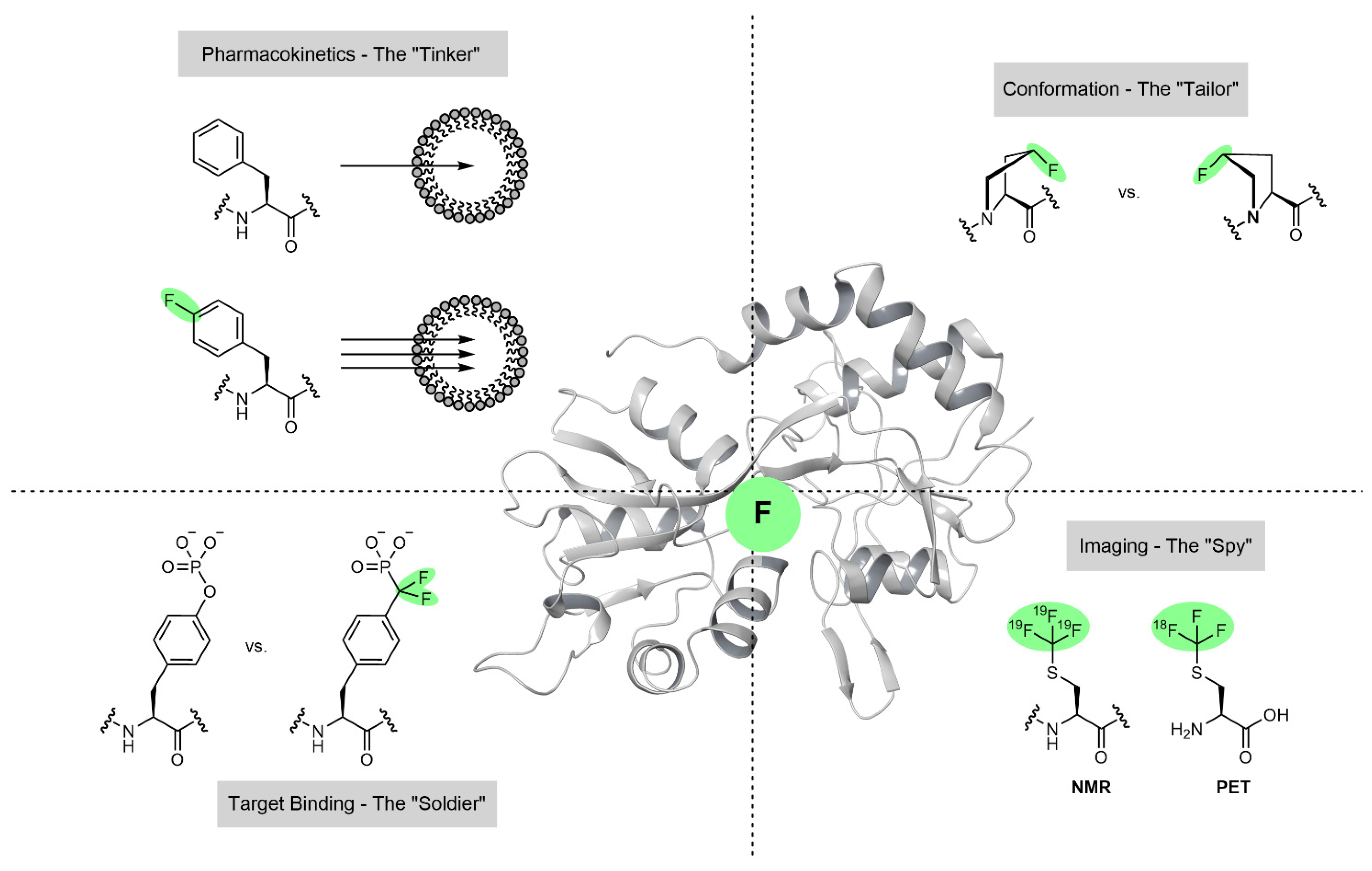
Tinker, Tailor, Soldier, Spy: The Diverse Roles That Fluorine Can Play within Amino Acid Side Chains
Abstract
1. Introduction
2. Synthetic Aspects
2.1. Strategies for the Synthesis of Side Chain-Fluorinated Amino Acids
2.2. Elaboration of Side Chain-Fluorinated Amino Acids into Peptides and Proteins
3. Fluorine, the “Spy”: Transmitting Intelligence on the Properties of Amino Acids, Peptides, and Proteins
3.1. 19F-Containing Amino Acids as NMR Tags
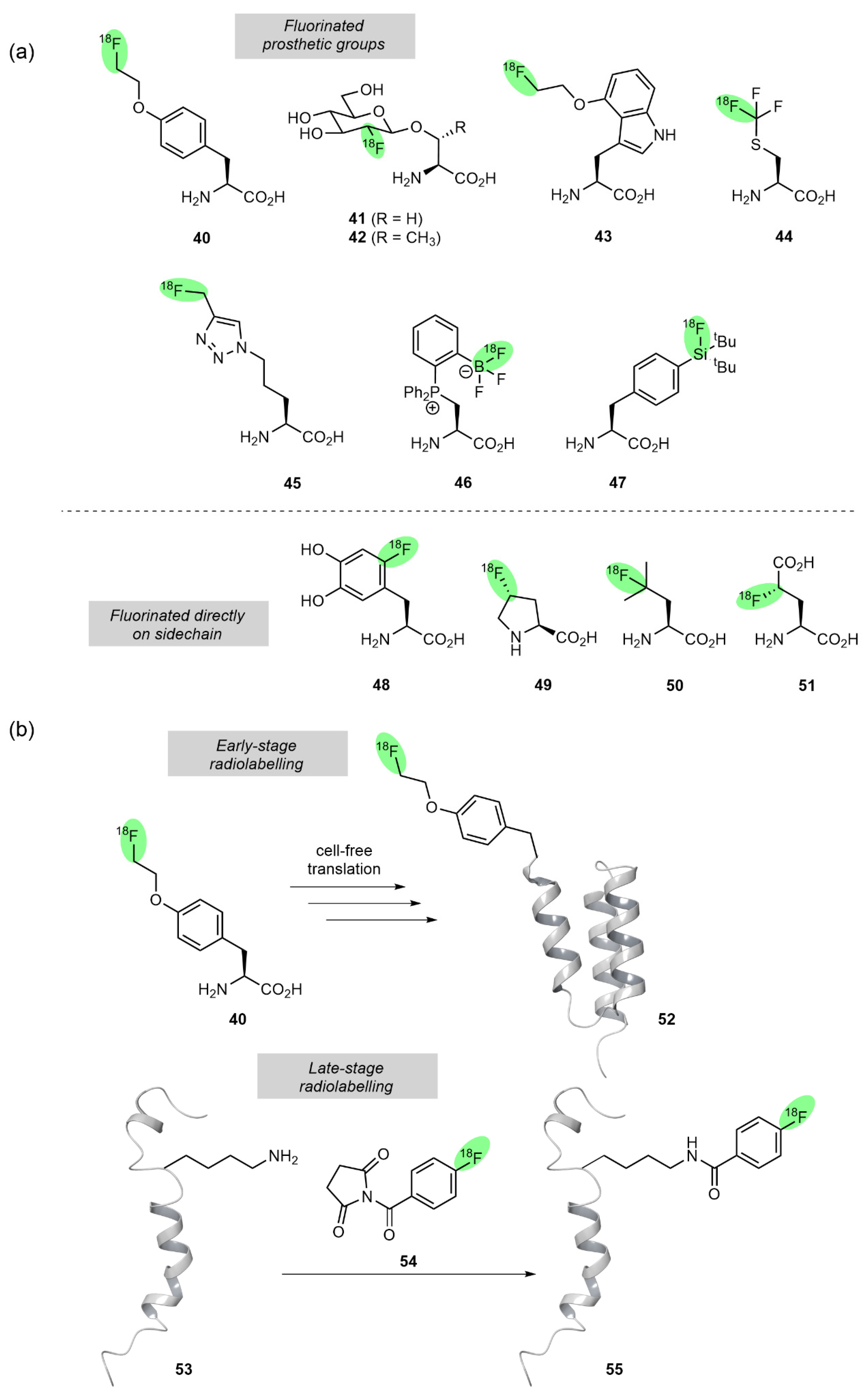
3.2. 18F-Labelled Amino Acids and Peptides as PET Tracers
4. Fluorine, the “Tinker”: Improving the Pharmacokinetic Properties of Amino Acids, Peptides, and Proteins
4.1. Hydrophobicity and Permeability
4.2. Stability towards Proteolysis
4.3. Resistance to P450 Oxidation
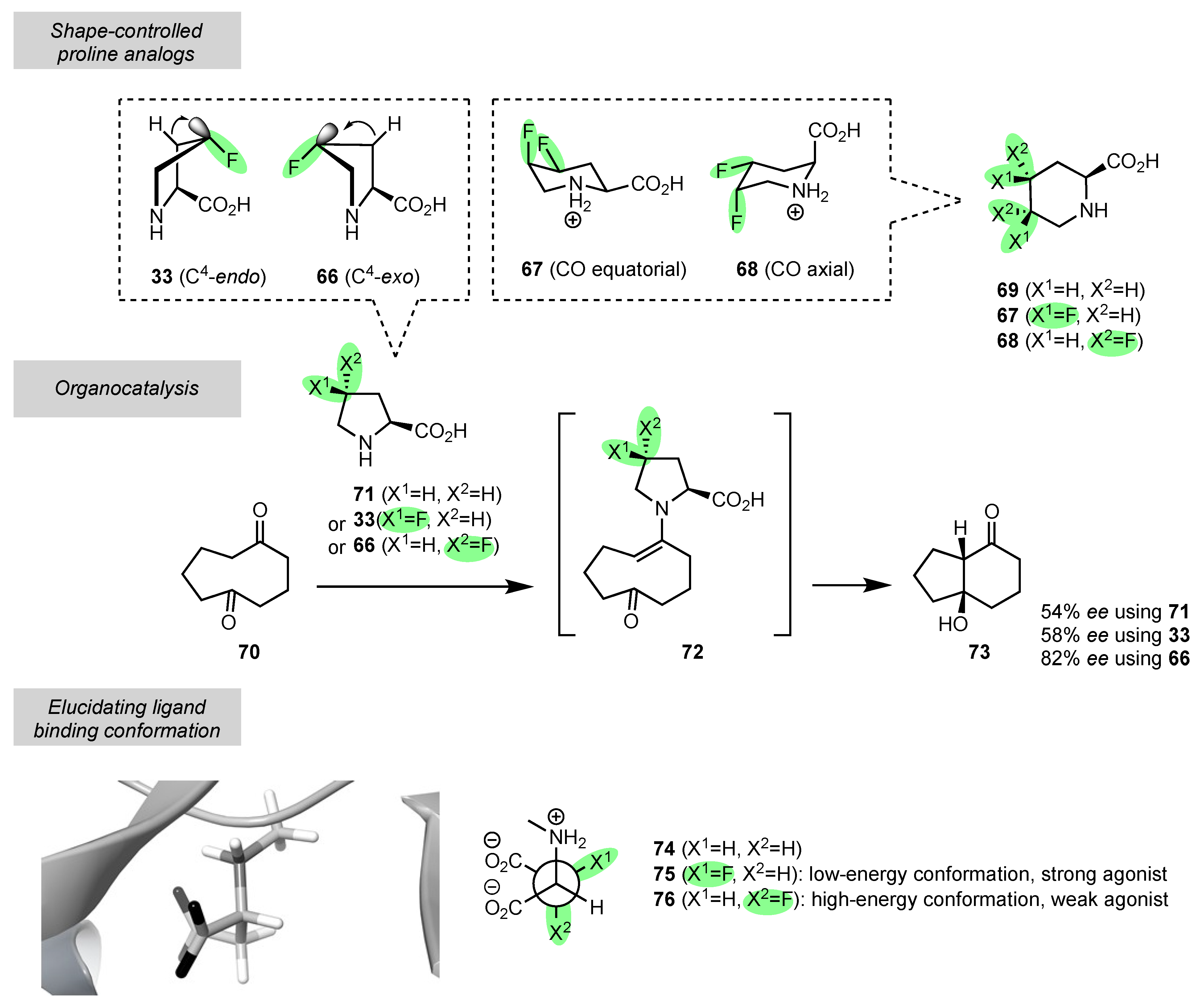
5. Fluorine, the “Tailor”: Folding Amino Acids, Peptides, and Proteins into Precise 3D Shapes
5.1. Conformational Control at the Individual Amino Acid Level
5.2. Peptide Secondary Structure
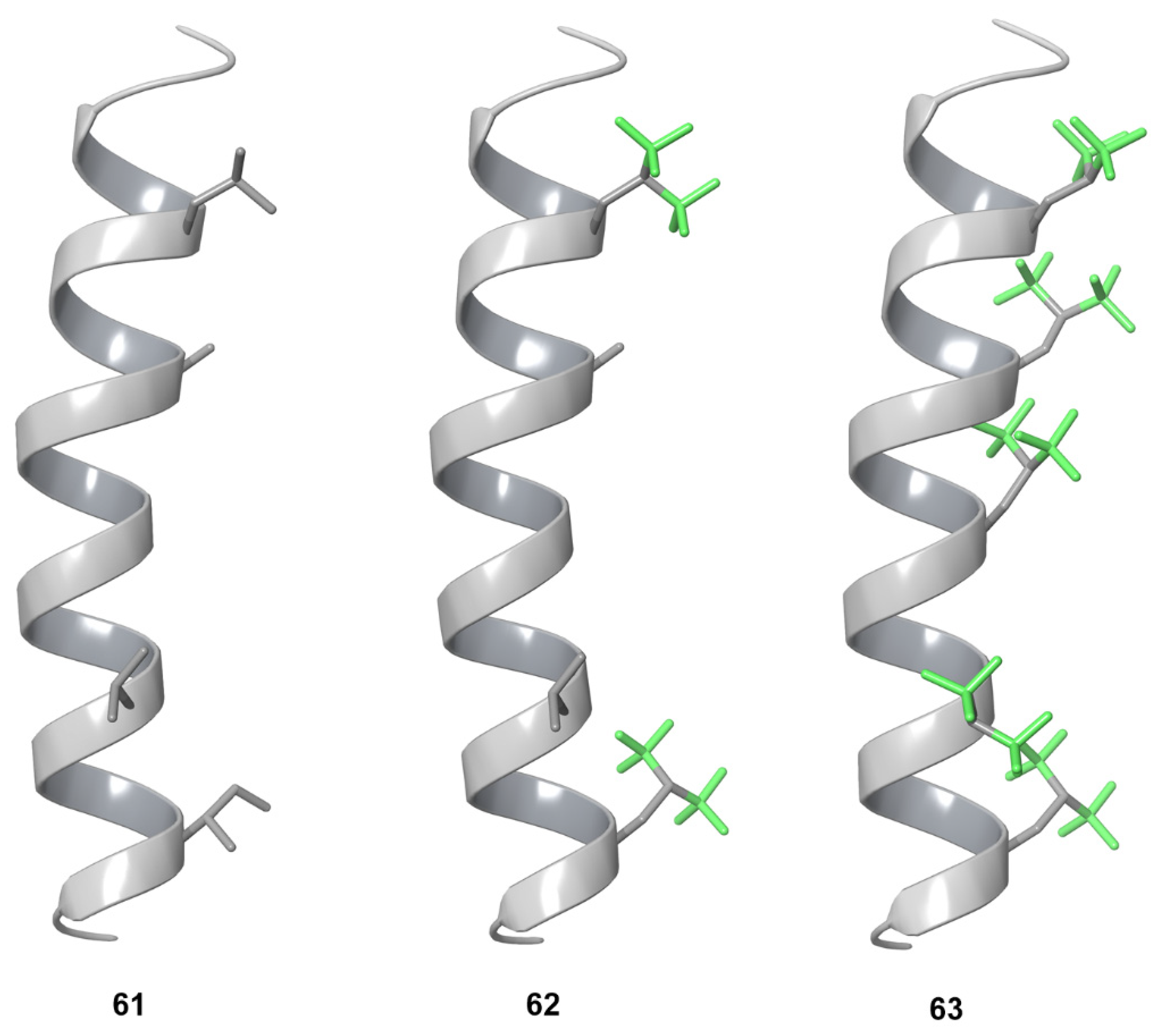
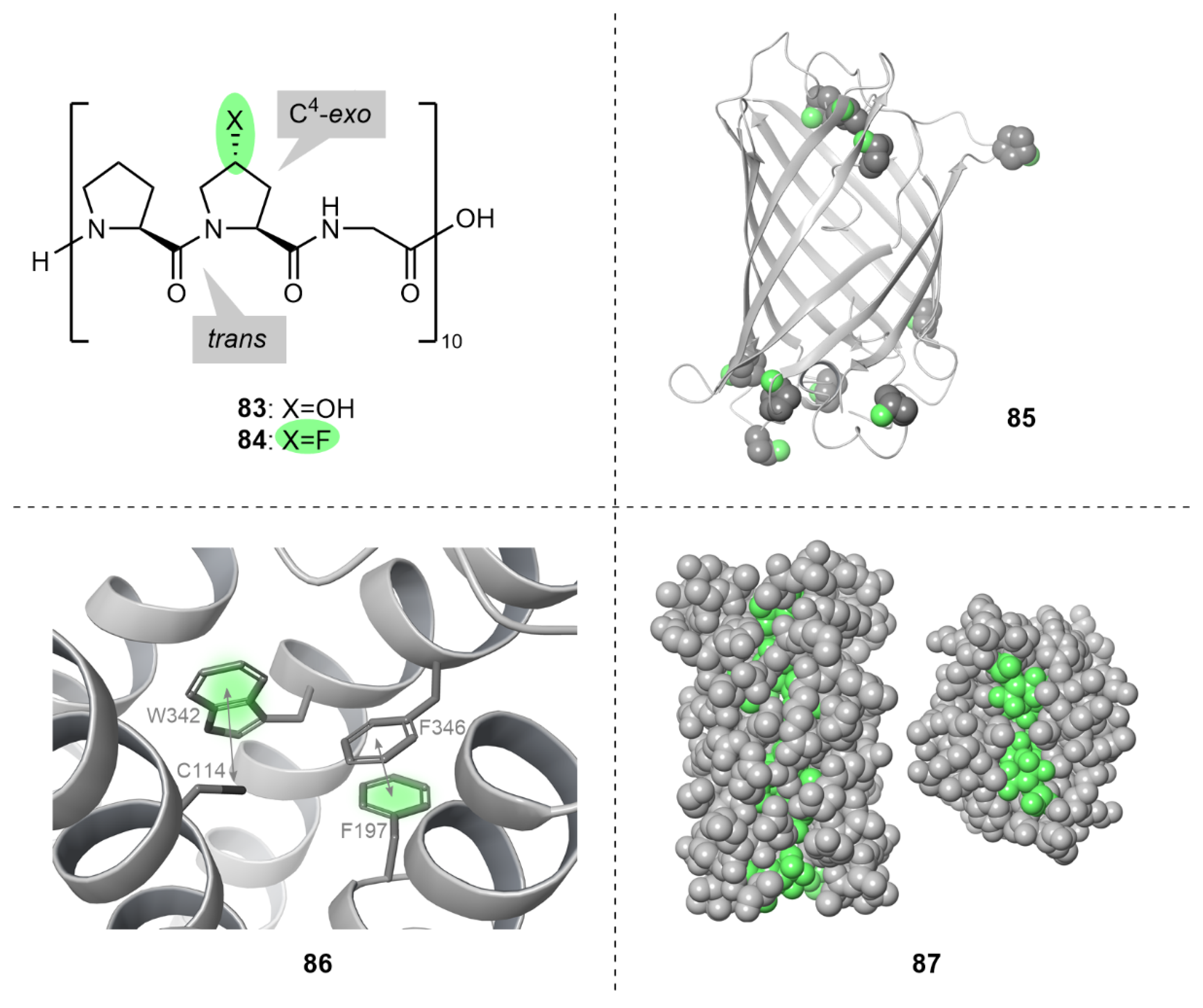
5.3. Protein Tertiary Structure
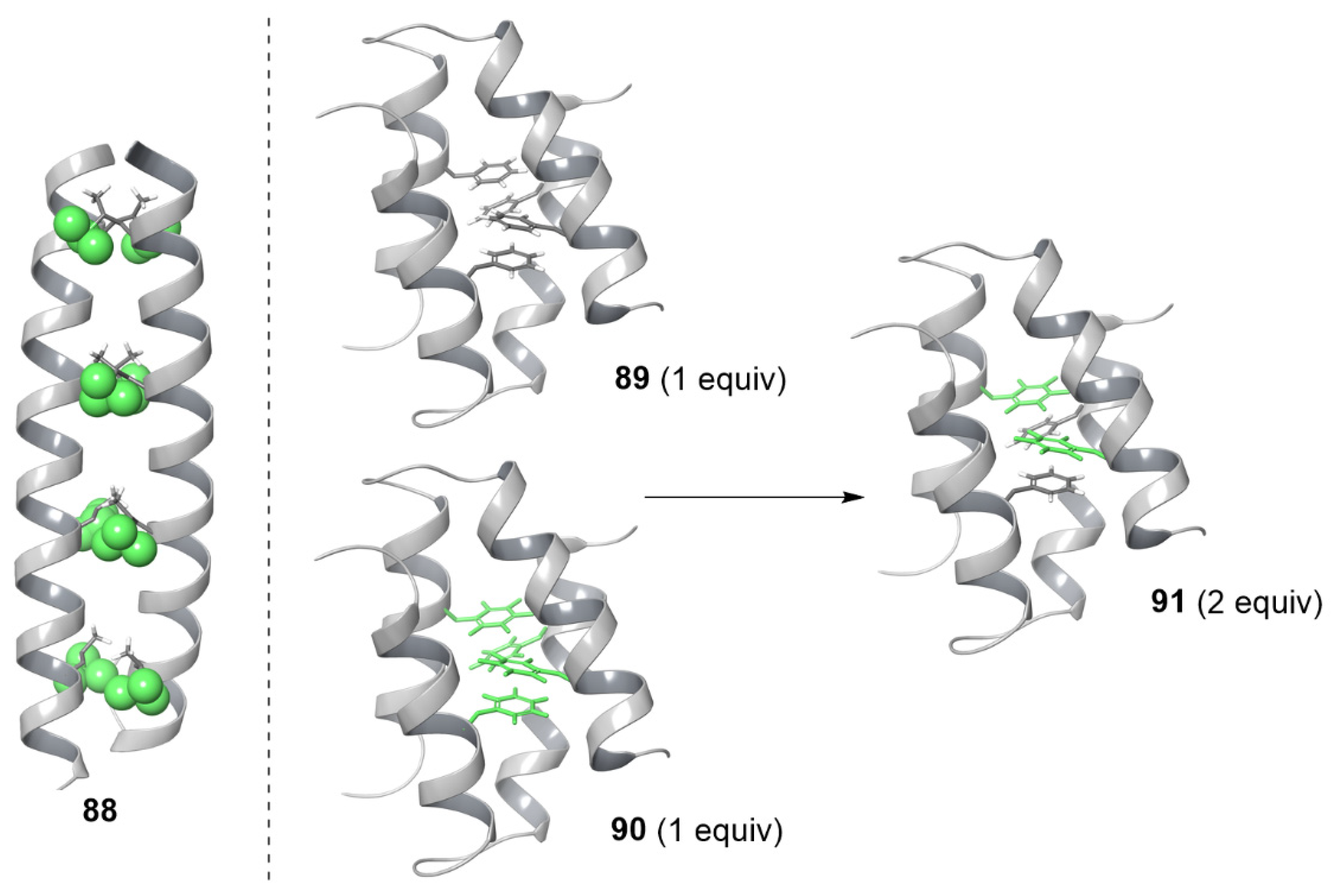
5.4. Protein Quaternary Structure
6. Fluorine, the “Soldier”: Guiding Amino Acids, Peptides, and Proteins to Hit Their Biological Targets
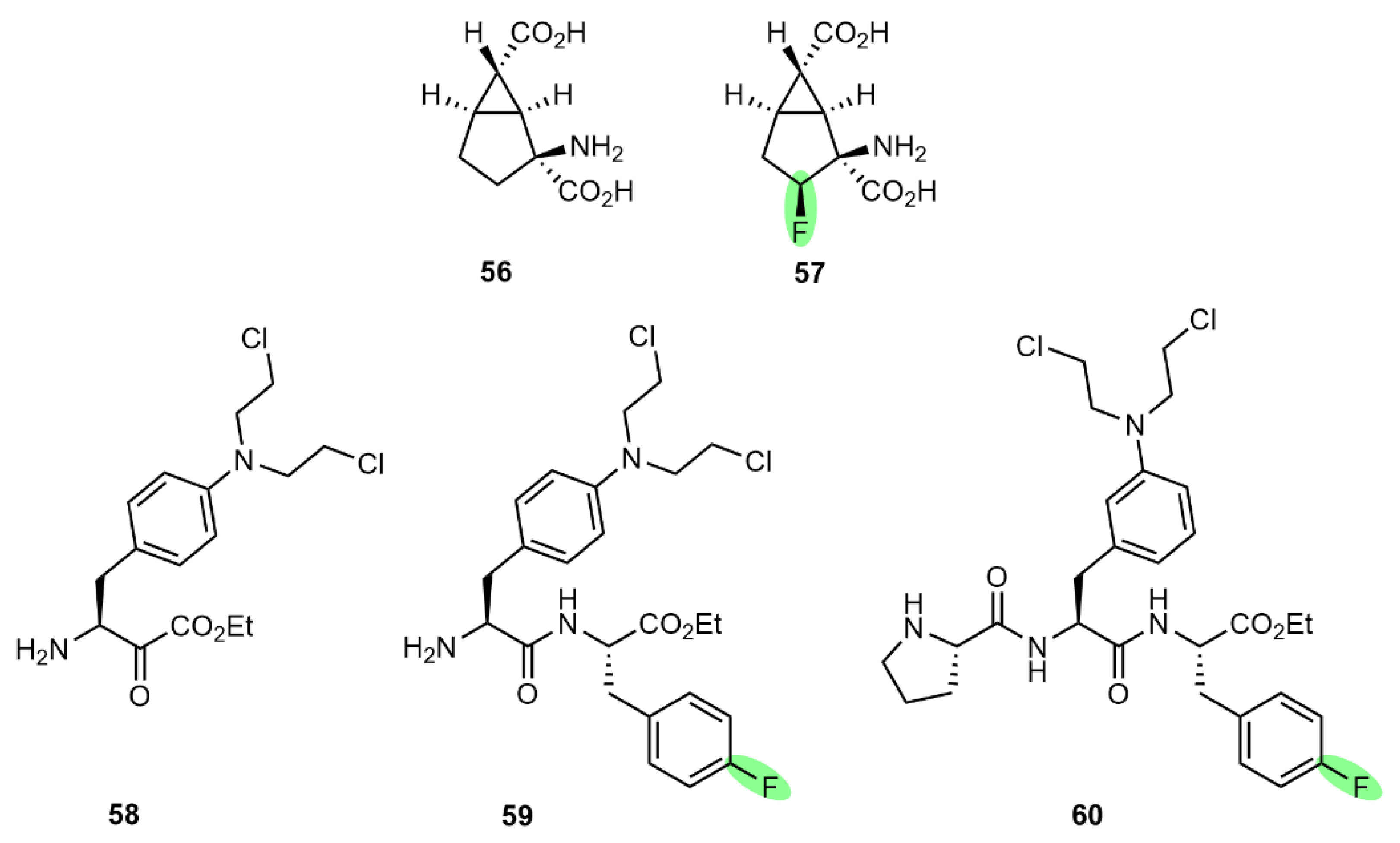
6.1. Mechanism-Based Enzyme Inhibitors
6.2. Enhancing Intermolecular Forces
6.3. Conformational Pre-Organization
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shah, P.; Westwell, A.D. The role of fluorine in medicinal chemistry. J. Enzyme Inhib. Med. Chem. 2007, 22, 527–540. [Google Scholar] [CrossRef]
- Kirk, K.L. Fluorination in medicinal chemistry: Methods, strategies, and recent developments. Org. Process Res. Dev. 2008, 12, 305–321. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, J.; Gu, Z.; Wang, S.; Zhu, W.; Acena, J.L.; Soloshonok, V.A.; Izawa, K.; Liu, H. Next generation of fluorine-containing pharmaceuticals, compounds currently in phase II–III clinical trials of major pharmaceutical companies: New structural trends and therapeutic areas. Chem. Rev. 2016, 116, 422–518. [Google Scholar] [CrossRef]
- Meanwell, N.A. Fluorine and fluorinated motifs in the design and application of bioisosteres for drug design. J. Med. Chem. 2018, 61, 5822–5880. [Google Scholar] [CrossRef]
- Inoue, M.; Sumii, Y.; Shibata, N. Contribution of Organofluorine Compounds to Pharmaceuticals. ACS Omega 2020, 5, 10633–10640. [Google Scholar] [CrossRef]
- Haufe, G.; Leroux, F. Fluorine in Life Sciences: Pharmacueticals, Medicinal Diagnostics, and Agrochemicals; Academic Press: London, UK, 2018. [Google Scholar]
- Johnson, B.M.; Shu, Y.Z.; Zhuo, X.; Meanwell, N.A. Metabolic and Pharmaceutical Aspects of Fluorinated Compounds. J. Med. Chem. 2020, 63, 6315–6386. [Google Scholar] [CrossRef]
- Gillis, E.P.; Eastman, K.J.; Hill, M.D.; Donnelly, D.J.; Meanwell, N.A. Applications of Fluorine in Medicinal Chemistry. J. Med. Chem. 2015, 58, 8315–8359. [Google Scholar] [CrossRef]
- Bohm, H.J.; Banner, D.; Bendels, S.; Kansy, M.; Kuhn, B.; Muller, K.; Obst-Sander, U.; Stahl, M. Fluorine in medicinal chemistry. ChemBioChem 2004, 5, 637–643. [Google Scholar] [CrossRef]
- Ogawa, Y.; Tokunaga, E.; Kobayashi, O.; Hirai, K.; Shibata, N. Current Contributions of Organofluorine Compounds to the Agrochemical Industry. iScience 2020, 23, 101467. [Google Scholar] [CrossRef]
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 2008, 7, 21–39. [Google Scholar] [CrossRef]
- Doak, B.C.; Zheng, J.; Dobritzsch, D.; Kihlberg, J. How Beyond Rule of 5 Drugs and Clinical Candidates Bind to Their Targets. J. Med. Chem. 2016, 59, 2312–2327. [Google Scholar] [CrossRef]
- Lagasse, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- DeGoey, D.A.; Chen, H.J.; Cox, P.B.; Wendt, M.D. Beyond the Rule of 5: Lessons learned from AbbVie’s drugs and compound collection. J. Med. Chem. 2018, 61, 2636–2651. [Google Scholar] [CrossRef] [PubMed]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The current state of peptide drug discovery: Back to the future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed]
- Craik, D.J.; Kan, M.W. How can we improve peptide drug discovery? Learning from the past. Expert Opin. Drug Discov. 2021, 16, 1399–1402. [Google Scholar] [CrossRef] [PubMed]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Mansour, F.; Hunter, L. Synthesis and applications of backbone-fluorinated amino acids. In Fluorine in Life Sciences: Pharmaceuticals, Medicinal Diagnostics, and Agrochemicals; Academic Press: Cambridge, MA, USA, 2019; pp. 325–347. [Google Scholar] [CrossRef]
- Yoder, N.C.; Kumar, K. Fluorinated amino acids in protein design and engineering. Chem. Soc. Rev. 2002, 31, 335–341. [Google Scholar] [CrossRef]
- Jäckel, C.; Koksch, B. Fluorine in Peptide Design and Protein Engineering. Eur. J. Org. Chem. 2005, 21, 4483–4503. [Google Scholar] [CrossRef]
- Salwiczek, M.; Nyakatura, E.K.; Gerling, U.I.; Ye, S.; Koksch, B. Fluorinated amino acids: Compatibility with native protein structures and effects on protein-protein interactions. Chem. Soc. Rev. 2012, 41, 2135–2171. [Google Scholar] [CrossRef]
- Buer, B.C.; Marsh, E.N.G. Design, synthesis, and study of fluorinated proteins. Methods Mol. Biol. 2014, 1216, 89–116. [Google Scholar] [CrossRef] [PubMed]
- Odar, C.; Winkler, M.; Wiltschi, B. Fluoro amino acids: A rarity in nature, yet a prospect for protein engineering. Biotechnol. J. 2015, 10, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.A.; Voller, J.S.; Budisa, N.; Koksch, B. Deciphering the Fluorine Code—The Many Hats Fluorine Wears in a Protein Environment. Acc. Chem. Res. 2017, 50, 2093–2103. [Google Scholar] [CrossRef]
- Mei, H.; Han, J.; White, S.; Graham, D.J.; Izawa, K.; Sato, T.; Fustero, S.; Meanwell, N.A.; Soloshonok, V.A. Tailor-Made Amino Acids and Fluorinated Motifs as Prominent Traits in Modern Pharmaceuticals. Chemistry 2020, 26, 11349–11390. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Davis, R.; Budisa, N. Biochemistry of fluoroprolines: The prospect of making fluorine a bioelement. Beilstein J. Org. Chem. 2021, 17, 439–460. [Google Scholar] [CrossRef]
- Akcay, G.; Kumar, K. A New Paradigm for Protein Design and Biological Self-Assembly. J. Fluor. Chem. 2009, 130, 1178–1182. [Google Scholar] [CrossRef][Green Version]
- Kukhar, V.P. Fluorine-containing amino acids. J. Fluor. Chem. 1994, 69, 199–205. [Google Scholar] [CrossRef]
- Taguchi, T.; Okada, M. Fluorinated cyclopropanes. J. Fluor. Chem. 2000, 105, 279–283. [Google Scholar] [CrossRef]
- Smits, R.; Cadicamo, C.D.; Burger, K.; Koksch, B. Synthetic strategies to α-trifluoromethyl and α-difluoromethyl substituted α-amino acids. Chem. Soc. Rev. 2008, 37, 1727–1739. [Google Scholar] [CrossRef]
- Sorochinsky, A.E.; Soloshonok, V.A. Asymmetric synthesis of fluorine-containing amines, amino alcohols, α- and β-amino acids mediated by chiral sulfinyl group. J. Fluor. Chem. 2010, 131, 127–139. [Google Scholar] [CrossRef]
- Qiu, X.L.; Qing, F.L. Recent Advances in the Synthesis of Fluorinated Amino Acids. Eur. J. Org. Chem. 2011, 2011, 3261–3278. [Google Scholar] [CrossRef]
- Moschner, J.; Stulberg, V.; Fernandes, R.; Huhmann, S.; Leppkes, J.; Koksch, B. Approaches to Obtaining Fluorinated α-Amino Acids. Chem. Rev. 2019, 119, 10718–10801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Gao, Y.; Hu, X.S.; Ji, C.B.; Liu, Y.L.; Yu, J.S. Recent Advances in Catalytic Enantioselective Synthesis of Fluorinated α- and β-Amino Acids. Adv. Synth. Catal. 2020, 362, 4763–4793. [Google Scholar] [CrossRef]
- Mykhailiuk, P.K. Fluorine-containing prolines: Synthetic strategies, applications, and opportunities. J. Org. Chem. 2022, 87, 6961–7005. [Google Scholar] [CrossRef]
- Mei, H.; Han, J.; Klika, K.D.; Izawa, K.; Sato, T.; Meanwell, N.A.; Soloshonok, V.A. Applications of fluorine-containing amino acids for drug design. Eur. J. Med. Chem. 2020, 186, 111826. [Google Scholar] [CrossRef]
- Mikami, K.; Fustero, S.; Sánchez-Roselló, M.; Aceña, J.; Soloshonok, V.; Sorochinsky, A. Synthesis of Fluorinated β-Amino Acids. Synthesis 2011, 19, 3045–3079. [Google Scholar] [CrossRef]
- Soloshonok, V.A.; Kukhar, V.P.; Galushko, S.V.; Svistunova, N.Y.; Avilov, D.V.; Kuz’mina, N.A.; Raevski, N.I.; Struchkov, Y.T.; Pysarevsky, A.P.; Belokon, Y.N. General method for the synthesis of enantiomerically pure β-hydroxy-α-amino acids, containing fluorine atoms in the side chains. Case of stereochemical distinction between methyl and trifluoromethyl groups. X-ray crystal and molecular structure of the nickel(II) complex of (2S,3S)-2(trifluoromethyl)threonine. J. Chem. Soc. Perkin Trans. 1 1993, 24, 3143–3155. [Google Scholar] [CrossRef]
- Damhaut, P.; Lemaire, C.; Plenevaux, A.; Brihaye, C.; Christiaens, L.; Comar, D. No-carrier-added aymmetric synthesis of α-methyl-α-amino acids labelled with fluorine-18. Tetrahedron 1997, 53, 5785–5796. [Google Scholar] [CrossRef]
- Fustero, S.; Navarro, A.; Pina, B.; García Soler, J.; Bartolomé, A.; Asensio, A.; Simón, A.; Bravo, P.; Fronza, G.; Volonterio, A.; et al. Enantioselective synthesis of fluorinated α-amino acids and derivative in combination with ring-closing metathesis: Intramolecular π-stacking interactions as a source of stereocontrol. Org. Lett. 2001, 3, 2621–2624. [Google Scholar] [CrossRef]
- Herbert, B.; Kim, I.H.; Kirk, K.L. Synthesis of 2-fluoro- and 6-fluoro-(2S,3R)-(3,4-dihydroxyphenyl)serine as potential in vivo precursors of fluorinated norepinephrines. J. Org. Chem. 2001, 66, 4892–4897. [Google Scholar] [CrossRef]
- Fustero, S.; Bartolomé, A.; Sanz-Cervera, J.F.; Sánchez-Roselló, M.; García Soler, J.; de Arellano, C.R.; Fuentes, A.S. Diastereoselective synthesis of fluorinated, seven-membered β-amino acids derivatives via ring-closing metathesis. Org. Lett. 2003, 5, 2523–2526. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-X.; Qin, Y.; Qing, F. Asymmetric synthesis of both enantiomers of anti-4,4,4-trifluorothreonine and 2-amino-4,4,4-trifluorobutanoic acid. J. Org. Chem. 2003, 68, 7544–7547. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, D.B.; de la Salud-Bea, R.; Jahng, W. Synthesis of quaternary amino acids bearing a (2’Z)-fluorovinyl α-branch: Potential PLP enzyme inactivators. Org. Lett. 2004, 6, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Otaka, A.; Mitsuyama, E.; Watanabe, J.; Watanabe, H.; Fujii, N. Synthesis of fluorine-containing bioisosteres corresponding to phosphoamino acids and dipeptide units. Biopolymers 2004, 76, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Zhu, S.; Jiang, H.; Zhu, S. Transition metal-catalyzed formation of CF3-substituted α,β-unsaturated alkene and the synthesis of α-trifluoromethyl substituted β-amino ester. Tetrahedron 2006, 62, 11760–11765. [Google Scholar] [CrossRef]
- Yajima, T.; Nagano, H. Photoinduced diastereoselective addition of perfluoroalkyl iodides to acrylic acid derivatives for the synthesis of fluorinated amino acids. Org. Lett. 2007, 9, 2513–2515. [Google Scholar] [CrossRef]
- Pigza, J.A.; Quach, T.; Molinski, T.F. Oxazoline-oxazinone oxidative rearrangement. divergent syntheses of (2S,3S)-4,4,4-trifluorovaline and (2S,4S)-5,5,5-Trifluoroleucine. J. Org. Chem. 2009, 74, 5510–5515. [Google Scholar] [CrossRef]
- Drège, E.; Guillaume, A.; Boukhedimi, N.; Marrot, J.; Troufflard, C.; Tran Huu-Dau, M.-E.; Joseph, D.; Delarue-Cochin, S. A facile and stereocontrolled synthesis of γ-substituted γ-fluoroglutamates by conjugate addition: Conflicting effect between fluorinated enaminoester and hinderered Michael acceptor. J. Org. Chem. 2010, 75, 7596–7604. [Google Scholar] [CrossRef]
- Aceña, J.L.; Sorochinsky, A.E.; Moriwaki, H.; Sato, T.; Soloshonok, V.A. Synthesis of fluorine-containing α-amino acids in enantiomerically pure form via homologation of Ni(II) complexes of glycine and alanine Schiff bases. J. Fluor. Chem. 2013, 155, 21–38. [Google Scholar] [CrossRef]
- Erdbrink, H.; Nyakatura, E.K.; Huhmann, S.; Gerling, U.I.M.; Lentz, D.; Koksch, B.; Czekelius, C. Synthesis of enantiomerically pure (2S,3S)-5,5,5-trifluoroisoleucine and (2R,3S)-5,5,5-trifluoro-allo-isoleucine. Beilstein J. Org. Chem. 2013, 9, 2009–2014. [Google Scholar] [CrossRef]
- Ivashkin, P.; Lemonnier, G.; Tora, A.S.; Pin, J.P.; Goudet, C.; Jubault, P.; Pannecoucke, X. Synthesis and studies on the mGluR agonist activity of FAP4 stereoisomers. Bioorg. Med. Chem. Lett. 2015, 25, 2523–2526. [Google Scholar] [CrossRef] [PubMed]
- Grigolato, L.; Brittain, W.D.G.; Hudson, A.S.; Czyzewska, M.M.; Cobb, S.L. Synthesis of pentafluorosulfanyl (SF5) containing aromatic amino acids. J. Fluor. Chem. 2018, 212, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-B.; Ren, X.; Zheng, B.-Q.; Ji, J.; Qiu, Z.-B.; Li, Y. A diastereoselective Mannich reaction of α-fluoroketones with ketimines: Construction of β-fluoroamine motifs with vicinal tetrasubstituted stereocenters. Tetrahedron Lett. 2018, 59, 2091–2094. [Google Scholar] [CrossRef]
- Ouchakour, L.; Ábrahámi, R.A.; Forró, E.; Haukka, M.; Fülöp, F.; Kiss, L. Stereocontrolled synthesis of fluorine-containing piperidine γ-amino acid derivatives. Eur. J. Org. Chem. 2019, 2019, 2202–2211. [Google Scholar] [CrossRef]
- Kirk, K.L.; Herbert, B.; Lu, S.F.; Jayachandran, B.; Padgett, W.L.; Olufunke, O.; Daly, J.W.; Haufe, G.; Laue, K.W. Chemical and biochemical approaches to the enantiomers of chiral fluorinated catecholamines and amino acids. In Asymmetric Fluoroorganic Chemistry; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1999; Volume 746, pp. 194–209. [Google Scholar]
- Zheng, H.; Comeforo, K.; Gao, J. Expanding the fluorous arsenal: Tetrafluorinated phenylalanines for protein design. J. Am. Chem. Soc. 2009, 131, 18–19. [Google Scholar] [CrossRef]
- Pace, C.J.; Zheng, H.; Mylvaganam, R.; Kim, D.; Gao, J. Stacked fluoroaromatics as supramolecular synthons for programming protein dimerization specificity. Angew. Chem. Int. Ed. 2012, 51, 103–107. [Google Scholar] [CrossRef]
- Buller, A.R.; Brinkmann-Chen, S.; Romney, D.K.; Herger, M.; Murciano-Calles, J.; Arnold, F.H. Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation. Proc. Natl. Acad. Sci. USA 2015, 112, 14599–14604. [Google Scholar] [CrossRef]
- Islam, M.N.; Hitchings, R.; Kumar, S.; Fontes, F.L.; Lott, J.S.; Kruh-Garcia, N.A.; Crick, D.C. Mechanism of Fluorinated Anthranilate-Induced Growth Inhibition in Mycobacterium tuberculosis. ACS Infect. Dis. 2019, 5, 55–62. [Google Scholar] [CrossRef]
- Dong, C.; Huang, F.; Deng, H.; Schaffrath, C.; Spencer, J.B.; O’Hagan, D.; Naismith, J.H. Crystal structure and mechanism of fluorinating enzyme. Nature 2004, 427, 561–565. [Google Scholar] [CrossRef]
- Doi, M.; Nishi, Y.; Kiritoshi, N.; Iwata, T.; Mago, M.; Nakano, H.; Uchiyama, S.; Nakazawa, T.; Wakamiya, T.; Kobayashi, Y. Simple and efficient syntheses if Boc- and Fmoc-protected 4(R)- and 4(S)-fluoroproline solely from 4(R)-hydroxyproline. Tetrahedron 2002, 58, 8453–8459. [Google Scholar] [CrossRef]
- Hajduch, J.; Cramer, J.C.; Kirk, K.L. An Enantioselective Synthesis of (S)-4-Fluorohistidine. J. Fluor. Chem. 2008, 129, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Ehrlichmann, W.; Uebele, M.; Machulla, H.J.; Reischl, G. Automated synthesis of n.c.a. [18F]FDOPA via nucleophilic aromatic substitution with [18F]fluoride. Appl. Radiat. Isot. 2009, 67, 1650–1653. [Google Scholar] [CrossRef]
- Kiss, L.; Forro, E.; Fustero, S.; Fulop, F. Regio- and diastereoselective fluorination of alicyclic β-amino acids. Org. Biomol. Chem. 2011, 9, 6528–6534. [Google Scholar] [CrossRef] [PubMed]
- Kiss, L.; Nonn, M.; Sillanpaa, R.; Fustero, S.; Fulop, F. Efficient regio- and stereoselective access to novel fluorinated β-aminocyclohexanecarboxylates. Beilstein J. Org. Chem. 2013, 9, 1164–1169. [Google Scholar] [CrossRef]
- Remete, A.; Nonn, M.; Fustero, S.; Fülöp, F.; Kiss, L. A Stereocontrolled Protocol to Highly Functionalized Fluorinated Scaffolds through a Fluoride Opening of Oxiranes. Molecules 2016, 21, 1493. [Google Scholar] [CrossRef] [PubMed]
- Alluri, S.R.; Riss, P.J. Stereospecific radiosynthesis of 3-fluoro amino acids: Access to enantiomerically pure radioligands for positron emission tomography. Org. Biomol. Chem. 2018, 16, 2219–2224. [Google Scholar] [CrossRef]
- Malashchuk, A.; Chernykh, A.V.; Dobrydnev, A.V.; Grygorenko, O.O. Fluorine-labelled spiro[3.3]heptane-derived building blocks: Is single fluorine the best? Eur. J. Org. Chem. 2021, 2021, 4897–4910. [Google Scholar] [CrossRef]
- Padmakshan, D.; Bennett, S.; Otting, G.; Easton, C. Stereocontrolled Synthesis of (S)-γ-Fluoroleucine. Synlett 2007, 2007, 1083–1084. [Google Scholar] [CrossRef]
- Molnar, I.G.; Tanzer, E.M.; Daniliuc, C.; Gilmour, R. Enantioselective aziridination of cyclic enals facilitated by the fluorine-iminium ion gauche effect. Chemistry 2014, 20, 794–800. [Google Scholar] [CrossRef]
- Tsushima, T.; Kawada, K.; Nishikawa, J.; Sato, T.; Tori, K.; Tsuji, T.; Misaki, S. Fluorine-containing amino acids and their derivatives. 3. Stereoselective synthesis and unusual conformational features of threo- and erythro-3-fluorophenylalanine. J. Org. Chem. 1984, 49, 1163–1169. [Google Scholar] [CrossRef]
- Labroo, V.M.; Hebel, D.; Kirk, K.L.; Cohen, L.A.; Lemieux, C.; Schiller, P.W. Direct electrophilic fluorination of tyrosine in dermorphin analogues and its effect on biological activity, receptor affinity and selectivity. Int. J. Peptide Protein Res. 1991, 37, 430–439. [Google Scholar] [CrossRef]
- Pedregal, C.; Prowse, W. Stereoselective synthesis of 2-amino-3-fluoro bicyclo[3.1.0]hexane-2,6-dicarboxylic acid. Bioorg. Med. Chem. 2002, 10, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, F.; Kirjavainen, A.K.; Forsback, S.; Krzyczmonik, A.; Keller, T.; Newington, I.M.; Glaser, M.; Luthra, S.K.; Solin, O.; Gouverneur, V. Organomediated enantioselective 18F fluorination for PET applications. Angew. Chem. Int. Ed. 2015, 54, 13366–13369. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Khangarot, R.K.; Stahl, L.; Veresmortean, C.; Pradhan, P.; Yang, L.; Zajc, B. Generating stereodiversity: Diastereoselective fluorination and highly diastereoselective epimerization of α-amino acid building blocks. Org. Lett. 2018, 20, 3574–3578. [Google Scholar] [CrossRef] [PubMed]
- Bandak, D.; Babii, O.; Vasiuta, R.; Komarov, I.V.; Mykhailiuk, P.K. Design and synthesis of novel 19F-amino acid: A promising 19F NMR label for peptide studies. Org. Lett. 2015, 17, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Hebel, D.; Kirk, K.L.; Cohen, L.A.; Labroo, V.M. First direct fluorination of tyrosine-containing biologically active peptides. Tetrahedron Lett. 1990, 31, 619–622. [Google Scholar] [CrossRef]
- Lu, D.F.; Liu, G.S.; Zhu, C.L.; Yuan, B.; Xu, H. Iron(II)-catalyzed intramolecular olefin aminofluorination. Org. Lett. 2014, 16, 2912–2915. [Google Scholar] [CrossRef]
- Zhu, R.Y.; Tanaka, K.; Li, G.C.; He, J.; Fu, H.Y.; Li, S.H.; Yu, J.Q. Ligand-enabled stereoselective β-C(sp3)-H fluorination: Synthesis of unnatural enantiopure anti-β-fluoro-α-amino acids. J. Am. Chem. Soc. 2015, 137, 7067–7070. [Google Scholar] [CrossRef]
- Miao, J.; Yang, K.; Kurek, M.; Ge, H. Palladium-catalyzed site-selective fluorination of unactivated C(sp3)-H bonds. Org. Lett. 2015, 17, 3738–3741. [Google Scholar] [CrossRef]
- Lévine-Pinto, H.; Bouabdallah, B.; Morgat, J.L.; Gourdji, D.; Fromageot, P. Specific and direct fluorination of an histidine-containing peptide: Thyroliberin. Biochem. Biophys. Res. Commun. 1981, 103, 1121–1130. [Google Scholar] [CrossRef]
- Bume, D.D.; Pitts, C.R.; Jokhai, R.T.; Lectka, T. Direct, visible light-sensitized benzylic C-H fluorination of peptides using dibenzosuberenone: Selectivity for phenylalanine-like residues. Tetrahedron 2016, 72, 6031–6036. [Google Scholar] [CrossRef]
- Glaser, R.W.; Sachse, C.; Durr, U.H.; Wadhwani, P.; Ulrich, A.S. Orientation of the antimicrobial peptide PGLa in lipid membranes determined from 19F-NMR dipolar couplings of 4-CF3-phenylglycine labels. J. Magn. Reson. 2004, 168, 153–163. [Google Scholar] [CrossRef]
- Schottelius, M.; Berger, S.; Poethko, T.; Schwaiger, M.; Wester, H.-J. Development of novel 68Ga- and 18F-labeled GnRH-I analogues with high GnRHR-targeting efficiency. Bioconjug. Chem. 2008, 19, 1256–1268. [Google Scholar] [CrossRef]
- Clark, G.A.; Baleja, J.D.; Kumar, K. Cross-strand interactions of fluorinated amino acids in β-hairpin constructs. J. Am. Chem. Soc. 2012, 134, 17912–17921. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peggion, C.; Biondi, B.; Battistella, C.; De Zotti, M.; Oancea, S.; Formaggio, F.; Toniolo, C. Spectroscopically labeled peptaibiotics. Synthesis and properties of selected trichogin GA IV analogs bearing a side-chain-monofluorinated aromatic amino acid for 19F-NMR analysis. Chem. Biodivers. 2013, 10, 904–919. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Afonin, S.; Kara, S.; Budisa, N.; Mykhailiuk, P.K.; Ulrich, A.S. γ-(S)-Trifluoromethyl proline: Evaluation as a structural substitute of proline for solid state 19F-NMR peptide studies. Org. Biomol. Chem. 2015, 13, 3171–3181. [Google Scholar] [CrossRef] [PubMed]
- Wodtke, R.; Ruiz-Gomez, G.; Kuchar, M.; Pisabarro, M.T.; Novotna, P.; Urbanova, M.; Steinbach, J.; Pietzsch, J.; Loser, R. Cyclopeptides containing the DEKS motif as conformationally restricted collagen telopeptide analogues: Synthesis and conformational analysis. Org. Biomol. Chem. 2015, 13, 1878–1896. [Google Scholar] [CrossRef]
- Kuchar, M.; Neuber, C.; Belter, B.; Bergmann, R.; Lenk, J.; Wodtke, R.; Kniess, T.; Steinbach, J.; Pietzsch, J.; Loser, R. Evaluation of fluorine-18-labeled α1(I)-N-telopeptide analogs as substrate-based radiotracers for PET imaging of melanoma-associated lysyl oxidase. Front. Chem. 2018, 6, 121. [Google Scholar] [CrossRef]
- Arias, M.; Aramini, J.M.; Riopel, N.D.; Vogel, H.J. Fluorine-19 NMR spectroscopy of fluorinated analogs of tritrpticin highlights a distinct role for Tyr residues in antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183260. [Google Scholar] [CrossRef]
- Corlett, A.; Sani, M.A.; Van Zuylekom, J.; Ang, C.S.; von Guggenberg, E.; Cullinane, C.; Blyth, B.; Hicks, R.J.; Roselt, P.D.; Thompson, P.E.; et al. A new turn in peptide-based imaging agents: Foldamers afford improved theranostics targeting cholecystokinin-2 receptor-positive cancer. J. Med. Chem. 2021, 64, 4841–4856. [Google Scholar] [CrossRef]
- O’Connor, N.K.; Rai, D.K.; Clark, B.R.; Murphy, C.D. Production of the novel lipopeptide antibiotic trifluorosurfactin via precursor-directed biosynthesis. J. Fluor. Chem. 2012, 143, 210–215. [Google Scholar] [CrossRef]
- O’Connor, N.K.; Hudson, A.S.; Cobb, S.L.; O’Neil, D.; Robertson, J.; Duncan, V.; Murphy, C.D. Novel fluorinated lipopeptides from Bacillus sp. CS93 via precursor-directed biosynthesis. Amino Acids 2014, 46, 2745–2752. [Google Scholar] [CrossRef] [PubMed]
- Leick, V. Effect of actinomycin D and DL-p-fluorophenylalanine on ribosome formation in Tetrahmena pyriformis. Eur. J. Biochem. 1969, 8, 215–220. [Google Scholar] [CrossRef]
- Takeuchi, T.; Prockop, D.J. Biosynthesis of abnormal collagens with amino acid analogues. 1. Incorporation of l-azetidine-2-carboxylic acid and cis-4-fluoro-l-proline into protocollagen and collagen. Biochim. Biophys. Acta (BBA) Protein Struct. 1969, 175, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Barker, C.; Lewis, D. Impaired regulation of aromatic amino acid synthesis in a mutant resistant to p-fluorophenylalanine. J. Gen. Microbiol. 1974, 82, 337–343. [Google Scholar] [CrossRef][Green Version]
- Hardy, C.; Binkley, S.B. The effect of p-fluorophenylalanine on nucleic acid biosynthesis and cell divisionin Escherichia coli. Biochemistry 1967, 6, 1892–1898. [Google Scholar] [CrossRef]
- Wang, P.; Tang, Y.; Tirrell, D.A. Incorporation of trifluoroisoleucine into proteins in vivo. J. Am. Chem. Soc. 2003, 125, 6900–6906. [Google Scholar] [CrossRef]
- Wang, P.; Fichera, A.; Kumar, K.; Tirrell, D.A. Alternative translations of a single RNA message: An identity switch of (2S,3R)-4,4,4-trifluorovaline between valine and isoleucine codons. Angew. Chem. Int. Ed. 2004, 43, 3664–3666. [Google Scholar] [CrossRef]
- Son, S.; Tanrikulu, I.C.; Tirrell, D.A. Stabilization of bzip peptides through incorporation of fluorinated aliphatic residues. ChemBioChem 2006, 7, 1251–1257. [Google Scholar] [CrossRef]
- Jackson, J.C.; Duffy, S.P.; Hess, K.R.; Mehl, R.A. Improving nature’s enzyme active site with genetically encoded unnatural amino acids. J. Am. Chem. Soc. 2006, 128, 11124–11127. [Google Scholar] [CrossRef]
- Rodriguez, E.A.; Lester, H.A.; Dougherty, D.A. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: Minimizing misacylation. RNA 2007, 13, 1703–1714. [Google Scholar] [CrossRef][Green Version]
- Ye, S.; Ann Berger, A.; Petzold, D.; Reimann, O.; Matt, B.; Koksch, B. Chemical aminoacylation of tRNAs with fluorinated amino acids for in vitro protein mutagenesis. Beilstein J. Org. Chem. 2010, 6, 40. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hoppmann, C.; Yang, B.; Wang, L. Using Protein-Confined Proximity to Determine Chemical Reactivity. J. Am. Chem. Soc. 2016, 138, 14832–14835. [Google Scholar] [CrossRef]
- Alleyne, C.; Amin, R.P.; Bhatt, B.; Bianchi, E.; Blain, J.C.; Boyer, N.; Branca, D.; Embrey, M.W.; Ha, S.N.; Jette, K.; et al. Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design. J. Med. Chem. 2020, 63, 13796–13824. [Google Scholar] [CrossRef]
- Ford, D.J.; Duggan, N.M.; Fry, S.E.; Ripoll-Rozada, J.; Agten, S.M.; Liu, W.; Corcilius, L.; Hackeng, T.M.; van Oerle, R.; Spronk, H.M.H.; et al. Potent Cyclic Peptide Inhibitors of FXIIa Discovered by mRNA Display with Genetic Code Reprogramming. J. Med. Chem. 2021, 64, 7853–7876. [Google Scholar] [CrossRef]
- Iskandar, S.E.; Bowers, A.A. mRNA Display Reaches for the Clinic with New PCSK9 Inhibitor. ACS Med. Chem. Lett. 2022, 13, 1379–1383. [Google Scholar] [CrossRef]
- Tucker, T.J.; Embrey, M.W.; Alleyne, C.; Amin, R.P.; Bass, A.; Bhatt, B.; Bianchi, E.; Branca, D.; Bueters, T.; Buist, N.; et al. A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors. J. Med. Chem. 2021, 64, 16770–16800. [Google Scholar] [CrossRef]
- Josephson, K.; Ricardo, A.; Szostak, J.W. mRNA display: From basic principles to macrocycle drug discovery. Drug Discov. Today 2014, 19, 388–399. [Google Scholar] [CrossRef]
- Newton, M.S.; Cabezas-Perusse, Y.; Tong, C.L.; Seelig, B. In vitro selection of peptides and proteins—Advantages of mRNA display. ACS Synth. Biol. 2020, 9, 181–190. [Google Scholar] [CrossRef]
- Nagumo, Y.; Fujiwara, K.; Horisawa, K.; Yanagawa, H.; Doi, N. PURE mRNA display for in vitro selection of single-chain antibodies. J. Biochem. 2016, 159, 519–526. [Google Scholar] [CrossRef]
- Sharaf, N.G.; Gronenborn, A.M. 19F-modified proteins and 19F-containing ligands as tools in solution NMR studies of protein interactions. Methods Enzymol. 2015, 565, 67–95. [Google Scholar] [CrossRef]
- Arntson, K.E.; Pomerantz, W.C. Protein-observed fluorine NMR: A bioorthogonal approach for small molecule discovery. J. Med. Chem. 2016, 59, 5158–5171. [Google Scholar] [CrossRef]
- Verhoork, S.J.M.; Killoran, P.M.; Coxon, C.R. Fluorinated Prolines as Conformational Tools and Reporters for Peptide and Protein Chemistry. Biochemistry 2018, 57, 6132–6143. [Google Scholar] [CrossRef]
- Divakaran, A.; Kirberger, S.E.; Pomerantz, W.C.K. SAR by (Protein-Observed) 19F NMR. Acc. Chem. Res. 2019, 52, 3407–3418. [Google Scholar] [CrossRef]
- Gimenez, D.; Phelan, A.; Murphy, C.D.; Cobb, S.L. 19F NMR as a tool in chemical biology. Beilstein J. Org. Chem. 2021, 17, 293–318. [Google Scholar] [CrossRef]
- Tsetlin, V.I.; Arseniev, A.S.; Utkin, Y.N.; Gurevich, A.Z.; Senyavina, L.B.; Bystrov, V.F.; Ivanov, V.T.; Ovchinnikov, Y.A. Conformational studies of Neurotoxin II from Naja naja oxiana. Eur. J. Biochem. 1979, 94, 337–346. [Google Scholar] [CrossRef]
- Liao, T.; Berlin, K.D. The use of p-fluorobenzenesulfonyl chloride as a reagent for studies of proteins by fluorine nuclear magnetic resonance. Anal. Biochem. 1985, 148, 365–375. [Google Scholar] [CrossRef]
- Ekanayake, K.B.; Mahawaththa, M.C.; Qianzhu, H.; Abdelkader, E.H.; George, J.; Ullrich, S.; Murphy, R.B.; Fry, S.E.; Johansen-Leete, J.; Payne, R.J.; et al. Probing Ligand Binding Sites on Large Proteins by Nuclear Magnetic Resonance Spectroscopy of Genetically Encoded Non-Canonical Amino Acids. J. Med. Chem. 2023, 66, 5289–5304. [Google Scholar] [CrossRef]
- Huang, Y.; Reddy, K.D.; Bracken, C.; Qiu, B.; Zhan, W.; Eliezer, D.; Boudker, O. Environmentally ultrasensitive fluorine probe to resolve protein conformational ensembles by 19F NMR and cryo-EM. J. Am. Chem. Soc. 2023, 145, 8583–8592. [Google Scholar] [CrossRef]
- Hattori, Y.; Heidenreich, D.; Ono, Y.; Sugiki, T.; Yokoyama, K.I.; Suzuki, E.I.; Fujiwara, T.; Kojima, C. Protein 19F-labeling using transglutaminase for the NMR study of intermolecular interactions. J. Biomol. NMR 2017, 68, 271–279. [Google Scholar] [CrossRef]
- Lian, C.; Le, H.; Montez, B.; Patterson, J.; Harrell, S.; Laws, D.; Matsumura, I.; Pearson, J.; Oldfield, E. Fluorine-19 nuclear magnetic resonance spectroscopic study of fluorophenylalanine- and fluorotyrptophan-labeled avian egg white lysozymes. Biochemistry 1994, 33, 5238–5245. [Google Scholar] [CrossRef]
- Duewel, H.S.; Daub, E.; Robinson, V.; Honek, J.F. Elucidation of solvent exposure, side-chain reactivity, and steric demands of the trifluoromethionine residue in a recombinant protein. Biochemistry 2001, 40, 13167–13176. [Google Scholar] [CrossRef] [PubMed]
- Dupureur, C.M.; Dominguez, M.A. The PD...(D/E)XK motif in restriction enzymes: A link between function and conformation. Biochemistry 2001, 40, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Frieden, C. The kinetics of side chain stabilization during protein folding. Biochemistry 2003, 42, 12439–12446. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Mercier, P.; Letourneau, P.J.; Sykes, B.D. Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies. Protein Sci. 2005, 14, 2447–2460. [Google Scholar] [CrossRef]
- Evanics, F.; Kitevski, J.L.; Bezsonova, I.; Forman-Kay, J.; Prosser, R.S. 19F NMR studies of solvent exposure and peptide binding to an SH3 domain. Biochim. Biophys. Acta (BBA) Gen. Subj. 2007, 1770, 221–230. [Google Scholar] [CrossRef]
- Li, C.; Lutz, E.A.; Slade, K.M.; Ruf, R.A.; Wang, G.F.; Pielak, G.J. 19F NMR studies of α-synuclein conformation and fibrillation. Biochemistry 2009, 48, 8578–8584. [Google Scholar] [CrossRef]
- Voloshchuk, N.; Zhu, A.Y.; Snydacker, D.; Montclare, J.K. Positional effects of monofluorinated phenylalanines on histone acetyltransferase stability and activity. Bioorg. Med. Chem. Lett. 2009, 19, 5449–5451. [Google Scholar] [CrossRef]
- Pomerantz, W.C.; Wang, N.; Lipinski, A.K.; Wang, R.; Cierpicki, T.; Mapp, A.K. Profiling the dynamic interfaces of fluorinated transcription complexes for ligand discovery and characterization. ACS Chem. Biol. 2012, 7, 1345–1350. [Google Scholar] [CrossRef]
- Mishra, N.K.; Urick, A.K.; Ember, S.W.; Schonbrunn, E.; Pomerantz, W.C. Fluorinated aromatic amino acids are sensitive 19F NMR probes for bromodomain-ligand interactions. ACS Chem. Biol. 2014, 9, 2755–2760. [Google Scholar] [CrossRef]
- Suzuki, Y.; Brender, J.R.; Soper, M.T.; Krishnamoorthy, J.; Zhou, Y.; Ruotolo, B.T.; Kotov, N.A.; Ramamoorthy, A.; Marsh, E.N.G. Resolution of oligomeric species during the aggregation of Aβ1–40 using 19F NMR. Biochemistry 2013, 52, 1903–1912. [Google Scholar] [CrossRef]
- Tressler, C.M.; Zondlo, N.J. (2S,4R)- and (2S,4S)-perfluoro-tert-butyl 4-hydroxyproline: Two conformationally distinct proline amino acids for sensitive application in 19F NMR. J. Org. Chem. 2014, 79, 5880–5886. [Google Scholar] [CrossRef]
- Tressler, C.M.; Zondlo, N.J. Perfluoro-tert-butyl hydroxyprolines as sensitive, conformationally responsive molecular probes: Detection of protein kinase activity by 19F NMR. ACS Chem. Biol. 2020, 15, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, W.; Langenhan, J.; Huhmann, S.; Moschner, J.; Chang, R.; Accorsi, M.; Seo, J.; Rademann, J.; Meijer, G.; Koksch, B.; et al. An Intrinsic Hydrophobicity Scale for Amino Acids and Its Application to Fluorinated Compounds. Angew. Chem. Int. Ed. 2019, 58, 8216–8220. [Google Scholar] [CrossRef] [PubMed]
- Larda, S.T.; Simonetti, K.; Al-Abdul-Wahid, M.S.; Sharpe, S.; Prosser, R.S. Dynamic equilibria between monomeric and oligomeric misfolded states of the mammalian prion protein measured by 19F NMR. J. Am. Chem. Soc. 2013, 135, 10533–10541. [Google Scholar] [CrossRef] [PubMed]
- Woll, M.G.; Hadley, E.B.; Mecozzi, S.; Gellman, S.H. Stabilizing and destabilizing effects of phenylalanine → F5-phenylalanine mutations on the folding of a small protein. J. Am. Chem. Soc. 2006, 128, 15932–15933. [Google Scholar] [CrossRef]
- Esteban-Martin, S.; Strandberg, E.; Salgado, J.; Ulrich, A.S. Solid state NMR analysis of peptides in membranes: Influence of dynamics and labeling scheme. Biochim. Biophys. Acta (BBA) Biomembr. 2010, 1798, 252–257. [Google Scholar] [CrossRef]
- Grage, S.L.; Xu, X.; Schmitt, M.; Wadhwani, P.; Ulrich, A.S. 19F-Labeling of peptides revealing long-range NMR distances in fluid membranes. J. Phys. Chem. Lett. 2014, 5, 4256–4259. [Google Scholar] [CrossRef]
- Kokhan, S.O.; Tymtsunik, A.V.; Grage, S.L.; Afonin, S.; Babii, O.; Berditsch, M.; Strizhak, A.V.; Bandak, D.; Platonov, M.O.; Komarov, I.V.; et al. Design, synthesis, and application of an optimized monofluorinated aliphatic label for peptide studies by solid-state 19F NMR spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 14788–14792. [Google Scholar] [CrossRef]
- Grage, S.L.; Sani, M.A.; Cheneval, O.; Henriques, S.T.; Schalck, C.; Heinzmann, R.; Mylne, J.S.; Mykhailiuk, P.K.; Afonin, S.; Komarov, I.V.; et al. Orientation and Location of the Cyclotide Kalata B1 in Lipid Bilayers Revealed by Solid-State NMR. Biophys. J. 2017, 112, 630–642. [Google Scholar] [CrossRef]
- Mikhailiuk, P.K.; Afonin, S.; Chernega, A.N.; Rusanov, E.B.; Platonov, M.O.; Dubinina, G.G.; Berditsch, M.; Ulrich, A.S.; Komarov, I.V. Conformationally rigid trifluoromethyl-substituted α-amino acid designed for peptide structure analysis by solid-state 19F NMR spectroscopy. Angew. Chem. Int. Ed. 2006, 45, 5659–5661. [Google Scholar] [CrossRef]
- Afonin, S.; Mikhailiuk, P.K.; Komarov, I.V.; Ulrich, A.S. Evaluating the amino acid CF3-bicyclopentylglycine as a new label for solid-state 19F-NMR structure analysis of membrane-bound peptides. J. Pept. Sci. 2007, 13, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, D.; Oda, K.; Inden, M.; Morikawa, S.; Inubushi, T.; Taniguchi, T.; Hijioka, M.; Kitamura, Y.; Tooyama, I. Fluorodopa is a Promising Fluorine-19 MRI Probe for Evaluating Striatal Dopaminergic Function in a Rat Model of Parkinson’s Disease. J. Neurosci. Res. 2017, 95, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, S.; Cavazzini, M.; Capuani, S.; Ciardello, A.; Pozzi, G. Synthesis and 19F NMR parameters of a perfluoro-tert-butoxy tagged l-DOPA analogue. J. Fluor. Chem. 2020, 237, 109596. [Google Scholar] [CrossRef]
- Tirotta, I.; Dichiarante, V.; Pigliacelli, C.; Cavallo, G.; Terraneo, G.; Bombelli, F.B.; Metrangolo, P.; Resnati, G. 19F magnetic resonance imaging (MRI): From design of materials to clinical applications. Chem. Rev. 2015, 115, 1106–1129. [Google Scholar] [CrossRef]
- Ishiwata, K.; Kubota, K.; Murakami, M.; Kubota, R.; Sasaki, T.; Ishii, S.; Seda, M. Re-evaluation of amino acid PET studies: Can the protein synthesis rates in brain and tumor tissues be easured in vivo? J. Nucl. Med. 1993, 34, 1936–1943. [Google Scholar]
- Makrides, V.; Bauer, R.; Weber, W.; Wester, H.J.; Fischer, S.; Hinz, R.; Huggel, K.; Opfermann, T.; Herzau, M.; Ganapathy, V.; et al. Preferred transport of O-(2-[18F]fluoroethyl)-d-tyrosine (D-FET) into the porcine brain. Brain Res. 2007, 1147, 25–33. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, S.J.; Yook, C.M.; Oh, S.J.; Ryu, J.S.; Lee, J.J. Biological evaluation of new [18F]F-labeled synthetic amino acid derivatives as oncologic radiotracers. J. Label. Compd. Radiopharm. 2016, 59, 404–410. [Google Scholar] [CrossRef]
- Langen, K.J.; Hamacher, K.; Weckesser, M.; Floeth, F.; Stoffels, G.; Bauer, D.; Coenen, H.H.; Pauleit, D. O-(2-[18F]fluoroethyl)-l-tyrosine: Uptake mechanisms and clinical applications. Nucl. Med. Biol. 2006, 33, 287–294. [Google Scholar] [CrossRef]
- Fedorova, O.S.; Kuznetsova, O.F.; Shatik, S.V.; Stepanova, M.A.; Belokon, Y.N.; Maleev, V.I.; Krasikova, R.N. 18F-labeled tyrosine derivatives: Synthesis and experimental studies on accumulation in tumors and abscesses. Russ. J. Bioorg. Chem. 2009, 35, 306–314. [Google Scholar] [CrossRef]
- Stegmayr, C.; Stoffels, G.; Filss, C.; Heinzel, A.; Lohmann, P.; Willuweit, A.; Ermert, J.; Coenen, H.H.; Mottaghy, F.M.; Galldiks, N.; et al. Current trends in the use of O-(2-[18F]fluoroethyl)-l-tyrosine ([18F]FET) in neurooncology. Nucl. Med. Biol. 2021, 92, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Franck, D.; Kniess, T.; Steinbach, J.; Zitzmann-Kolbe, S.; Friebe, M.; Dinkelborg, L.M.; Graham, K. Investigations into the synthesis, radiofluorination and conjugation of a new [18F]fluorocyclobutyl prosthetic group and its in vitro stability using a tyrosine model system. Bioorg. Med. Chem. 2013, 21, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Betts, H.M.; Milicevic Sephton, S.; Tong, C.; Awais, R.O.; Hill, P.J.; Perkins, A.C.; Aigbirhio, F.I. Synthesis, in vitro evaluation, and radiolabeling of fluorinated puromycin analogues: Potential candidates for PET imaging of protein synthesis. J. Med. Chem. 2016, 59, 9422–9430. [Google Scholar] [CrossRef] [PubMed]
- Chiotellis, A.; Muller, A.; Weyermann, K.; Leutwiler, D.S.; Schibli, R.; Ametamey, S.M.; Kramer, S.D.; Mu, L. Synthesis and preliminary biological evaluation of O-2((2-[18F]fluoroethyl)methylamino)ethyltyrosine ([18F]FEMAET) as a potential cationic amino acid PET tracer for tumor imaging. Amino Acids 2014, 46, 1947–1959. [Google Scholar] [CrossRef]
- Maschauer, S.; Pischetsrieder, M.; Kuwert, T.; Prante, O. Utility of 1,3,4,6-tetra-O-acetyl-2-deoxy-2-[18F]fluoro-glucopyranoside for no-carrier-added 18F-glycosylation of amino acids. J. Label. Compd. Radiopharm. 2005, 48, 701–719. [Google Scholar] [CrossRef]
- Chiotellis, A.; Muller, A.; Mu, L.; Keller, C.; Schibli, R.; Kramer, S.D.; Ametamey, S.M. Synthesis and biological evaluation of 18F-labeled Fluoroethoxy tryptophan analogues as potential PET tumor imaging agents. Mol. Pharm. 2014, 11, 3839–3851. [Google Scholar] [CrossRef]
- Kiesewetter, D.O.; Gao, H.; Ma, Y.; Niu, G.; Quan, Q.; Guo, N.; Chen, X. 18F-radiolabeled analogs of exendin-4 for PET imaging of GLP-1 in insulinoma. Eur. J. Nucl. Med. Mol. Imaging 2012, 39, 463–473. [Google Scholar] [CrossRef]
- Liu, S.; Ma, H.; Zhang, Z.; Lin, L.; Yuan, G.; Tang, X.; Nie, D.; Jiang, S.; Yang, G.; Tang, G. Synthesis of enantiopure 18F-trifluoromethyl cysteine as a structure-mimetic amino acid tracer for glioma imaging. Theranostics 2019, 9, 1144–1153. [Google Scholar] [CrossRef]
- Baguet, T.; Bouton, J.; Janssens, J.; Pauwelyn, G.; Verhoeven, J.; Descamps, B.; Van Calenbergh, S.; Vanhove, C.; De Vos, F. Radiosynthesis, in vitro and preliminary biological evaluation of [18F]2-amino-4-((2-((3-fluorobenzyl)oxy)benzyl)(2-((3-(fluoromethyl)benzyl)oxy)benzyl)amino)butanoic acid, a novel alanine serine cysteine transporter 2 inhibitor-based positron emission tomography tracer. J. Label. Compd. Radiopharm. 2020, 63, 442–455. [Google Scholar] [CrossRef]
- Iovkova, L.; Könning, D.; Wängler, B.; Schirrmacher, R.; Schoof, S.; Arndt, H.D.; Jurkschat, K. SiFA-Modified phenylalanine: A key compound for the efficient synthesis of 18F-labelled peptides. Eur. J. Inorg. Chem. 2011, 2011, 2238–2246. [Google Scholar] [CrossRef]
- Bernard, J.; Malacea-Kabbara, R.; Clemente, G.S.; Burke, B.P.; Eymin, M.J.; Archibald, S.J.; Juge, S. o-Boronato- and o-trifluoroborato-phosphonium salts supported by l-α-amino acid side chain. J. Org. Chem. 2015, 80, 4289–4298. [Google Scholar] [CrossRef]
- Fuchtner, F.; Steinbach, J. Efficient synthesis of the 18F-labelled 3-O-methyl-6-[18F]fluoro-l-DOPA. Appl. Radiat. Isot. 2003, 58, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, R.N.; Zaitsev, V.V.; Ametamey, S.M.; Kuznetsova, O.F.; Fedorova, O.S.; Mosevich, I.K.; Belokon, Y.N.; Vyskocil, S.; Shatik, S.V.; Nader, M.; et al. Catalytic enantioselective synthesis of 18F-fluorinated α-amino acids under phase-transfer conditions using (S)-NOBIN. Nucl. Med. Biol. 2004, 31, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Haase, C.; Bergmann, R.; Fuechtner, F.; Hoepping, A.; Pietzsch, J. L-type amino acid transporters LAT1 and LAT4 in cancer: Uptake of 3-O-methyl-6-18F-fluoro-l-DOPA in human adenocarcinoma and squamous cell carcinoma in vitro and in vivo. J. Nucl. Med. 2007, 48, 2063–2071. [Google Scholar] [CrossRef]
- Chondrogiannis, S.; Grassetto, G.; Marzola, M.C.; Rampin, L.; Massaro, A.; Bellan, E.; Ferretti, A.; Mazza, A.; Al-Nahhas, A.; Rubello, D. 18F-DOPA PET/CT biodistribution consideration in 107 consecutive patients with neuroendocrine tumours. Nucl. Med. Commun. 2012, 33, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Li, C.T.; Palotti, M.; Holden, J.E.; Oh, J.; Okonkwo, O.; Christian, B.T.; Bendlin, B.B.; Buyan-Dent, L.; Harding, S.J.; Stone, C.K.; et al. A dual-tracer study of extrastriatal 6-[18F]fluoro-m-tyrosine and 6-[18F]-fluoro-l-DOPA uptake in Parkinson’s disease. Synapse 2014, 68, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Hanaoka, H.; Higuchi, T.; Tsushima, Y. Selective synthesis of l-2-[18F]fluoro-alpha-methylphenylalanine via copper-mediated 18F-fluorination of (mesityl)(aryl)iodonium salt. J. Label. Compd. Radiopharm. 2020, 63, 368–375. [Google Scholar] [CrossRef]
- Wester, H.; Herz, M.; Senekowitsch-Schmidtke, R.; Schwaiger, M.; Stöcklin, G.; Hamacher, K. Preclinical evaluation of 4-[18F]fluoroprolines: Diastereomeric effect on metabolism and uptake in mice. Nucl. Med. Biol. 1999, 26, 259–265. [Google Scholar] [CrossRef]
- Yu, W.; Williams, L.; Camp, V.M.; Malveaux, E.; Olson, J.J.; Goodman, M.M. Stereoselective synthesis and biological evaluation of syn-1-amino-3-[18F]fluorocyclobutyl-1-carboxylic acid as a potential positron emission tomography brain tumor imaging agent. Bioorg. Med. Chem. 2009, 17, 1982–1990. [Google Scholar] [CrossRef]
- Koglin, N.; Mueller, A.; Berndt, M.; Schmitt-Willich, H.; Toschi, L.; Stephens, A.W.; Gekeler, V.; Friebe, M.; Dinkelborg, L.M. Specific PET imaging of xC− transporter activity using a 18F-labeled glutamate derivative reveals a dominant pathway in tumor metabolism. Clin. Cancer Res. 2011, 17, 6000–6011. [Google Scholar] [CrossRef]
- Schuster, D.M.; Nanni, C.; Fanti, S.; Oka, S.; Okudaira, H.; Inoue, Y.; Sörensen, J.; Owenius, R.; Choyke, P.; Turkbey, B.; et al. Anti-1-amino-3-18F-fluorocyclobutane-1-carboxylic acid: Physiologic uptake patterns, incidental findings, and variants that may simulate disease. J. Nucl. Med. 2014, 55, 1986–1992. [Google Scholar] [CrossRef]
- Bouhlel, A.; Zhou, D.; Li, A.; Yuan, L.; Rich, K.M.; McConathy, J. Synthesis, radiolabeling, and biological evaluation of (R)- and (S)-2-amino-5-[18F]fluoro-2-methylpentanoic acid ((R)-, (S)-[18F]FAMPe) as potential positron emission tomography tracers for brain tumors. J. Med. Chem. 2015, 58, 3817–3829. [Google Scholar] [CrossRef]
- Dunphy, M.P.S.; Harding, J.J.; Venneti, S.; Zhang, H.; Burnazi, E.M.; Bromberg, J.; Omuro, A.M.; Hsieh, J.J.; Mellinghoff, I.K.; Staton, K.; et al. in vivo PET Assay of tumor glutamine flux and metabolism: In-human trial of 18F-(2S,4R)-4-fluoroglutamine. Radiology 2018, 287, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Čolović, M.; Yang, H.; Merkens, H.; Colpo, N.; Bénard, F.; Schaffer, P. The effect of chirality on the application of 5-[18F]fluoro-aminosuberic acid ([18F]FASu) for oxidative stress imaging. Mol. Imaging Biol. 2019, 22, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Liu, S.; Liu, Y.; Sun, Y.; Cheng, X.; Huang, Y.; Yang, Z.; Wu, Z. Synthesis and biological evaluation of [18F](2S,4S)4-(3-fluoropropyl) arginine as a tumor imaging agent. Eur. J. Med. Chem. 2019, 183, 111730. [Google Scholar] [CrossRef]
- Liu, S.; Wu, R.; Sun, Y.; Ploessl, K.; Zhang, Y.; Liu, Y.; Wu, Z.; Zhu, L.; Kung, H.F. Design, synthesis and evaluation of a novel glutamine derivative (2S,4R)-2-amino-4-cyano-4-[18F]fluorobutanoic acid. New. J. Chem. 2020, 44, 9109–9117. [Google Scholar] [CrossRef]
- Pickel, T.C.; Voll, R.J.; Yu, W.; Wang, Z.; Nye, J.A.; Bacsa, J.; Olson, J.J.; Liebeskind, L.S.; Goodman, M.M. Synthesis, radiolabeling, and biological evaluation of the cis stereoisomers of 1-amino-3-fluoro-4-(fluoro-18F)cyclopentane-1-carboxylic Acid as PET imaging agents. J. Med. Chem. 2020, 63, 12008–12022. [Google Scholar] [CrossRef] [PubMed]
- Pickel, T.C.; Pashikanti, G.; Voll, R.J.; Yu, W.; Zhang, Z.; Nye, J.A.; Bacsa, J.; Olson, J.J.; Liebeskind, L.S.; Goodman, M.M. Synthesis, radiolabeling, and biological evaluation of the trans-stereoisomers of 1-amino-3-(fluoro-18F)-4-fluorocyclopentane-1-carboxylic acid as PET imaging agents. ACS Pharmacol. Transl. Sci. 2021, 4, 1195–1203. [Google Scholar] [CrossRef]
- Lacan, G.; Satyamurthy, N.; Barrio, J.R. Synthesis of stereo (R and S) and geometric (E and Z) isomers of [18F]fluoro-β-fluoromethylene-m-tyrosine derivatives: In vivo Probes of central dopaminergic function. Nucl. Med. Biol. 1999, 26, 359–363. [Google Scholar] [CrossRef]
- Hanaoka, H.; Ohshima, Y.; Yamaguchi, A.; Suzuki, H.; Ishioka, N.S.; Higuchi, T.; Arano, Y.; Tsushima, Y. Novel 18F-labeled α-methyl-phenylalanine derivative with high tumor accumulation and ideal pharmacokinetics for tumor-specific imaging. Mol. Pharm. 2019, 16, 3609–3616. [Google Scholar] [CrossRef]
- Nodwell, M.B.; Yang, H.; Merkens, H.; Malik, N.; Colovic, M.; Bjorn, W.; Martin, R.E.; Benard, F.; Schaffer, P.; Britton, R. 18F-Branched-chain amino acids: Structure-activity relationships and PET imaging potential. J. Nucl. Med. 2019, 60, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Yanai, A.; Harada, R.; Iwata, R.; Yoshikawa, T.; Ishikawa, Y.; Furumoto, S.; Ishida, T.; Yanai, K. Site-specific labeling of F-18 proteins using a supplemented cell-free protein synthesis system and O-2-[18F]fluoroethyl-l-tyrosine: [18F]FET-HER2 affibody molecule. Mol. Imaging Biol. 2019, 21, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Yin, D.; Li, G.; Wang, M.; Li, S.; Zheng, M.; Cai, H.; Wang, Y. Radiolabeling and in vitro and in vivo characterization of [18F]FB-[R8,15,21, L17]-VIP as a PET imaging agent for tumor overexpressed VIP receptors. Chem. Biol. Drug Des. 2006, 68, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Liu, Z.; Miao, Z.; Liu, H.; Subbarayan, M.; Chin, F.T.; Zhang, L.; Gambhir, S.S.; Cheng, Z. PET of malignant melanoma using 18F-labeled metallopeptides. J. Nucl. Med. 2009, 50, 1865–1872. [Google Scholar] [CrossRef]
- Rojas, S.; Nolis, P.; Gispert, J.D.; Spengler, J.; Albericio, F.; Herance, J.R.; Abad, S. Efficient cysteine labelling of peptides with N-succinimidyl 4-[18F]fluorobenzoate: Stability study and in vivo biodistribution in rats by positron emission tomography (PET). RSC Adv. 2013, 3, 8028–8036. [Google Scholar] [CrossRef]
- Yang, Y.; Richter, S.; Wuest, F.; Doschak, M.R. Synthesis and structural identification of fluorine-18 labeled parathyroid hormone. J. Label. Compd. Radiopharm. 2015, 58, 453–457. [Google Scholar] [CrossRef]
- Perreault, A.; Knight, J.C.; Wang, M.; Way, J.; Wuest, F. 18F-Labeled wild-type annexin V: Comparison of random and site-selective radiolabeling methods. Amino Acids 2016, 48, 65–74. [Google Scholar] [CrossRef]
- Yang, X.; Mease, R.C.; Pullambhatla, M.; Lisok, A.; Chen, Y.; Foss, C.A.; Wang, Y.; Shallal, H.; Edelman, H.; Hoye, A.T.; et al. [18F]Fluorobenzoyllysinepentanedioic acid carbamates: New scaffolds for positron emission tomography (PET) imaging of prostate-specific membrane antigen (PSMA). J. Med. Chem. 2016, 59, 206–218. [Google Scholar] [CrossRef]
- Gillman, K.W.; Starrett, J.E., Jr.; Parker, M.F.; Xie, K.; Bronson, J.J.; Marcin, L.R.; McElhone, K.E.; Bergstrom, C.P.; Mate, R.A.; Williams, R.; et al. Discovery and evaluation of BMS-708163, a potent, selective and orally bioavailable γ-secretase inhibitor. ACS Med. Chem. Lett. 2010, 1, 120–124. [Google Scholar] [CrossRef]
- Nakazato, A.; Kumagai, T.; Sakagami, K.; Yoshikawa, R.; Suzuki, Y.; Chaki, S.; Ito, H.; Taguchi, T.; Nakanishi, S.; Okuyama, S. Synthesis, SARs and pharmacological characterization of 2-amino-3 or 6-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid derivatives as potent, selective, and orally active group II metabotropic glutamate receptor agonists. J. Med. Chem. 2000, 43, 4893–4909. [Google Scholar] [CrossRef]
- Larsson, R.; Dhar, S.; Ehrsson, H.; Nygren, P.; Lewensohn, R. Comparison of the cytotoxic activity of melphalan with l-prolyl-m-l-sarcolysyl-l-p-fluorophenylalanine in human tumour cell lines and primary cultures of tumour cells from patients. Br. J. Cancer 1998, 78, 328–335. [Google Scholar] [CrossRef]
- Gullbo, J.; Dhar, S.; Luthman, K.; Ehrsson, H.; Lewensohn, R.; Nygren, P.; Larsson, R. Antitumor activity of the alkylating oligopeptides J1 (l-melphalanyl-p-l-fluorophenylalanine ethyl ester) and P2 (l-prolyl-m-l-sarcolysyl-p-l-fluorophenylalanine ethyl ester): Comparison with melphalan. Anti-Cancer Drugs 2003, 14, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Vine, W.H.; Hsieh, K.; Marshall, G.R. Synthesis of fluorine-containing peptides. Analogues of angiotensin II containing hexafluorovaline. J. Med. Chem. 1981, 24, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Gottler, L.M.; Lee, H.Y.; Shelburne, C.E.; Ramamoorthy, A.; Marsh, E.N.G. Using fluorous amino acids to modulate the biological activity of an antimicrobial peptide. ChemBioChem 2008, 9, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Asante, V.; Mortier, J.; Schluter, H.; Koksch, B. Impact of fluorination on proteolytic stability of peptides in human blood plasma. Bioorg. Med. Chem. 2013, 21, 3542–3546. [Google Scholar] [CrossRef] [PubMed]
- Huhmann, S.; Koksch, B. Fine-tuning the proteolytic stability of peptides with fluorinated amino acids. Eur. J. Org. Chem. 2018, 2018, 3667–3679. [Google Scholar] [CrossRef]
- Koksch, B.; Sewald, N.; Hofmann, H.; Burger, K.; Jakubke, H. Proteolytically stable peptides by incorporation of α-Tfm amino acids. J. Pept. Sci. 1997, 3, 157–167. [Google Scholar] [CrossRef]
- Krowarsch, D.; Cierpicki, T.; Jelen, F.; Otlewski, J. Canonical protein inhibitors of serine proteases. Cell. Mol. Life Sci. 2003, 60, 2427–2444. [Google Scholar] [CrossRef]
- Budisa, N.; Wenger, W.; Wiltschi, B. Residue-specific global fluorination of Candida antarctica lipase B in Pichia pastoris. Mol. Biosyst. 2010, 6, 1630–1639. [Google Scholar] [CrossRef]
- Piekielna, J.; Perlikowska, R.; do-Rego, J.C.; do-Rego, J.L.; Cerlesi, M.C.; Calo, G.; Kluczyk, A.; Lapinski, K.; Tomboly, C.; Janecka, A. Synthesis of mixed opioid affinity cyclic endomorphin-2 analogues with fluorinated phenylalanines. ACS Med. Chem. Lett. 2015, 6, 579–583. [Google Scholar] [CrossRef]
- Huhmann, S.; Stegemann, A.K.; Folmert, K.; Klemczak, D.; Moschner, J.; Kube, M.; Koksch, B. Position-dependent impact of hexafluoroleucine and trifluoroisoleucine on protease digestion. Beilstein J. Org. Chem. 2017, 13, 2869–2882. [Google Scholar] [CrossRef]
- Middendorp, S.J.; Wilbs, J.; Quarroz, C.; Calzavarini, S.; Angelillo-Scherrer, A.; Heinis, C. Peptide Macrocycle Inhibitor of Coagulation Factor XII with Subnanomolar Affinity and High Target Selectivity. J. Med. Chem. 2017, 60, 1151–1158. [Google Scholar] [CrossRef]
- Laskowski, M.; Kato, I. Protein inhibitors of proteinases. Ann. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Gauthier, J.Y.; Chauret, N.; Cromlish, W.; Desmarais, S.; Duong, L.T.; Falgueyret, J.P.; Kimmel, D.B.; Lamontagne, S.; Leger, S.; LeRiche, T.; et al. The discovery of odanacatib (MK-0822), a selective inhibitor of cathepsin K. Bioorg. Med. Chem. Lett. 2008, 18, 923–928. [Google Scholar] [CrossRef]
- Hunter, L. The C-F bond as a conformational tool in organic and biological chemistry. Beilstein J. Org. Chem. 2010, 6, 38. [Google Scholar] [CrossRef]
- O’Hagan, D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem. Soc. Rev. 2008, 37, 308–319. [Google Scholar] [CrossRef]
- Staas, D.D.; Savage, K.L.; Sherman, V.L.; Shimp, H.L.; Lyle, T.A.; Tran, L.O.; Wiscount, C.M.; McMasters, D.R.; Sanderson, P.E.; Williams, P.D.; et al. Discovery of potent, selective 4-fluoroproline-based thrombin inhibitors with improved metabolic stability. Bioorg. Med. Chem. 2006, 14, 6900–6916. [Google Scholar] [CrossRef]
- Holzberger, B.; Obeid, S.; Welte, W.; Diederichs, K.; Marx, A. Structural insights into the potential of 4-fluoroproline to modulate biophysical properties of proteins. Chem. Sci. 2012, 3, 2924–2931. [Google Scholar] [CrossRef]
- Borgogno, A.; Ruzza, P. The impact of either 4-R-hydroxyproline or 4-R-fluoroproline on the conformation and SH3m-cort binding of HPK1 proline-rich peptide. Amino Acids 2013, 44, 607–614. [Google Scholar] [CrossRef]
- Catherine, C.; Oh, S.J.; Lee, K.-H.; Min, S.-E.; Won, J.-I.; Yun, H.; Kim, D.-M. Engineering thermal properties of elastin-like polypeptides by incorporation of unnatural amino acids in a cell-free protein synthesis system. Biotechnol. Bioprocess. Eng. 2015, 20, 417–422. [Google Scholar] [CrossRef]
- Dietz, D.; Kubyshkin, V.; Budisa, N. Applying γ-substituted prolines in the foldon peptide: Polarity contradicts preorganization. ChemBioChem 2015, 16, 403–406. [Google Scholar] [CrossRef]
- Rienzo, M.; Rocchi, A.R.; Threatt, S.D.; Dougherty, D.A.; Lummis, S.C. Perturbation of Critical Prolines in Gloeobacter violaceus Ligand-gated Ion Channel (GLIC) Supports Conserved Gating Motions among Cys-loop Receptors. J. Biol. Chem. 2016, 291, 6272–6280. [Google Scholar] [CrossRef]
- Patrick, D.A.; Gillespie, J.R.; McQueen, J.; Hulverson, M.A.; Ranade, R.M.; Creason, S.A.; Herbst, Z.M.; Gelb, M.H.; Buckner, F.S.; Tidwell, R.R. Urea derivatives of 2-aryl-benzothiazol-5-amines: A new class of potential drugs for human African trypanosomiasis. J. Med. Chem. 2017, 60, 957–971. [Google Scholar] [CrossRef]
- Chandler, C.L.; List, B. Catalytic, asymmetric transannular aldolizations: Total synthesis of (+)-hirsutene. J. Am. Chem. Soc. 2008, 130, 6737–6739. [Google Scholar] [CrossRef]
- Díaz, J.; Goodman, J.M. Proline-catalyzed aldol reactions of cyclic diketones: Fluorine modifies pathways as well as transition states. Tetrahedron 2010, 66, 8021–8028. [Google Scholar] [CrossRef]
- Yap, D.Q.J.; Cheerlavancha, R.; Lowe, R.; Wang, S.; Hunter, L. Investigation of cis- and trans-4-Fluoroprolines as Enantioselective Catalysts in a Variety of Organic Transformations. Aust. J. Chem. 2015, 68, 44–49. [Google Scholar] [CrossRef][Green Version]
- Hofman, G.J.; Ottoy, E.; Light, M.E.; Kieffer, B.; Kuprov, I.; Martins, J.C.; Sinnaeve, D.; Linclau, B. Minimising conformational bias in fluoroprolines through vicinal difluorination. Chem. Commun. 2018, 54, 5118–5121. [Google Scholar] [CrossRef]
- Testa, A.; Lucas, X.; Castro, G.V.; Chan, K.H.; Wright, J.E.; Runcie, A.C.; Gadd, M.S.; Harrison, W.T.A.; Ko, E.J.; Fletcher, D.; et al. 3-Fluoro-4-hydroxyprolines: Synthesis, conformational analysis, and stereoselective recognition by the VHL E3 ubiquitin ligase for targeted protein degradation. J. Am. Chem. Soc. 2018, 140, 9299–9313. [Google Scholar] [CrossRef]
- Singh, S.; Martinez, C.-M.; Calvet-Vitale, S.; Prasad, A.K.; Prangé, T.; Dalko, P.I.; Dhimane, H. Synthesis and conformational analysis of fluorinated pipecolic acids. Synlett 2012, 23, 2421–2425. [Google Scholar] [CrossRef][Green Version]
- Chen, S.; Ruan, Y.; Lu, J.L.; Hunter, L.; Hu, X.G. Diastereoselective synthesis and conformational analysis of 4,5-difluoropipecolic acids. Org. Biomol. Chem. 2020, 18, 8192–8198. [Google Scholar] [CrossRef]
- Vance, K.M.; Simorowski, N.; Traynelis, S.F.; Furukawa, H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat. Commun. 2011, 2, 294. [Google Scholar] [CrossRef]
- Chia, P.W.; Livesey, M.R.; Slawin, A.M.; van Mourik, T.; Wyllie, D.J.; O’Hagan, D. 3-Fluoro-N-methyl-D-aspartic acid (3F-NMDA) stereoisomers as conformational probes for exploring agonist binding at NMDA receptors. Chemistry 2012, 18, 8813–8819. [Google Scholar] [CrossRef]
- Mykhailiuk, P.K.; Kubyshkin, V.; Bach, T.; Budisa, N. Peptidyl-prolyl model study: How does the electronic effect influence the amide bond conformation? J. Org. Chem. 2017, 82, 8831–8841. [Google Scholar] [CrossRef]
- Horng, J.C.; Raines, R.T. Stereoelectronic effects on polyproline conformation. Protein Sci. 2006, 15, 74–83. [Google Scholar] [CrossRef]
- Newberry, R.W.; Raines, R.T. 4-Fluoroprolines: Conformational analysis and effects on the stability and folding of peptides and proteins. Top. Heterocycl. Chem. 2017, 48, 1–25. [Google Scholar] [CrossRef]
- Chiu, H.-P.; Suzuki, Y.; Gullickson, D.; Ahmad, R.; Kokona, B.; Fairman, R.; Cheng, R.P. Helix propensity of highly fluorinated amino acids. J. Am. Chem. Soc. 2006, 128, 15556–15557. [Google Scholar] [CrossRef]
- Gerling, U.I.M.; Salwiczek, M.; Cadicamo, C.D.; Erdbrink, H.; Czekelius, C.; Grage, S.L.; Wadhwani, P.; Ulrich, A.S.; Behrends, M.; Haufe, G.; et al. Fluorinated amino acids in amyloid formation: A symphony of size, hydrophobicity and α-helix propensity. Chem. Sci. 2014, 5, 819–830. [Google Scholar] [CrossRef]
- Lim, D.S.; Lin, J.-H.; Welch, J.T. The synthesis and characterization of a pentafluorosulfanylated peptide. Eur. J. Org. Chem. 2012, 21, 3946–3954. [Google Scholar] [CrossRef]
- Eberhardt, E.S.; Panasik, N.; Raines, R.T. Inductive effects of the energetics of prolyl peptide bond isomerization: Implications for collagen folding and stability. J. Am. Chem. Soc. 1996, 118, 12261–12266. [Google Scholar] [CrossRef]
- Holmgren, S.K.; Taylor, K.M.; Bretscher, L.E.; Raines, R.T. Code for collagen’s stability deciphered. Nature 1998, 392, 666–667. [Google Scholar] [CrossRef]
- Limapichat, W.; Lester, H.A.; Dougherty, D.A. Chemical scale studies of the Phe-Pro conserved motif in the cys loop of Cys loop receptors. J. Biol. Chem. 2010, 285, 8976–8984. [Google Scholar] [CrossRef] [PubMed]
- Rubini, M.; Scharer, M.A.; Capitani, G.; Glockshuber, R. (4R)- and (4S)-fluoroproline in the conserved cis-prolyl peptide bond of the thioredoxin fold: Tertiary structure context dictates ring puckering. ChemBioChem 2013, 14, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Mosesso, R.; Dougherty, D.A.; Lummis, S.C.R. Probing Proline Residues in the Prokaryotic Ligand-Gated Ion Channel, ELIC. Biochemistry 2018, 57, 4036–4043. [Google Scholar] [CrossRef]
- Mosesso, R.; Dougherty, D.A.; Lummis, S.C.R. Proline residues in the transmembrane/extracellular domain interface loops have different behaviors in 5-HT3 and nACh receptors. ACS Chem. Neurosci. 2019, 10, 3327–3333. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Lingel, A.; Pluckthun, A. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 2001, 276, 16548–16554. [Google Scholar] [CrossRef]
- Moroder, L.; Budisa, N. Synthetic biology of protein folding. ChemPhysChem 2010, 11, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Wynn, R.; Harkins, P.C.; Richars, F.M.; Fox, R.O. Comparison of straight chain and cyclic unnatural amino acids embedded in the core of staphylococcal nuclease. Protein Sci. 1997, 6, 1621–1626. [Google Scholar] [CrossRef]
- Cornilescu, G.; Hadley, E.B.; Woll, M.G.; Markley, J.L.; Gellman, S.H.; Cornilescu, C.C. Solution structure of a small protein containing a fluorinated side chain in the core. Protein Sci. 2006, 16, 14–19. [Google Scholar] [CrossRef]
- Daeffler, K.N.; Lester, H.A.; Dougherty, D.A. Functionally important aromatic-aromatic and sulfur-π interactions in the D2 dopamine receptor. J. Am. Chem. Soc. 2012, 134, 14890–14896. [Google Scholar] [CrossRef]
- Zhong, W.; Gallivan, J.P.; Zhang, Y.; Li, L.; Lester, H.A.; Dougherty, D.A. From ab initio quantum mechanics to molecular neurobiology: A cation-π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. USA 1998, 95, 12088–12093. [Google Scholar] [CrossRef]
- Beene, D.L.; Brandt, G.S.; Zhong, W.; Zacharias, N.M.; Lester, H.A.; Dougherty, D.A. Cation-π interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry 2002, 41, 10262–10269. [Google Scholar] [CrossRef]
- Lummis, S.C.; Beene, D.L.; Harrison, N.J.; Lester, H.A.; Dougherty, D.A. A cation-π binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 2005, 12, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Marsh, E.N.G. Towards the nonstick egg: Designing fluorous proteins. Chem. Biol. 2000, 7, R153–R157. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Lee, K.-H.; Al-Hashimi, H.M.; Marsh, E.N.G. Modulating protein structure with fluorous amino acids: Increased stability and native-like structure conferred on a 4-helix bundle protein by hexafluoroleucine. J. Am. Chem. Soc. 2006, 128, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Buer, B.C.; de la Salud-Bea, R.; Al Hashimi, H.M.; Marsh, E.N.G. Engineering protein stability and specificity using fluorous amino acids: The importance of packing effects. Biochemistry 2009, 48, 10810–10817. [Google Scholar] [CrossRef] [PubMed]
- Marsh, E.N.G. Fluorinated proteins: From design and synthesis to structure and stability. Acc. Chem. Res. 2014, 47, 2878–2886. [Google Scholar] [CrossRef]
- Gottler, L.M.; de la Salud-Bea, R.; Marsh, E.N.G. The fluorous effect in proteins: Properties of α4F6, a 4-α-helix bundle protein with a fluorocarbon core. Biochemistry 2008, 47, 4480–4490. [Google Scholar] [CrossRef]
- Buer, B.C.; Meagher, J.L.; Stuckey, J.A.; Marsh, E.N.G. Structural basis for the enhanced stability of highly fluorinated proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 4810–4815. [Google Scholar] [CrossRef]
- Buer, B.C.; Meagher, J.L.; Stuckey, J.A.; Marsh, E.N.G. Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains. Protein Sci. 2012, 21, 1705–1715. [Google Scholar] [CrossRef][Green Version]
- Robalo, J.R.; Huhmann, S.; Koksch, B.; Vila Verde, A. The Multiple Origins of the Hydrophobicity of Fluorinated Apolar Amino Acids. Chem 2017, 3, 881–897. [Google Scholar] [CrossRef]
- Lee, K.; Lee, H.; Slutsky, M.M.; Anderson, J.T.; Marsh, E.N.G. Fluorous effect in proteins: De novo design and characterization of a four-α-helix bundle protein containing hexafluoroleucine. Biochemistry 2004, 43, 16277–16284. [Google Scholar] [CrossRef] [PubMed]
- Bilgiçer, B.; Fichera, A.; Kumar, K. A coiled coil with a fluorous core. J. Am. Chem. Soc. 2001, 123, 4393–4399. [Google Scholar] [CrossRef]
- Bilgiçer, B.; Xing, X.; Kumar, K. Programmed self-sorting of coiled coils with leucine and hexafluoroleucine cores. J. Am. Chem. Soc. 2001, 123, 11815–11816. [Google Scholar] [CrossRef] [PubMed]
- Renner, C.; Alefelder, S.; Bae, J.H.; Budisa, N.; Huber, R.; Moroder, L. Fluoroprolines as tools for protein design and engineering. Angew. Chem. Int. Ed. 2001, 40, 923–925. [Google Scholar] [CrossRef]
- Tang, Y.; Ghirlanda, G.; Petka, W.A.; Nakajima, T.; DeGrado, W.F.; Tirrell, D.A. Fluorinated coiled-coil proteins prepared in vivo display enhanced thermal and chemical stability. Angew. Chem. Int. Ed. 2001, 40, 1494–1496. [Google Scholar] [CrossRef]
- Tang, Y.; Ghirlanda, G.; Vaidehi, N.; Kua, J.; Mainz, D.T.; Goddard, W.A.; DeGrado, W.F.; Tirrell, D.A. Stabilization of coiled-coil peptide omains by introduction of trifluoroleucine. Biochemistry 2001, 40, 2790–2796. [Google Scholar] [CrossRef]
- Tang, Y.; Tirrell, D.A. Biosynthesis of a highly stable coiled-coil protein containing hexafluoroleucine in an engineered bacterial host. J. Am. Chem. Soc. 2001, 123, 11089–11090. [Google Scholar] [CrossRef]
- Jäckel, C.; Seufert, W.; Thust, S.; Koksch, B. Evaluation of the molecular interactions of fluorinated amino acids with native polypeptides. ChemBioChem 2004, 5, 717–720. [Google Scholar] [CrossRef]
- Salwiczek, M.; Koksch, B. Effects of fluorination on the folding kinetics of a heterodimeric coiled coil. ChemBioChem 2009, 10, 2867–2870. [Google Scholar] [CrossRef]
- Salwiczek, M.; Samsonov, S.; Vagt, T.; Nyakatura, E.; Fleige, E.; Numata, J.; Colfen, H.; Pisabarro, M.T.; Koksch, B. Position-dependent effects of fluorinated amino acids on the hydrophobic core formation of a heterodimeric coiled coil. Chemistry 2009, 15, 7628–7636. [Google Scholar] [CrossRef]
- Gottler, L.M.; de la Salud-Bea, R.; Shelburne, C.E.; Ramamoorthy, A.; Marsh, E.N.G. Using fluorous amino acids to probe the effects of changing hydrophobicity on the physical and biological properties of the β-hairpin antimicrobial peptide protegrin-1. Biochemistry 2008, 47, 9243–9250. [Google Scholar] [CrossRef] [PubMed]
- Niemz, A.; Tirrell, D.A. Self-association and mebrane-binding behaviour of melittins containing trifluoroleucine. J. Am. Chem. Soc. 2001, 123, 7407–7413. [Google Scholar] [CrossRef] [PubMed]
- Bilgiçer, B.; Kumar, K. De novo design of defined helical bundles in membrane environments. Proc. Natl. Acad. Sci. USA 2004, 101, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Cejas, M.A.; Kinney, W.A.; Chen, C.; Vinter, J.G.; Almond, H.R.; Balss, K.M.; Maryanoff, C.A.; Schmidt, U.; Breslav, M.; Mahan, A.; et al. Thrombogenic collagen-mimetic peptides: Self-assembly of triple helix-based fibrils drive by hydrophobic interactions. Proc. Natl. Acad. Sci. USA 2008, 105, 8513–8518. [Google Scholar] [CrossRef] [PubMed]
- Yuvienco, C.; More, H.T.; Haghpanah, J.S.; Tu, R.S.; Montclare, J.K. Modulating Supramolecular Assemblies and Mechanical Properties of Engineered Protein Materials by Fluorinated Amino Acids. Biomacromolecules 2012, 13, 2273–2278. [Google Scholar] [CrossRef]
- Kralj, S.; Bellotto, O.; Parisi, E.; Garcia, A.M.; Iglesias, D.; Semeraro, S.; Deganutti, C.; D’Andrea, P.; Vargiu, A.V.; Geremia, S.; et al. Heterochirality and Halogenation Control Phe-Phe Hierarchical Assembly. ACS Nano 2020, 14, 16951–16961. [Google Scholar] [CrossRef]
- Aviv, M.; Cohen-Gerassi, D.; Orr, A.A.; Misra, R.; Arnon, Z.A.; Shimon, L.J.W.; Shacham-Diamand, Y.; Tamamis, P.; Adler-Abramovich, L. Modification of a Single Atom Affects the Physical Properties of Double Fluorinated Fmoc-Phe Derivatives. Int. J. Mol. Sci. 2021, 22, 9634. [Google Scholar] [CrossRef]
- Zheng, H.; Gao, J. Highly specific heterodimerization mediated by quadrupole interactions. Angew. Chem. Int. Ed. 2010, 49, 8635–8639. [Google Scholar] [CrossRef]
- Silverman, R.B.; Abeles, R.H. Inactivation of pyridoxal phosphate dependent enzymes by mono-and polyhaloalanines. Biochemistry 1976, 15, 4718–4723. [Google Scholar] [CrossRef]
- Metcalf, B.W.; Bey, P.; Danzin, C.; Jung, M.J.; Casara, P.; Vevert, J.P. Catalytic irreversible inhibition of mammalian ornithine decarboxylase (E.C.4.1.1.17) by substrate and product analogs. J. Am. Chem. Soc. 1977, 100, 2551–2553. [Google Scholar] [CrossRef]
- Pan, Y.; Qiu, J.; Silverman, R.B. Design, synthesis, and biological activity of a difluoro-substituted, conformationally rigid vigabatrin analogue as a potent γ-aminobutyric acid aminotransferase inhibitor. J. Med. Chem. Lett 2003, 46, 5292–5293. [Google Scholar] [CrossRef] [PubMed]
- LoGiudice, N.; Le, L.; Abuan, I.; Leizorek, Y.; Roberts, S.C. Alpha-difluoromethylornithine, an irreversible inhibitor of polyamine biosynthesis, as a therapeutic strategy against hyperproliferative and infectious diseases. Med. Sci. 2018, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Ojemalm, K.; Higuchi, T.; Lara, P.; Lindahl, E.; Suga, H.; von Heijne, G. Energetics of side-chain snorkeling in transmembrane helices probed by nonproteinogenic amino acids. Proc. Natl. Acad. Sci. USA 2016, 113, 10559–10564. [Google Scholar] [CrossRef] [PubMed]
- Leppkes, J.; Dimos, N.; Loll, B.; Hohmann, T.; Dyrks, M.; Wieseke, A.; Keller, B.G.; Koksch, B. Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin. RSC Chem. Biol. 2022, 3, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, J.A.; Christianson, D.W. The contribution of halogen atoms to protein-ligand interactions. Int. J. Biol. Macromol. 1992, 14, 193–197. [Google Scholar] [CrossRef]
- Kalindjian, S.B.; Buck, I.M.; Davies, J.M.R.; Dunstone, D.J.; Hudson, M.L.; Low, C.M.R.; McDonald, I.M.; Pether, M.J.; Steel, K.I.M.; Tozer, M.J.; et al. Non-peptide cholecystokinin-B/gastrin receptor antagonists based on bicyclic, heteroaromatic skeletons. J. Med. Chem. 1996, 39, 1806–1815. [Google Scholar] [CrossRef]
- Hoyt, S.B.; London, C.; Gorin, D.; Wyvratt, M.J.; Fisher, M.H.; Abbadie, C.; Felix, J.P.; Garcia, M.L.; Li, X.; Lyons, K.A.; et al. Discovery of a novel class of benzazepinone NaV1.7 blockers: Potential treatments for neuropathic pain. Bioorg. Med. Chem. Lett. 2007, 17, 4630–4634. [Google Scholar] [CrossRef]
- Piepenbrink, K.H.; Borbulevych, O.Y.; Sommese, R.F.; Clemens, J.; Armstrong, K.M.; Desmond, C.; Do, P.; Baker, B.M. Fluorine substitutions in an antigenic peptide selectively modulate T-cell receptor binding in a minimally perturbing manner. Biochem. J. 2009, 423, 353–361. [Google Scholar] [CrossRef]
- Müller, K.; Faeh, C.; Diederich, F. Fluorine in pharmaceuticals: Looking beyond intuition. Science 2007, 317, 1881–1886. [Google Scholar] [CrossRef]
- Andersen, O.S.; Greathouse, D.V.; Providence, L.L.; Becker, M.D.; Koeppe, R.E. Importance of tryptophan dipoles for protein function: 5-Fluorination of tryptophans in gramicidin A channels. J. Am. Chem. Soc. 1998, 120, 5142–5146. [Google Scholar] [CrossRef]
- Morikubo, N.; Fukuda, Y.; Ohtake, K.; Shinya, N.; Kiga, D.; Sakamoto, K.; Asanuma, M.; Hirota, H.; Yokoyama, S.; Hoshino, T. Cation-π interaction in the polyolefin cyclization cascade uncovered by incorporating unnaturalamino acids into the catalytic sites of squalene cyclase. J. Am. Chem. Soc. 2006, 128, 13184–13194. [Google Scholar] [CrossRef]
- Pless, S.A.; Millen, K.S.; Hanek, A.P.; Lynch, J.W.; Lester, H.A.; Lummis, S.C.; Dougherty, D.A. A cation-π interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 2008, 28, 10937–10942. [Google Scholar] [CrossRef]
- He, T.; Gershenson, A.; Eyles, S.J.; Lee, Y.-J.; Liu, W.R.; Wang, J.; Gao, J.; Roberts, M.F. Fluorinated aromatic amino acids distinguish cation-π interactions from membrane insertion. J. Biol. Chem. 2015, 290, 19334–19342. [Google Scholar] [CrossRef]
- Ahern, C.A.; Eastwood, A.L.; Lester, H.A.; Dougherty, D.A.; Horn, R. A cation-π interaction between extracellular TEA and an aromatic residue in potassium channels. J. Gen. Physiol. 2006, 128, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, V.P.; Eastwood, A.L.; Dougherty, D.A.; Horn, R.; Ahern, C.A. A cation-π interaction discriminates among sodium channels that are either sensitive or resistant to tetrodotoxin block. J. Biol. Chem. 2007, 282, 8044–8051. [Google Scholar] [CrossRef] [PubMed]
- Granados, A.; Olmo, A.D.; Peccati, F.; Billard, T.; Sodupe, M.; Vallribera, A. Fluorous l-carbidopa precursors: Highly enantioselective synthesis and computational prediction of bioactivity. J. Org. Chem. 2018, 83, 303–313. [Google Scholar] [CrossRef]
- Jin, C.; Wei, L.; Ohgaki, R.; Tominaga, H.; Xu, M.; Okuda, S.; Okanishi, H.; Kawamoto, Y.; He, X.; Nagamori, S.; et al. Interaction of Halogenated Tyrosine/Phenylalanine Derivatives with Organic Anion Transporter 1 in the Renal Handling of Tumor Imaging Probes. J. Pharmacol. Exp. Ther. 2020, 375, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.F.; Armstrong, R.N. Proton configuration in the ground state and transition state of a glutathione transferase-catalyzed reaction inferred from the properties of tetradeca(3-fluorotyrosyl)glutathione transferase. J. Am. Chem. Soc. 1996, 118, 2295–2296. [Google Scholar] [CrossRef]
- Xiao, G.; Parsons, J.F.; Armstrong, R.N.; Gilliland, G.L. Crystal structure of tetradeca-(3-fluorotyrosyl)-glutathione transferase. J. Am. Chem. Soc. 1997, 119, 9325–9326. [Google Scholar] [CrossRef]
- Narjes, F.; Koehler, K.F.; Koch, U.; Gerlach, B.; Colarusso, S.; Steinkühler, C.; Brunetti, M.; Altamura, S.; De Francesco, R.; Matassa, V.G. A designed P1 cysteine mimetic for covalent and non-covalent inhibitors of HCV NS3 protease. Bioorg. Med. Chem. Lett. 2002, 12, 701–704. [Google Scholar] [CrossRef]
- Han, W.; Hu, Z.; Jiang, X.; Wasserman, Z.R.; Decicco, C.P. Glycine α-ketoamides as HCV NS3 protease inhibitors. Bioorg. Med. Chem. Lett. 2003, 13, 1111–1114. [Google Scholar] [CrossRef]
- Zheng, B.; D’Andrea, S.V.; Sun, L.-Q.; Wang, A.X.; Chen, Y.; Hrnciar, P.; Friborg, J.; Falk, P.; Hernandez, D.; Yu, F.; et al. Potent inhibitors of hepatitis C virus NS3 protease: Employment of a difluoromethyl group as a hydrogen-bond donor. ACS Med. Chem. Lett. 2018, 9, 143–148. [Google Scholar] [CrossRef]
- Lemonnier, G.; Lion, C.; Quirion, J.C.; Pin, J.P.; Goudet, C.; Jubault, P. α-Amino-β-fluorocyclopropanecarboxylic acids as a new tool for drug development: Synthesis of glutamic acid analogs and agonist activity towards metabotropic glutamate receptor 4. Bioorg. Med. Chem. 2012, 20, 4716–4726. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, L.; Otaka, A.; Smyth, M.S.; Roller, P.P.; Burke, T.R.; den Hertog, J.; Zhang, Z. Why is phosphonodifluoromethyl phenylalanine a more potent inhibitory moiety than phosphonomethyl phenylalanine towards protein-tyrosine phosphatases. Biochem. Biophys. Res. Commun. 1995, 216, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Isenegger, P.G.; Josephson, B.; Gaunt, B.; Davy, M.J.; Gouverneur, V.; Baldwin, A.J.; Davis, B.G. Posttranslational, site-directed photochemical fluorine editing of protein sidechains to probe residue oxidation state via 19F-nuclear magnetic resonance. Nat. Protoc. 2023, 18, 1543–1562. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miles, S.A.; Nillama, J.A.; Hunter, L. Tinker, Tailor, Soldier, Spy: The Diverse Roles That Fluorine Can Play within Amino Acid Side Chains. Molecules 2023, 28, 6192. https://doi.org/10.3390/molecules28176192
Miles SA, Nillama JA, Hunter L. Tinker, Tailor, Soldier, Spy: The Diverse Roles That Fluorine Can Play within Amino Acid Side Chains. Molecules. 2023; 28(17):6192. https://doi.org/10.3390/molecules28176192
Chicago/Turabian StyleMiles, Samantha A., Joshua Andrew Nillama, and Luke Hunter. 2023. "Tinker, Tailor, Soldier, Spy: The Diverse Roles That Fluorine Can Play within Amino Acid Side Chains" Molecules 28, no. 17: 6192. https://doi.org/10.3390/molecules28176192
APA StyleMiles, S. A., Nillama, J. A., & Hunter, L. (2023). Tinker, Tailor, Soldier, Spy: The Diverse Roles That Fluorine Can Play within Amino Acid Side Chains. Molecules, 28(17), 6192. https://doi.org/10.3390/molecules28176192






