Sabra, S.A.; El-meligy, M.A.; Mohy-Eldin, M.S. Wound Dressing Membranes based on Immobilized Anisaldehyde onto (Chitosan-GA-Gelatin) Copolymer: In-vitro and in-vivo evaluations. Int. J. Biol. Macromol. 2022, 211, 94–106. [Google Scholar] [CrossRef]
- Guo, K.; Wang, H.; Li, S.; Zhang, H.; Li, S.; Zhu, H.; Yang, Z.; Zhang, L.; Chang, P.; Zheng, X. Collagen-Based Thiol-Norbornene Photoclick Bio-Ink with Excellent Bioactivity and Printability. ACS Appl. Mater. Interfaces 2021, 13, 7037–7050. [Google Scholar] [CrossRef]
- Furtado, M.; Chen, L.; Chen, Z.; Chen, A.; Cui, W. Development of Fish Collagen in Tissue Regeneration and Drug Delivery. Eng. Regen. 2022, 3, 217–231. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Liu, A.; Wang, W. Improved Thermal-stability and Mechanical Properties of Type I Collagen by Crosslinking with Casein, Keratin and Soy Protein Isolate Using Transglutaminase. Int. J. Biol. Macromol. 2017, 98, 292–301. [Google Scholar] [CrossRef]
- Bakilan, F.; Armagan, O.; Ozgen, M.; Tascioglu, F.; Bolluk, O.; Alatas, O. Effects of Native Type II Collagen Treatment on Knee Osteoarthritis: A Randomized Controlled Trial. Eurasian J. Med. 2016, 48, 95–101. [Google Scholar] [CrossRef]
- Al-Shaer, A.; Lyons, A.; Ishikawa, Y.; Hudson, B.G.; Boudko, S.P.; Forde, N.R. Sequence-dependent Mechanics of Collagen Reflect its Structural and Functional Organization. Biophys. J. 2021, 120, 4013–4028. [Google Scholar] [CrossRef]
- Zhang, X.; Ookawa, M.; Tan, Y.; Ura, K.; Adachi, S.; Takagi, Y. Biochemical Characterisation and Assessment of Fibril-forming Ability of Collagens Extracted from Bester Sturgeon Huso huso × Acipenser ruthenus. Food Chem. 2014, 160, 305–312. [Google Scholar] [CrossRef]
- Romijn, E.I.; Finnøy, A.; Lilledahl, M.B. Analyzing the Feasibility of Discriminating between Collagen Types I and II using Polarization-resolved Second Harmonic Generation. J. Biophotonics 2019, 12, e201800090. [Google Scholar] [CrossRef]
- Padhi, S.; Chourasia, R.; Kumari, M.; Singh, S.P.; Rai, A.K. Production and Characterization of Bioactive Peptides from Rice Beans using Bacillus Subtilis. Bioresour. Technol. 2022, 351, 126932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sun, A.; Li, W.; Liu, T.; Su, Z. Mass Spectrometric Analysis of Enzymatic Digestion of Denatured Collagen for Identification of Collagen Type. J. Chromatogr. A 2006, 1114, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, H.; Hu, Z.; Chin, Y.; Zhang, X.; Chen, J.; Liu, D.; Hu, Y. Comparative Proteomics Analysis of Three Commercial Tuna Species through SWATH-MS based Mass Spectrometry and Chemometrics. Food Control 2022, 141, 109162. [Google Scholar] [CrossRef]
- Stephenson, B. A Modified Picro-Sirius Red (PSR) Staining Procedure with Polarization Microscopy for Identifying Collagen in Archaeological Residues. J. Archaeol. Sci. 2015, 61, 235–243. [Google Scholar] [CrossRef]
- Zhang, J.; Jeevithan, E.; Bao, B.; Wang, S.; Gao, K.; Zhang, C.; Wu, W. Structural Characterization, in-vivo Acute Systemic Toxicity Assessment and in-vitro Intestinal Absorption Properties of Tilapia (Oreochromis niloticus) Skin Acid and Pepsin Solublilized Type I Collagen. Process Biochem. 2016, 51, 2017–2025. [Google Scholar] [CrossRef]
- Hwang, J.-H.; Mizuta, S.; Yokoyama, Y.; Yoshinaka, R. Purification and Characterization of Molecular Species of Collagen in the Skin of Skate (Raja kenojei). Food Chem. 2007, 100, 921–925. [Google Scholar] [CrossRef]
- Hernández-Ruiz, K.L.; López-Cervantes, J.; Sánchez-Machado, D.I.; Campas-Baypoli, O.N.; Quintero-Guerrero, A.A.; de Lourdes Grijalva-Delgado, M.; Chávez-Almanza, A.F. Collagen Peptide Fractions from Tilapia (Oreochromis aureus Steindachner, 1864) Scales: Chemical Characterization and Biological Activity. Food Biosci. 2023, 53, 102658. [Google Scholar] [CrossRef]
- Abe, Y.; Krimm, S. Normal Vibrations of Crystalline Polyglycine I. Biopolymers 1972, 11, 1817–1839. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Hou, H.; Li, B.; Zhang, Y. Characterization of Acid- and Pepsin-soluble Collagen Extracted from the Skin of Nile Tilapia (Oreochromis niloticus). Int. J. Biol. Macromol. 2017, 99, 8–14. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Duan, S.; Jiang, Z.; Chen, S.; Sun, W.; Liu, X.; Sun, Z.; Li, Y.; Yan, M. Role of Chlorogenic Acid and Procyanidin in the Modification of Self-assembled Fibrillar Gel Prepared from Tilapia Collagen. Polym. Degrad. Stab. 2022, 206, 110177. [Google Scholar] [CrossRef]
- Zhu, S.; Gu, Z.; Xiong, S.; An, Y.; Liu, Y.; Yin, T.; You, J.; Hu, Y. Fabrication of a Novel Bio-inspired Collagen–polydopamine Hydrogel and Insights into the Formation Mechanism for Biomedical Applications. RSC Adv. 2016, 6, 66180–66190. [Google Scholar] [CrossRef]
- Gao, X.; He, J.; Chen, J.; Zheng, Y.; Li, Y.; Ye, T. Double-spotted Pufferfish (Takifugu bimaculatus) Skin Collagen: Preparation, Structure, Cytocompatibility, Rheological, and Functional Properties. Arab. J. Chem. 2023, 16, 104402. [Google Scholar] [CrossRef]
- Li, T.; Wu, C.-e.; Meng, X.; Fan, G.; Cao, Y.; Ying, R.; Tang, Y. Structural Characterization and Antioxidant Activity of a Glycoprotein Isolated from Camellia Oleifera Abel Seeds against D-galactose-induced Oxidative Stress in Mice. J. Funct. Foods 2020, 64, 103594. [Google Scholar] [CrossRef]
- Wang, L.; An, X.; Yang, F.; Xin, Z.; Zhao, L.; Hu, Q. Isolation and Characterisation of Collagens from the Skin, Scale and Bone of Deep-sea Redfish (Sebastes mentella). Food Chem. 2008, 108, 616–623. [Google Scholar] [CrossRef]
- Ikoma, T.; Kobayashi, H.; Tanaka, J.; Walsh, D.; Mann, S. Physical Properties of Type I Collagen Extracted from Fish Scales of Pagrus Major and Oreochromis Niloticas. Int. J. Biol. Macromol. 2003, 32, 199–204. [Google Scholar] [CrossRef]
- Pati, F.; Adhikari, B.; Dhara, S. Isolation and Characterization of Fish Scale Collagen of Higher Thermal Stability. Bioresour. Technol. 2010, 101, 3737–3742. [Google Scholar] [CrossRef]
- Yano, S.; Yamaguchi, K.; Shibata, M.; Ifuku, S.; Teramoto, N. Photocrosslinked Fish Collagen Peptide/Chitin Nanofiber Composite Hydrogels from Marine Resources: Preparation, Mechanical Properties, and an In Vitro Study. Polymers 2023, 15, 682. [Google Scholar] [CrossRef]
- Klabukov, I.; Tenchurin, T.; Shepelev, A.; Baranovskii, D.; Mamagulashvili, V.; Dyuzheva, T.; Krasilnikova, O.; Balyasin, M.; Lyundup, A.; Krasheninnikov, M.; et al. Biomechanical Behaviors and Degradation Properties of Multilayered Polymer Scaffolds: The Phase Space Method for Bile Duct Design and Bioengineering. Biomedicines 2023, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Haq, M.; Chun, B.-S. Characterization of Marine Derived Collagen Extracted from the By-products of Bigeye Tuna (Thunnus obesus). Int. J. Biol. Macromol. 2019, 135, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Asadi, A.; Monroe, M.R.; Douglas, E.P. pH Effects on Collagen Fibrillogenesis in Vitro: Electrostatic Interactions and Phosphate Binding. Mater. Sci. Eng. C 2009, 29, 1643–1649. [Google Scholar] [CrossRef]
- Yan, M.; Li, B.; Zhao, X.; Qin, S. Effect of Concentration, pH and Ionic Strength on the Kinetic Self-assembly of Acid-soluble Collagen from Walleye Pollock (Theragra chalcogramma) skin. Food Hydrocoll. 2012, 29, 199–204. [Google Scholar] [CrossRef]
- Yi, L.; Dong, N.; Yun, Y.; Deng, B.; Ren, D.; Liu, S.; Liang, Y. Chemometric Methods in Data Processing of Mass Spectrometry-based Metabolomics: A Review. Anal. Chim. Acta 2016, 914, 17–34. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Muyonga, J.H.; Cole, C.G.B.; Duodu, K.G. Characterisation of Acid Soluble Collagen from Skins of Young and Adult Nile Perch (Lates niloticus). Food Chem. 2004, 85, 81–89. [Google Scholar] [CrossRef]
- Buckley, M.; Fraser, S.; Herman, J.; Melton, N.D.; Mulville, J.; Pálsdóttir, A.H. Species Identification of Archaeological Marine Mammals Using Collagen Fingerprinting. J. Archaeol. Sci. 2014, 41, 631–641. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteom. 2012, 11, O111.016717. [Google Scholar] [CrossRef]
| Peptide Sequence | m/z | Protein Source | |
|---|---|---|---|
| C-TI/C-SK | data | data | |
| 1 | GPSGPQGAVGATGPK | 640.8333 | Type I procollagen alpha 1 chain OS = Okamejei kenojei GN = SkCOL1A1 PE = 2 SV = 1 |
| 2 | PAMPVPGPMGPMGPR | 746.3671 | Uncharacterized protein (Fragment) OS = Oreochromis niloticus GN = col1a1 PE = 4 SV = 1 |
| 3 | SPAMPVPGPMGPMGPR | 789.8831 | Uncharacterized protein (Fragment) OS = Oreochromis niloticus GN = col1a1 PE = 4 SV = 1 |
| 4 | GESGPSGPAGPAGPAGVR | 760.8762 | Uncharacterized protein OS = Oreochromis niloticus GN = LOC100694532 PE = 4 SV = 1 |
| 5 | SSGPPVPGPIGPMGPR | 751.8928 | Uncharacterized protein OS = Oreochromis niloticus GN = LOC100694532 PE = 4 SV = 1 |
| 6 | GLTGPIGVPGPPGAQGEK | 816.4412 | Uncharacterized protein OS = Oreochromis niloticus GN = LOC100694532 PE = 4 SV = 1 |
| 7 | GLAGPQGPR | 426.7379 | Uncharacterized protein (Fragment) OS = Anolis carolinensis GN = COL1A2 PE = 4 SV = 1 |
| 8 | GLSGDPGVQGIK | 564.3064 | Uncharacterized protein OS = Gasterosteus aculeatus PE = 4 SV = 1 |
| CI/CII | |||
| 1 | GPTGEIGATGLAGAR | 664.3519 | Collagen type I alpha 2 OS = Oreochromis niloticus GN = COL1A2 PE = 2 SV = 1 |
| 2 | GVLGLTGMR | 452.2577 | Type I procollagen alpha 1 chain OS = Okamejei kenojei GN = SkCOL1A1 PE = 2 SV = 1 |
| 3 | LGLTGMR | 374.2127 | Type I procollagen alpha 1 chain OS = Okamejei kenojei GN = SkCOL1A1 PE = 2 SV = 1 |
| 4 | GEPGAAGPAGPSGPMGPR | 781.8726 | Type I procollagen alpha 1 chain OS = Okamejei kenojei GN = SkCOL1A1 PE = 2 SV = 1 |
| 5 | SSGPPVPGPIGPMGPR | 751.8928 | Uncharacterized protein OS = Oreochromis niloticus GN = LOC100694532 PE = 4 SV = 1 |
| 6 | GLSGDPGVQGIK | 564.3064 | Uncharacterized protein OS = Gasterosteus aculeatus PE = 4 SV = 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, J.; Zhang, Q.; Fan, Y.; Zhang, H.; Ahmad, K.; Hou, H. Distribution, Typical Structure and Self-Assembly Properties of Collagen from Fish Skin and Bone. Molecules 2023, 28, 6529. https://doi.org/10.3390/molecules28186529
Zhang X, Wang J, Zhang Q, Fan Y, Zhang H, Ahmad K, Hou H. Distribution, Typical Structure and Self-Assembly Properties of Collagen from Fish Skin and Bone. Molecules. 2023; 28(18):6529. https://doi.org/10.3390/molecules28186529
Chicago/Turabian StyleZhang, Xuening, Jie Wang, Qian Zhang, Yan Fan, Hongwei Zhang, Khurshid Ahmad, and Hu Hou. 2023. "Distribution, Typical Structure and Self-Assembly Properties of Collagen from Fish Skin and Bone" Molecules 28, no. 18: 6529. https://doi.org/10.3390/molecules28186529
APA StyleZhang, X., Wang, J., Zhang, Q., Fan, Y., Zhang, H., Ahmad, K., & Hou, H. (2023). Distribution, Typical Structure and Self-Assembly Properties of Collagen from Fish Skin and Bone. Molecules, 28(18), 6529. https://doi.org/10.3390/molecules28186529







