Isolation and Identification of Phytocompounds from Maytenus dhofarensis and Their Biological Potentials
Abstract
1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
Solvents and Reagents
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Computational Studies of Compounds 1 and 3
3.5. Biological Assays
3.5.1. Homogenous Time-Resolved Fluorescence (HTRF) Kinase Assay
3.5.2. Antioxidant Assay Activity Using 2,2′-Diphenyl-1-picrylhydrazyl (DPPH) Radical-Scavenging Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Simmons, M.P.; Savolainen, V.; Clevinger, C.C.; Archer, R.H.; Davis, J.I. Phylogeny of the Celastraceae inferred from 26S nuclear ribosomal DNA, phytochrome B, rbcL, atpB, and morphology. Mol. Phylogenetics Evol. 2001, 19, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.G.; Delle Monache, G.; Delle Monache, F.; Marini-Bettolo, G.B. Chuchuhuasha—A drug used in folk medicine in the Amazonian and Andean areas. A chemical study of Maytenus laevis. J. Ethnopharmacol. 1982, 5, 73–77. [Google Scholar] [CrossRef]
- Veloso, C.; Oliveira, M.; Rodrigues, V.; Oliveira, C.; Duarte, L.; Teixeira, M.; Ferreira, A.; Perez, A. Evaluation of the effects of extracts of Maytenus imbricata (Celastraceae) on the treatment of inflammatory and metabolic dysfunction induced by high-refined carbohydrate diet. Inflammopharmacology 2019, 27, 539–548. [Google Scholar] [PubMed]
- Malaník, M.; Treml, J.; Rjašková, V.; Tížková, K.; Kaucká, P.; Kokoška, L.; Kubatka, P.; Šmejkal, K. Maytenus macrocarpa (Ruiz & Pav.) Briq.: Phytochemistry and Pharmacological Activity. Molecules 2019, 24, 2288. [Google Scholar] [PubMed]
- de Figueiredo, P.T.; Silva, E.W.; Cordeiro, L.V.; Barros, R.P.; Lima, E.; Scotti, M.T.; da Silva, M.S.; Tavares, J.F.; Costa, V.C.d.O. Lupanes and friedelanes, the first chemical constituents of the aerial parts of Maytenus erythroxylon Reissek. Phytochem. Lett. 2021, 45, 19–24. [Google Scholar] [CrossRef]
- Taddeo, V.A.; Castillo, U.G.; Martínez, M.L.; Menjivar, J.; Jiménez, I.A.; Núñez, M.J.; Bazzocchi, I.L. Development and validation of an HPLC-PDA method for biologically active quinonemethide triterpenoids isolated from Maytenus chiapensis. Medicines 2019, 6, 36. [Google Scholar] [CrossRef]
- Zhang, L.; Ji, M.-Y.; Qiu, B.; Li, Q.-Y.; Zhang, K.-Y.; Liu, J.-C.; Dang, L.-S.; Li, M.-H. Phytochemicals and biological activities of species from the genus Maytenus. Med. Chem. Res. 2020, 29, 575–606. [Google Scholar] [CrossRef]
- De Sousa, G.F.; de Aguilar, M.G.; Takahashi, J.A.; Alves, T.M.; Kohlhoff, M.; Vieira Filho, S.A.; Silva, G.D.; Duarte, L.P. Flavonol triglycosides of leaves from Maytenus robusta with acetylcholinesterase inhibition. Phytochem. Lett. 2017, 19, 34–38. [Google Scholar]
- Pino, L.L.; García, T.H.; Delgado-Roche, L.; Rodeiro, I.; Hernández, I.; Vilegas, W.; Spengler, I. Polyphenolic profile by FIA/ESI/IT/MSn and antioxidant capacity of the ethanolic extract from the barks of Maytenus cajalbanica (Borhidi & O. Muñiz) Borhidi & O. Muñiz. Nat. Prod. Res. 2020, 34, 1481–1485. [Google Scholar]
- Olivaro, C.; Escobal, M.; de Souza, G.; Mederos, A. Chemical characterisation and in vitr o anthelmintic activity of phenolic-rich extracts from the leaves and branches of Maytenus ilicifolia, a native plant from South America. Nat. Prod. Res. 2022, 36, 3168–3172. [Google Scholar] [CrossRef]
- Vazdekis, N.E.; Chavez, H.; Estevez-Braun, A.; Ravelo, A.G. Triterpenoids and a lignan from the aerial parts of Maytenus apurimacensis. J. Nat. Prod. 2009, 72, 1045–1048. [Google Scholar] [CrossRef]
- Okoye, F.B.C.; Agbo, M.O.; Nworu, C.S.; Nwodo, N.J.; Esimone, C.O.; Osadebe, P.O.; Proksch, P. New neolignan glycoside and an unusual benzoyl malic acid derivative from Maytenus senegalensis leaves. Nat. Prod. Res. 2015, 29, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Perestelo, N.R.; Jiménez, I.A.; Tokuda, H.; Vázquez, J.T.; Ichiishi, E.; Bazzocchi, I.L. Absolute configuration of dihydro-β-agarofuran sesquiterpenes from Maytenus jelskii and their potential antitumor-promoting effects. J. Nat. Prod. 2016, 79, 2324–2331. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Enos, J.; Muñoz-Núñez, E.; Gutiérrez, M.; Quiroz-Carreño, S.; Pastene-Navarrete, E.; Céspedes Acuña, C. Dyhidro-β-agarofurans natural and synthetic as acetylcholinesterase and COX inhibitors: Interaction with the peripheral anionic site (AChE-PAS), and anti-inflammatory potentials. J. Enzyme Inhib. Med. Chem. 2022, 37, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Núñez, M.J.; Guadaño, A.; Jiménez, I.A.; Ravelo, A.G.; González-Coloma, A.; Bazzocchi, I.L. Insecticidal Sesquiterpene Pyridine Alkaloids from Maytenus c hiapensis. J. Nat. Prod. 2004, 67, 14–18. [Google Scholar] [CrossRef]
- Santos, V.A.F.F.M.; Regasini, L.O.; Nogueira, C.u.R.; Passerini, G.D.; Martinez, I.; Bolzani, V.S.; Graminha, M.r.A.; Cicarelli, R.M.; Furlan, M. Antiprotozoal sesquiterpene pyridine alkaloids from Maytenus ilicifolia. J. Nat. Prod. 2012, 75, 991–995. [Google Scholar] [CrossRef]
- Din, A.U.; Siddiqui, B.S. Royleanine A, an Antitumor Dihydro-β-agarofuran Sesquiterpene Pyridine Alkaloid from Maytenus royleanus. J. Braz. Chem. Soc. 2022, 33, 1386–1391. [Google Scholar]
- Wang, C.; Li, C.-J.; Yang, J.-Z.; Ma, J.; Chen, X.-G.; Hou, Q.; Zhang, D.-M. Anti-inflammatory sesquiterpene derivatives from the leaves of Tripterygium wilfordii. J. Nat. Prod. 2013, 76, 85–90. [Google Scholar] [CrossRef]
- González, A.G.; Jiménez, I.A.; Ravelo, A.G.; Sazatornil, J.G.; Bazzocchi, I.L. New sesquiterpenes with antifeedant activity from Maytenus canariensis (Celastraceae). Tetrahedron 1993, 49, 697–702. [Google Scholar] [CrossRef]
- Wibowo, M.; Wang, Q.; Holst, J.; White, J.M.; Hofmann, A.; Davis, R.A. Dihydro-β-agarofurans from the roots of the Australian endemic rainforest tree Maytenus bilocularis act as leucine transport inhibitors. Phytochem 2018, 148, 71–77. [Google Scholar] [CrossRef]
- Touré, S.; Nirma, C.; Falkowski, M.; Dusfour, I.; Boulogne, I.; Jahn-Oyac, A.; Coke, M.; Azam, D.; Girod, R.; Moriou, C. Aedes aegypti larvicidal sesquiterpene alkaloids from Maytenus oblongata. J. Nat. Prod. 2017, 80, 384–390. [Google Scholar] [PubMed]
- Zhu, Y.D.; Miao, Z.H.; Ding, J.; Zhao, W.M. Cytotoxic Dihydroagarofuranoid Sesquiterpenes from the Seeds of Celastrus orbiculatus. J. Nat. Prod. 2008, 71, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.J.; Wang, M.G.; Zhu, J.B.; Zhou, W.M.; Hu, Z.N.; Ji, Z.Q. Five New Insecticidal Sesquiterpenoids from Celastrus angulatus. J. Nat. Prod. 2001, 64, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Bazzocchi, I.L.; Nunez, M.J.; Pardo, L.; Castanys, S.; Campillo, M.; Jimenez, I.A. Biological Evaluation, Structure–Activity Relationships, and Three-Dimensional Quantitative Structure–Activity Relationship Studies of Dihydro-β-agarofuran Sesquiterpenes as Modulators of P-Glycoprotein-Dependent Multidrug Resistance. J. Med. Chem. 2007, 50, 4808–4817. [Google Scholar]
- Callies, O.; Sanchez-Canete, M.P.; Gamarro, F.; Jimenez, I.A.; Castanys, S.; Bazzocchi, I.L. Restoration of Chemosensitivity in P-Glycoprotein-Dependent Multidrug-Resistant Cells by Dihydro-β-agarofuran Sesquiterpenes from Celastrus vulcanicola. J. Nat. Prod. 2015, 78, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Callies, O.; Sanchez-Canete, M.P.; Gamarro, F.; Jimenez, I.A.; Castanys, S.; Bazzocchi, I.L. Optimization by Molecular Fine Tuning of Dihydro-β-agarofuran Sesquiterpenoids as Reversers of P-Glycoprotein-Mediated Multidrug Resistance. J. Med. Chem. 2016, 59, 1880–1890. [Google Scholar]
- Chen, J.J.; Chou, T.H.; Peng, C.F.; Chen, I.S.; Yang, S.Z. Antitubercular Dihydroagarofuranoid Sesquiterpenes from the Roots of Microtropis fokienensis. J. Nat. Prod. 2007, 70, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, W.; Gong, Q.; Zhang, H.; Zhao, W.M. Neuroprotective Dihydro-β-agarofuran-Type Sesquiterpenes from the Seeds of Euonymus maackii. J. Nat. Prod. 2019, 82, 3096–3103. [Google Scholar] [CrossRef]
- Luo, Y.; Pu, X.; Luo, G.; Zhou, M.; Ye, Q.; Liu, Y.; Gu, J.; Qi, H.; Li, G.; Zhang, G. Nitrogen-Containing Dihydro-β-agarofuran Derivatives from Tripterygium wilfordii. J. Nat. Prod. 2014, 77, 1650–1657. [Google Scholar]
- Gutierrez-Nicolas, F.; Oberti, J.C.; Ravelo, A.G.; EstevezBraun, A. β-Agarofurans and Sesquiterpene Pyridine Alkaloids from Maytenus spinosa. J. Nat. Prod. 2014, 77, 1853–1863. [Google Scholar]
- Corsino, J.; da Silva Bolzani, V.; Pereira, A.M.S.; França, S.C.; Furlan, M. Bioactive sesquiterpene pyridine alkaloids from Maytenus aquifolium. Phytochemistry 1998, 48, 137–140. [Google Scholar]
- Malebo, H.M.; Wiketye, V.; Katani, S.J.; Kitufe, N.A.; Nyigo, V.A.; Imeda, C.P.; Ogondiek, J.W.; Sunguruma, R.; Mhame, P.P.; Massaga, J.J. In vivo antiplasmodial and toxicological effect of Maytenus senegalensis traditionally used in the treatment of malaria in Tanzania. Malar. J. 2015, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.; Serrano, R.; Silva, O. Maytenus heterophylla and Maytenus senegalensis, two traditional herbal medicines. J. Nat. Sci. Biol. Med. 2011, 2, 59. [Google Scholar]
- Abreu-Naranjo, R.; Arteaga-Crespo, Y.; Bravo-Sanchez, L.R.; Pérez-Quintana, M.L.; García-Quintana, Y. Response surface methodology for optimisation of total polyphenol content and antioxidant activity of extracts from Maytenus macrocarpa bark by means of ultrasound-assisted extraction. Wood Sci. Technol. 2018, 52, 1359–1376. [Google Scholar] [CrossRef]
- Niero, R.; Faloni de Andrade, S.; Cechinel Filho, V. A review of the ethnopharmacology, phytochemistry and pharmacology of plants of the Maytenus genus. Curr. Pharm. Des. 2011, 17, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Veloso, C.C.; Soares, G.L.; Perez, A.C.; Rodrigues, V.G.; Silva, F.C. Pharmacological potential of Maytenus species and isolated constituents, especially tingenone, for treatment of painful inflammatory diseases. Rev. Bras. Farmacogn. 2017, 27, 533–540. [Google Scholar] [CrossRef]
- Moo-Puc, J.A.; Martín-Quintal, Z.; Mirón-López, G.; Moo-Puc, R.E.; Quijano, L.; Mena-Rejón, G.J. Isolation and antitrichomonal activity of the chemical constituents of the leaves of Maytenus phyllanthoides Benth. (Celastraceae). Quim. Nova 2014, 37, 85–88. [Google Scholar] [CrossRef]
- Anthony, G.; Miller, M.M. Plants of Dhofar the Southern Region of Oman Traditional, Economic and Medicinal Uses; The Office of the Adviser for Conservation of the Environment, Diwan of Royal Court: Muscat, Oman, 1988; Volume 1. [Google Scholar]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09 Revision E.01.; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Degorce, F.; Card, A.; Soh, S.; Trinquet, E.; Knapik, G.P.; Xie, B. HTRF: A technology tailored for drug discovery–a review of theoretical aspects and recent applications. Curr. Chem. Genom. 2009, 3, 22. [Google Scholar] [CrossRef]
- Koul, H.K.; Pal, M.; Koul, S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer 2013, 4, 342–359. [Google Scholar] [CrossRef]
- Bulavin, D.V.; Fornace, A.J., Jr. p38 MAP kinase’s emerging role as a tumor suppressor. Adv. Cancer Res. 2004, 92, 95–118. [Google Scholar] [PubMed]
- Clayden, J.; Greeves, N.; Warren, S. Organic Chemistry; Oxford University Press Inc.: New York, NY, USA, 2012; p. 796. [Google Scholar]
- Braca, A.; De Tommasi, N.; Di Bari, L.; Pizza, C.; Politi, M.; Morelli, I. Antioxidant principles from bauhinia t arapotensis. J. Nat. Prod. 2001, 64, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Ferrigni, N.; McLaughlin, J.; Powell, R.; Smith, C., Jr. Use of potato disc and brine shrimp bioassays to detect activity and isolate piceatannol as the antileukemic principle from the seeds of Euphorbia lagascae. J. Nat. Prod. 1984, 47, 347–352. [Google Scholar] [CrossRef] [PubMed]
- MarvinView (Version 17.2.6.0) Developed by ChemAxon. 2017. Available online: http://www.chemaxon.com (accessed on 10 June 2023).
- Yuan-Yuan, H.; Lu, C.; Guo-Xu, M.; Xu-Dong, X.; Xue-Gong, J.; Fu-Sheng, D.; Xue-Jian, L.; Jing-Quan, Y. A Review on Phytochemicals of the Genus Maytenus and Their Bioactive Studies. Molecules 2021, 26, 4563. [Google Scholar]
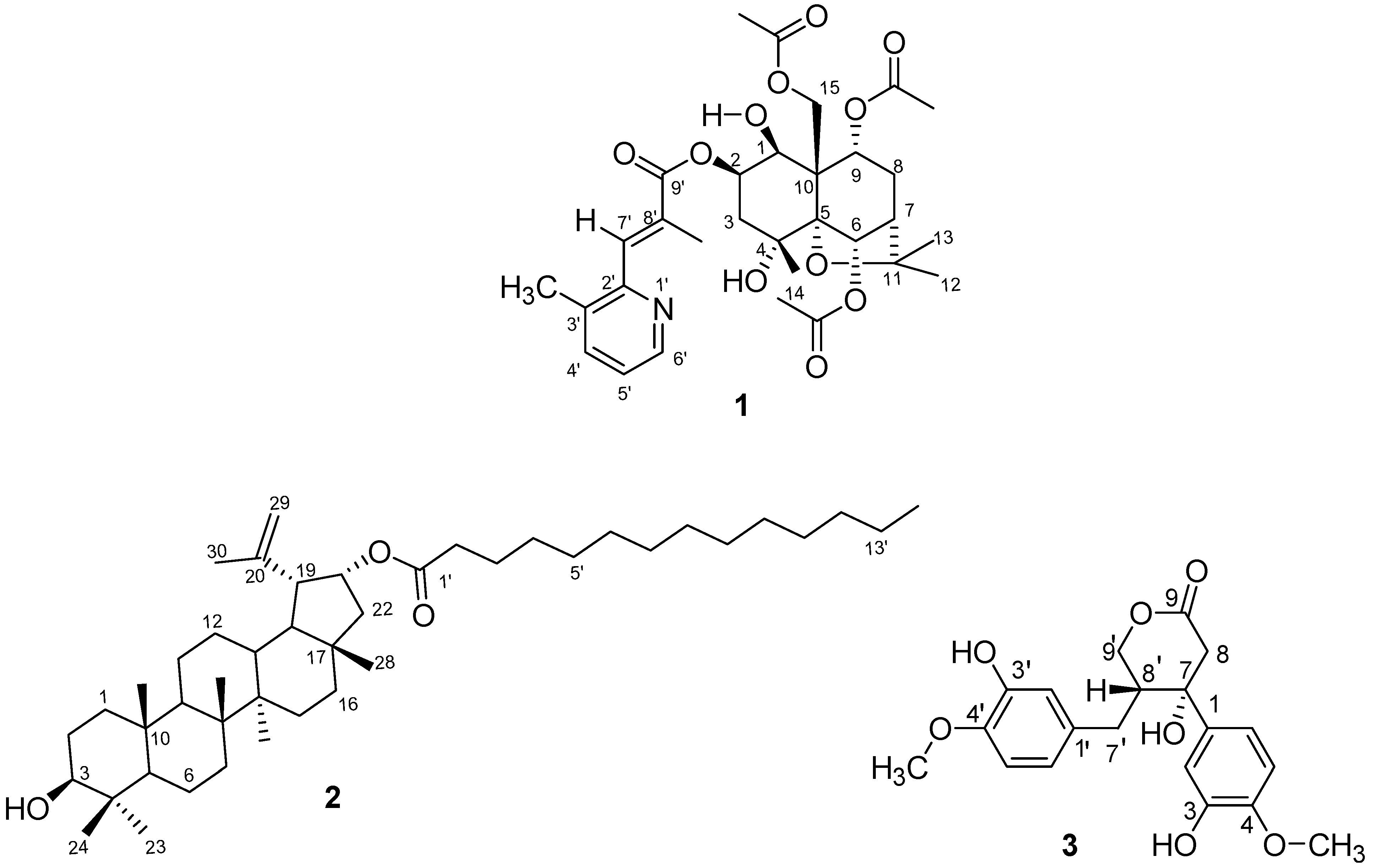
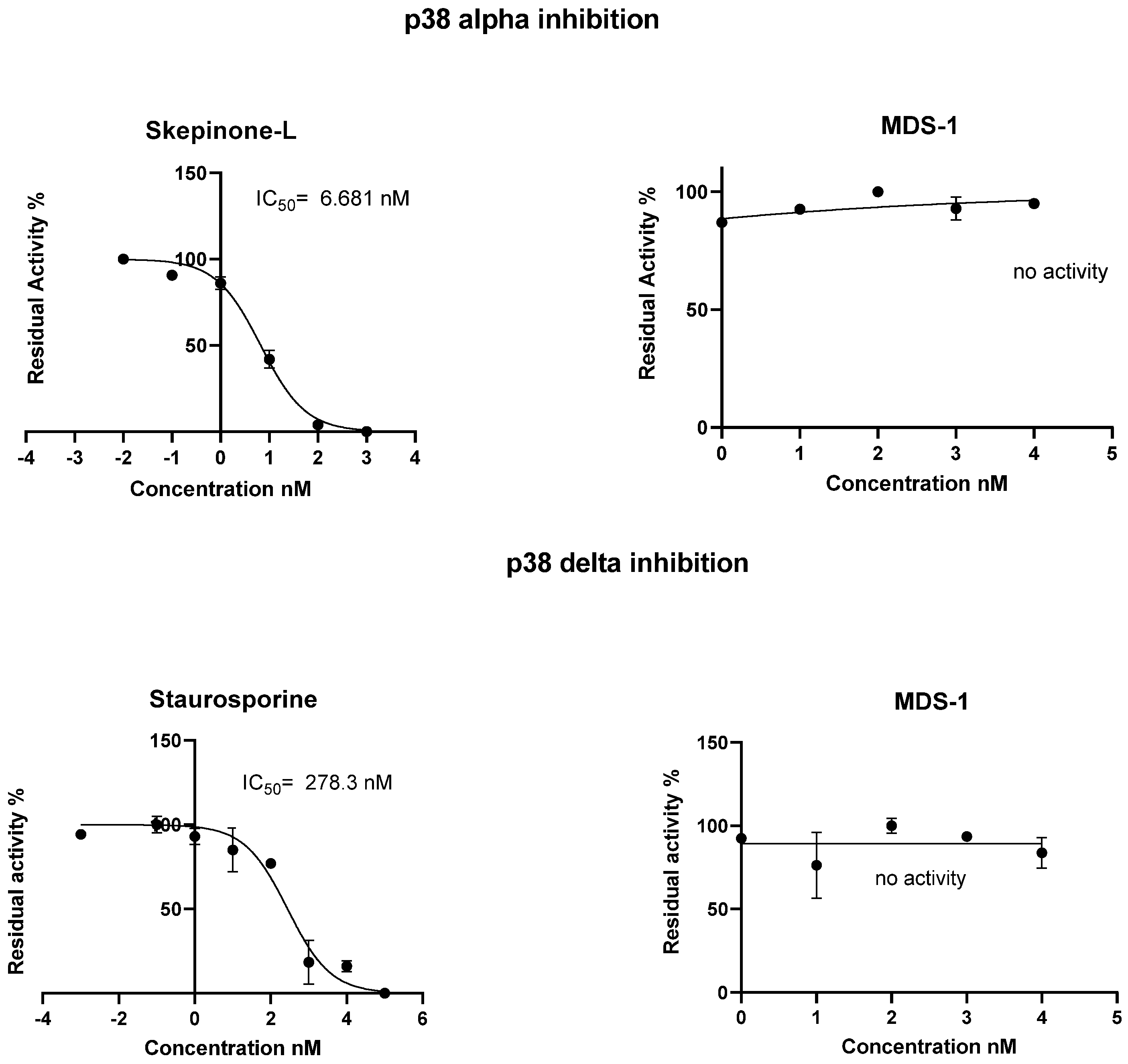
| Position | δC (ppm) | δH (ppm) (J in Hz) | HMBC (1H → 13C) |
|---|---|---|---|
| 1 | 70.6 | 5.44, d (3.2) | 2, 9′, 10, 15 |
| 2 | 68.3 | 5.53, m | - |
| 3 | 42.3 | Ha, 2.02, dd (3.2, 15.1) Hb, 2.18, dd (5.3, 16.0) | - |
| 4 | 69.9 | - | - |
| 5 | 91.1 | - | - |
| 6 | 78.7 | 6.22, s | 5, 7, 8, 10, 11, 16 |
| 7 | 49.2 | 2.35, m | - |
| 8 | 34.6 | Ha, 2.19, m Hb, 2.60, ddd (3.8, 11.1, 15.9) | - |
| 9 | 68.2 | 5.28, d (7.2) | 5, 7, 8, 10, 15 |
| 10 | 54.9 | - | - |
| 11 | 84.7 | - | - |
| 12 | 29.5 | 1.57, s | 7, 11, 13 |
| 13 | 25.7 | 1.52, s | 7, 11 |
| 14 | 24.9 | 1.49, s | 3, 5 |
| 15 | 65.5 | Ha, 4.92, d (12.8) Hb, 4.39, d (13.6) | 5, 9 5, 10 |
| 2` | 166.1 | - | - |
| 3` | 127.5 | - | - |
| 4` | 133.4 | 7.58, dd (7.4, 2.5) | 1′ |
| 5` | 128.7 | 7.48, dd (8.1, 7.6) | 3′ |
| 6` | 130.2 | 8.18, dd (8.3, 1.2) | 2′, 4′, 6′ |
| 7` | 140.2 | 6.93, dq (7.0, 1.3) | 2′, 10′, 11′ |
| 8` | 129.8 | - | - |
| 9` | 170.7 | - | - |
| 10` | 11.9 | 1.81, s | - |
| 11` | 14.7 | 1.85, s | 9` |
| OAc-6 | CH3: 20.6 | 1.78, s | - |
| C=O:169.5 | - | ||
| OAc-9 | CH3: 21.4 | 2.28, s | - |
| C=O:166.5 | - | - | |
| OAc-15 | CH3: 21.2 | 2.08, s | - |
| C=O:169.8 | - | - | |
| OH | - | 3.10 | 5, 7, 14 |
| Position | δC (ppm) | δH (ppm) (J in Hz) | HMBC (1H → 13C) |
|---|---|---|---|
| 1a | 38.5, CH2 | 0.98 a | |
| 1b | 1.61 a | ||
| 2 | 23.9, CH2 | 1.62 a, m | |
| 3 | 81.6, CH | 4.53, dd (14.0, 7.0) | 2, 4, 23, 24 |
| 4 | 37.9, C | ||
| 5 | 55.6, CH | 0.85 a | |
| 6 | 21.1, CH2 | 1.36 a | |
| 7 | 41.8, CH2 | 2.41, 2.39, dd (14.0, 7.0) | |
| 8 | 43.1, C | ||
| 9 | 50.5, CH | 1.29 a, m | |
| 10 | 37.2, C | ||
| 11 | 25.6, CH2 | 1.32 | |
| 12 | 27.6, CH2 | 0.88, t (7) | |
| 13 | 38.2, CH | 1.61 a, m | |
| 14 | 40.9, C | ||
| 15 | 34.3, CH2 | 1.35 a, m | |
| 16 | 40.1, CH2 | 1.15–1.30 a, m | |
| 17 | 42.9, C | ||
| 18 | 48.1, CH | 1.35 a, m | |
| 19 | 48.41, CH | 2.38 dd (14.0, 7.0) | 1′, 13, 18, 20, 21, 22, 29, 30 |
| 20 | 151.1, C | ||
| 21 | 68.4, CH | 3.99, m | |
| 22a | 35.7, CH2 | 1.42 a, m | |
| 22b | 1.32 a, m | ||
| 23 | 28.2, CH3 | 1.03, s | |
| 24 | 18.3, CH3 | 0.78, s | |
| 25 | 16.1, CH3 | 0.84, s | |
| 26 | 16.3, CH3 | 0.85, s | |
| 27 | 14.3, CH3 | 0.94, s | |
| 28 | 16.9, CH3 | 0.87, s | |
| 29a | 109.5, CH2 | 4.69, s | |
| 29b | 4.57, s | ||
| 30 | 18.1, CH3 | 1.68, s | |
| 1′ | 173.0, C | ||
| 2′ | 36.7, CH2 | 2.45, t (7) | |
| 3′ | 25.2, CH2 | 1.62, m | |
| 4′ | 29.5, CH2 | 1.18–1.28 a, m | |
| 5′ | 29.6, CH2 | 1.18–1.28 a, m | |
| 6′ | 29.7, CH2 | 1.18–1.28 a, m | |
| 7′ | 29.9, CH2 | 1.18–1.28 a, m | |
| 8′ | 29.8, CH2 | 1.18–1.28 a m | |
| 9′ | 29.8, CH2 | 1.18–1.28 a, m | |
| 10′ | 29.8, CH2 | 1.18–1.28 a, m | |
| 11′ | 29.7, CH2 | 1.18–1.28 a, m | |
| 12′ | 32.1, CH2 | 1.32 a, m | |
| 13′ | 22.8, CH2 | 0.79 | |
| 14′ | 14.7, CH3 | 0.88, t (7.0) | |
| OH | 2.94, br |
| Position | δC (ppm) | δH (ppm) (J in Hz) | HMBC (1H → 13C) |
|---|---|---|---|
| 1 | 130.7, C | ||
| 2 | 114.7, CH | 6.69, (br s) | |
| 3 | 144.7, C | ||
| 4 | 147.0, C | ||
| 5 | 113.1, CH | 6.63, d (7.9) | |
| 6 | 123.6, CH | 6.84, d (7.9) | |
| 7 | 76.9, C | ||
| 8a | 42.4, CH2 | 3.10, d (13.7) | 1, 2, 6, 7, 8′, 9′ |
| 8b | 2.91, d (13.7) | 1, 2, 6, 7, 8′, 9′ | |
| 9 | 179.1, C | ||
| 1′ | 126.5, C | ||
| 2′ | 111.9, CH | 6.60, (br s) | |
| 3′ | 147.0, C | ||
| 4′ | 145.4, C | ||
| 5′ | 114.9, CH | 6.82, d (7.9) | |
| 6′ | 121.9, CH | 6.62, d (7.9) | |
| 7′ a | 31.9, CH2 | 2.59, dd (9.4, 4.6) | 1′, 2′, 6′, 8′, 9′, 7, 8 |
| 7′ b | 2.49, dd (9.1, 4.5) | 1′, 2′, 6′, 8′, 9′, 7, 8 | |
| 8′ | 44.2, CH | 2.52, m | 1, 7, 8, 9, 2′, 6′, 7′, 9′ |
| 9′ a | 70.7, CH2 | 4.04, dd (8.9, 6.7) | 7, 9, 7′, 8′ |
| 9′ b | 3.99, dd (8.9, 7.8) | 7, 9, 7′, 8′ | |
| 4-OCH3 | 56.4, CH3 | 3.85, s | 3, 4 |
| 4′-OCH3 | 56.3, CH3 | 3.64, s | 3′, 4′ |
| OH | 5.5–5.8, (broad) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Rubaiai, F.; Al-Shariqi, Z.Z.; Al-Shabibi, K.S.; Husband, J.; Al-Hattali, A.M.; Goettert, M.; Laufer, S.; Baqi, Y.; Hassan, S.I.; Fatope, M.O. Isolation and Identification of Phytocompounds from Maytenus dhofarensis and Their Biological Potentials. Molecules 2023, 28, 6077. https://doi.org/10.3390/molecules28166077
Al-Rubaiai F, Al-Shariqi ZZ, Al-Shabibi KS, Husband J, Al-Hattali AM, Goettert M, Laufer S, Baqi Y, Hassan SI, Fatope MO. Isolation and Identification of Phytocompounds from Maytenus dhofarensis and Their Biological Potentials. Molecules. 2023; 28(16):6077. https://doi.org/10.3390/molecules28166077
Chicago/Turabian StyleAl-Rubaiai, Fatma, Zakiya Zahran Al-Shariqi, Khalsa S. Al-Shabibi, John Husband, Asmaa M. Al-Hattali, Marcia Goettert, Stefan Laufer, Younis Baqi, Syed Imran Hassan, and Majekodunmi O. Fatope. 2023. "Isolation and Identification of Phytocompounds from Maytenus dhofarensis and Their Biological Potentials" Molecules 28, no. 16: 6077. https://doi.org/10.3390/molecules28166077
APA StyleAl-Rubaiai, F., Al-Shariqi, Z. Z., Al-Shabibi, K. S., Husband, J., Al-Hattali, A. M., Goettert, M., Laufer, S., Baqi, Y., Hassan, S. I., & Fatope, M. O. (2023). Isolation and Identification of Phytocompounds from Maytenus dhofarensis and Their Biological Potentials. Molecules, 28(16), 6077. https://doi.org/10.3390/molecules28166077






