Identification of Xanthine Oxidase Inhibitors from Celery Seeds Using Affinity Ultrafiltration–Liquid Chromatography–Mass Spectrometry
Abstract
1. Introduction
2. Results
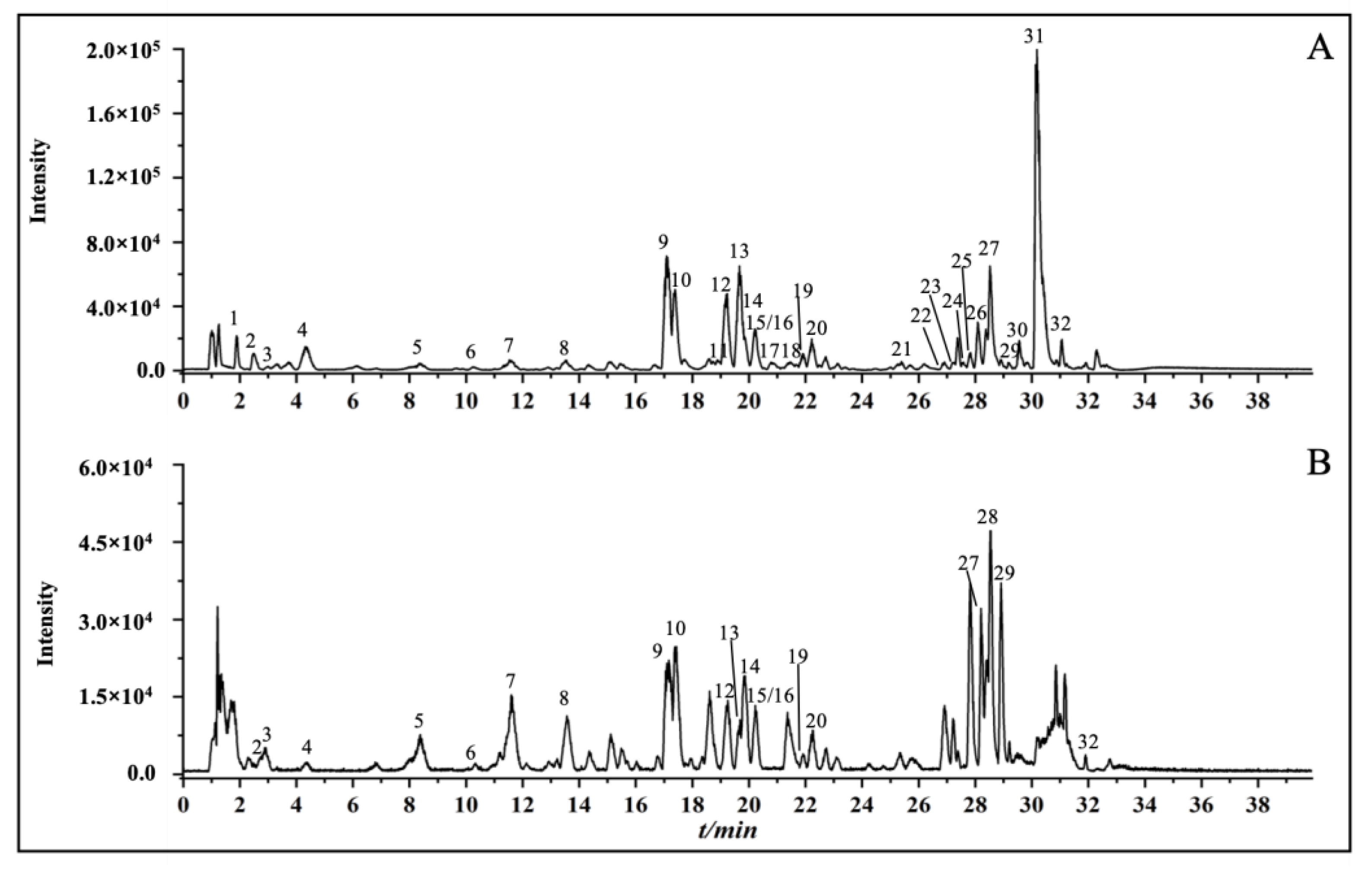
2.1. Chemical Profiles of Celery Seed Extracts
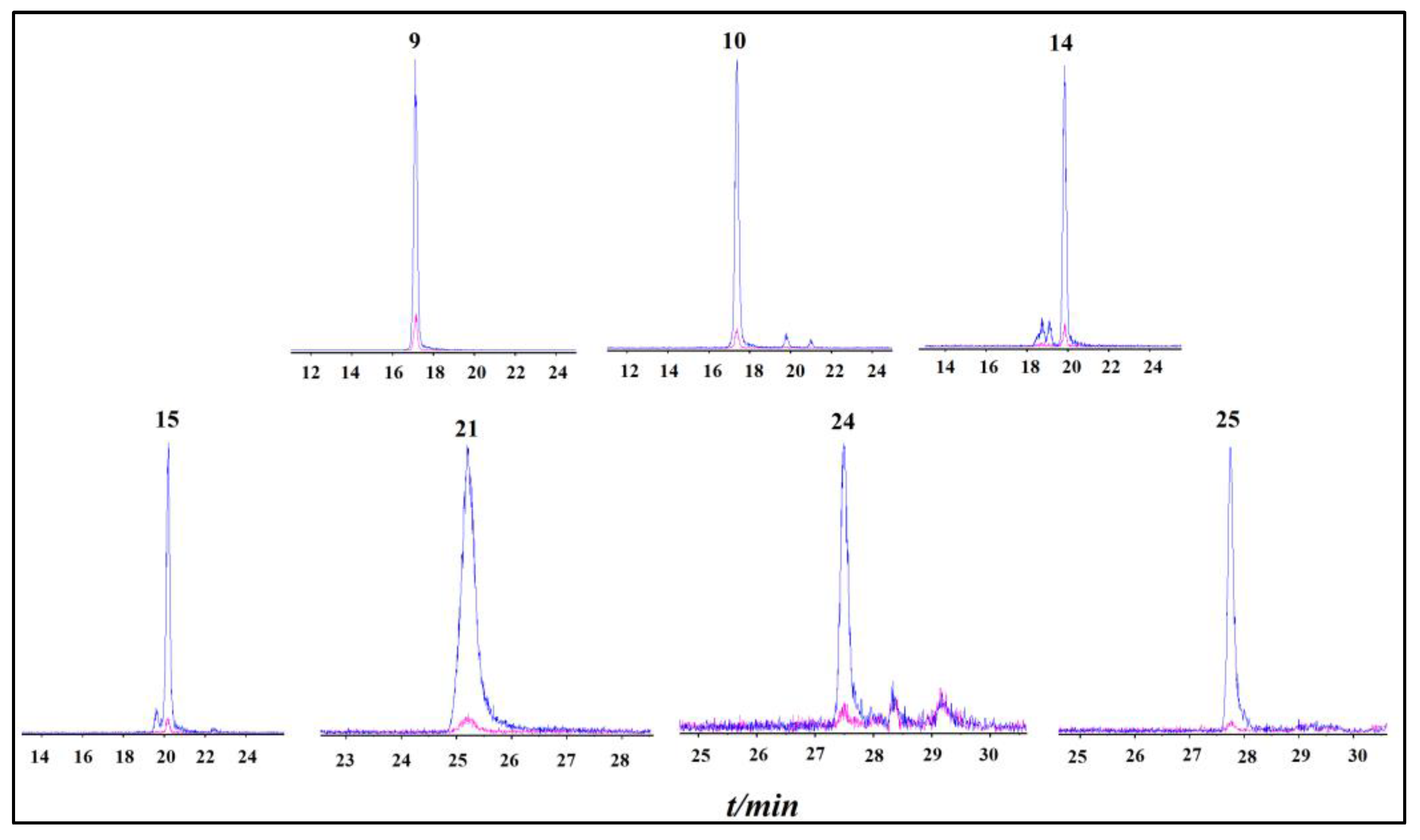
2.2. Identification of XOD-Binding Compounds from Celery Seed Extracts
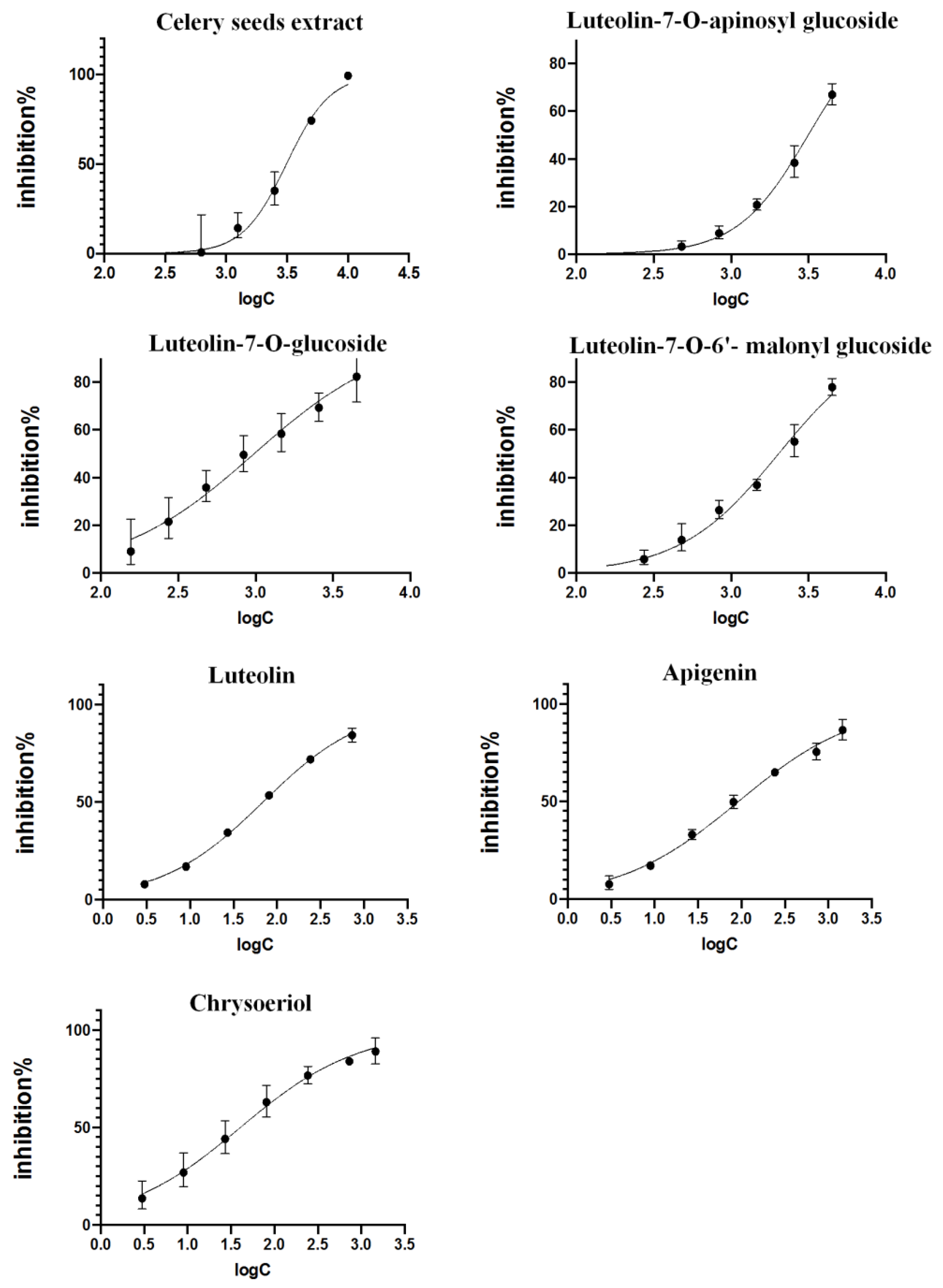
2.3. Validation of Inhibitory Activities
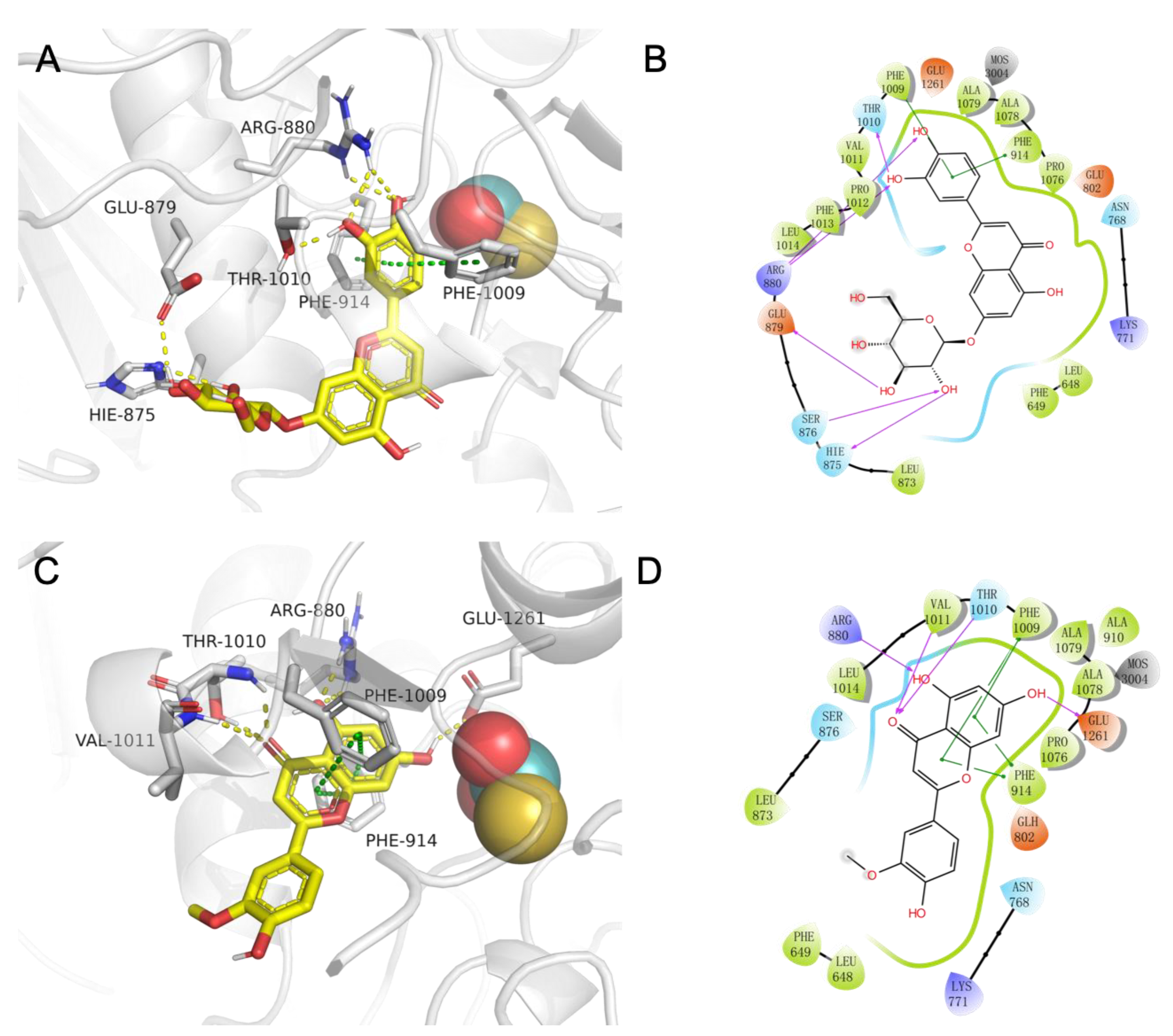
2.4. The Study of Molecular Recognition
3. Discussion
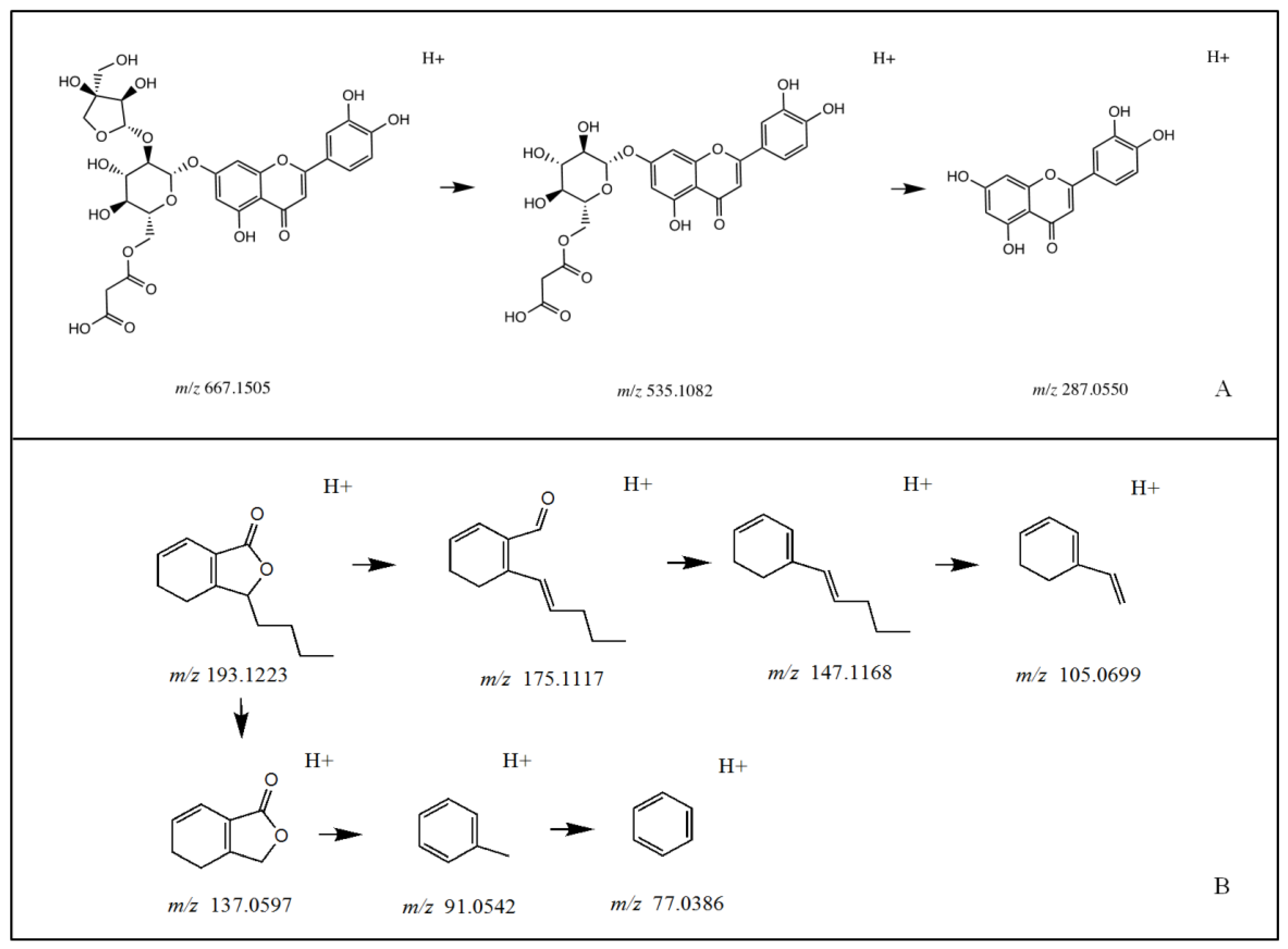
3.1. Identification of Chemical Constituents in Celery Seed Extracts
3.2. Identification of XOD-Binding Compounds from Celery Seed Extracts
3.3. Validation of Inhibitory Activities
3.4. The Study of Molecular Recognition
3.5. UF–LC–MS Assay for Screening Novel XOD Inhibitors
4. Materials and Methods
4.1. Materials and Reagents
4.2. Instruments
4.3. Chromatographic Analysis
4.4. Mass Spectrometry Conditions
4.5. Preparation of Celery Seed Extract
4.6. XOD Activity Assay
4.7. Screening for XOD Inhibitors Using UF–LC–MS
4.8. Molecular Docking
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Teng, Y.; Wang, G.B.; Wang, B.X.; Ma, A.J.; Wang, C.L. Assessment of the correlation between aquatic purine and human gout. J. Food Saf. Qual. 2019, 10, 7207–7211. [Google Scholar]
- Ruoff, G.; Edwards, N.L. Overview of serum uric acid treatment targets in gout: Why less than 6 mg/dL? Postgrad. Med. 2016, 128, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, X.; Wang, J.; Liu, H.; Kwong, J.S.; Chen, H.; Li, L.; Chung, S.C.; Shah, A.; Chen, Y.; et al. Diagnosis and treatment for hyperuricemia and gout: A systematic review of clinical practice guidelines and consensus statements. BMJ Open 2019, 9, e026677. [Google Scholar] [CrossRef]
- Kanemitsu, T.; Tsurudome, Y.; Kusunose, N.; Oda, M.; Matsunaga, N.; Koyanagi, S.; Ohdo, S. Periodic variation in bile acids controls circadian changes in uric acid via regulation of xanthine oxidase by the orphan nuclear receptor PPARα. J. Biol. Chem. 2017, 292, 21397–21406. [Google Scholar] [CrossRef]
- Thu, H.N.; Van, P.N.; Minh, K.N.; Thi, T.L. Optimization of extraction conditions of flavonoids from celery seed using response surface methodology. J. Food Meas. Charact. 2021, 15, 134–143. [Google Scholar]
- Maruyama, T.; Abbaskhan, A.; Choudhary, M.I.; Tsuda, Y.; Goda, Y.; Farille, M.; Reduron, J.P. Botanical origin of Indian celery seed (fruit). J. Nat. Med. 2009, 63, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Zorga, J.; Kunicka-Styczyńska, A.; Gruska, R.; Śmigielski, K. Ultrasound-Assisted Hydrodistillation of Essential Oil from Celery Seeds (Apium graveolens L.) and Its Biological and Aroma Profiles. Molecules 2020, 25, 5322. [Google Scholar] [CrossRef]
- Sowbhagya, H.B. Chemistry, technology, and nutraceutical functions of celery (Apium graveolens L.): An overview. Crit. Rev. Food Sci. Nutr. 2014, 54, 389–398. [Google Scholar] [CrossRef]
- Tashakori-Sabzevar, F.; Razavi, B.M.; Imenshahidi, M.; Daneshmandi, M.; Fatehi, H.; Sarkarizi, Y.E.; Mohajeri, S.A. Evaluation of mechanism for antihypertensive and vasorelaxant effects of hexanic and hydroalcoholic extracts of celery seed in normotensive and hypertensive rats. Rev. Bras. Farmacogn. 2016, 26, 619–626. [Google Scholar] [CrossRef]
- Li, S.P.; Li, L.Z.; Yan, H.; Jiang, X.; Hu, W.W.; Han, N.; Wang, D. Anti-gouty arthritis and anti-hyperuricemia properties of celery seed extracts in rodent models. Mol. Med. Rep. 2019, 20, 4623–4633. [Google Scholar] [CrossRef]
- Xu, N. Active Constituents and Anti-Gout Research of Apium Graveolens Seeds; Naval Medical University: Shanghai, China, 2012. [Google Scholar]
- Thu Hang, N.; Viet Hoang, L.; Van Phuong, N. Spectrum-effect relationship between high-performance thin-layer chromatography data and xanthine oxidase inhibitory activity of celery seed extract. Biomed. Chromatogr. 2021, 35, e5181. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhang, X.J.; Tian, X.; Wu, G.H. Pharmaceutical applications of affinity-ultrafiltration mass spectrometry: Recent advances and future prospects. J. Pharm. Biomed. Anal. 2016, 131, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Jiang, H.; Xu, C.; Tong, S.; Yan, J. Screening and identification of α-glucosidase inhibitors from Shenqi Jiangtang Granule by ultrafiltration liquid chromatography and mass spectrometry. J. Sep. Sci. 2018, 41, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Liao, X.; Chen, Z. Screening and characterization of potential α-glucosidase inhibitors from Cercis chinensis Bunge fruits using ultrafiltration coupled with HPLC-ESI-MS/MS. Food Chem. 2022, 372, 131316. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Yu, R.R.; Ai, N.; Fan, X.H. Rapid screening natural-origin lipase inhibitors from hypolipidemic decoctions by ultrafiltration combined with liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2015, 104, 67–74. [Google Scholar] [CrossRef]
- Wang, S.Q.; Huai, J.X.; Shang, Y.; Xie, L.L.; Cao, X.T.; Liao, J.; Zhang, T.; Dai, R.H. Screening for natural inhibitors of 5-lipoxygenase from Zi-shen pill extract by affinity ultrafiltration coupled with ultra performance liquid chromatography-mass spectrometry. J. Ethnopharmacol. 2020, 254, 112733. [Google Scholar] [CrossRef]
- Tao, Y.; Cai, H.; Li, W.D.; Cai, B.C. Ultrafiltration coupled with high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry for screening lipase binders from different extracts of Dendrobium officinale. Anal. Bioanal. Chem. 2015, 407, 6081–6093. [Google Scholar] [CrossRef]
- Lei, X.Q.; Li, G.; Cheng, L.; Wang, X.L.; Meng, F.Y. Identification of Ligustici Rhizoma et Radix and its adulterants based on their chemical constituents by UHPLC-Q/TOF-MS combined with data mining. J. Pharm. Biomed. Anal. 2018, 154, 123–137. [Google Scholar] [CrossRef]
- Lin, L.Z.; Lu, S.; Harnly, J.M. Detection and quantification of glycosylated flavonoid malonates in celery, Chinese celery, and celery seed by LC-DAD-ESI/MS. J. Agric. Food Chem. 2007, 55, 1321–1326. [Google Scholar] [CrossRef]
- Qu, C.C.; Wang, Y.; Zhang, B.; Lin, Z.J. Advances in pharmacology research of celery seed. China J. Tradit. Chin. Med. Pharm. 2019, 34, 5295–5299. [Google Scholar]
- Xu, F.; Zhang, L.; Zhao, X.; Zhou, Q.L.; Liu, G.X.; Yang, X.W.; Yang, D.H.; Cai, S.Q. Eleven absorbed constituents and 91 metabolites of Chuanxiong Rhizoma Decoction in rats. World J. Tradit. Chin. Med. 2021, 7, 33–46. [Google Scholar]
- Li, J. Study on Quality of Celery Seed and Other Common Uygur Herbs; Xinjiang Medical University: Ürümqi, China, 2013. [Google Scholar]
- Xu, N.; Chen, H.S. Quantitative analysis of multi-components with single marker in simultaneous determination of 3-n-butylphthalide, sedenenolide, and sedanolide in celery seed extract. Acad. J. Second Mil. Med. Univ. 2013, 34, 195–198. [Google Scholar] [CrossRef]
- Lyu, Q.; Kuo, T.H.; Sun, C.; Chen, K.; Hsu, C.C.; Li, X. Comprehensive structural characterization of phenolics in litchi pulp using tandem mass spectral molecular networking. Food Chem. 2019, 282, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Pool, E.; Ramos-Díaz, A.L.; Lizardi-Jiménez, M.A.; Pech-Cohuo, S.; Ayora-Talavera, T.; Cuevas-Bernardino, J.C.; García-Cruz, U.; Pacheco, N. Effect of solvent polarity on the Ultrasound Assisted extraction and antioxidant activity of phenolic compounds from habanero pepper leaves (Capsicum chinense) and its identification by UPLC-PDA-ESI-MS/MS. Ultrason. Sonochem. 2021, 76, 105658. [Google Scholar] [CrossRef]
- Guo, Z.Q.; Feng, Y.L.; Wang, J.H. A new strategy for non-targeted screening of phthalate metabolites using liquid chromatography-high resolution mass spectrometry. Int. J. Mass Spectrom. 2019, 443, 46–52. [Google Scholar] [CrossRef]
- Gu, D.Y.; Lv, J.L.; Gulinar, S.; HajiAkber, A. Determination of 3-n-butylphthalide in celery seed by HPLC. Lishizhen Med. Mater. Med. Res. 2007, 18, 513–514. [Google Scholar]
- Zhang, H.; Liu, C.; Wang, M.; Sui, Y. Metabolic profiling of senkyunolide A and identification of its metabolites in hepatocytes by ultra-high-performance liquid chromatography combined with diode-array detector and high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8894. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, M.; Jiang, B.; Yu, J.; Hao, Q.; Liu, W.; Hu, Z.; Zhang, Y.; Song, C. Extraction optimization, structural characterization and potential alleviation of hyperuricemia by flavone glycosides from celery seeds. Food Funct. 2022, 13, 9832–9846. [Google Scholar] [CrossRef]
- Wen, K.; Fang, X.; Yang, J.; Yao, Y.; Nandakumar, K.S.; Salem, M.L.; Cheng, K. Recent Research on Flavonoids and their Biomedical Applications. Curr. Med. Chem. 2021, 28, 1042–1066. [Google Scholar] [CrossRef]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Ali Shah, S.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial Effects of Flavonoids and Their Structure-Activity Relationship Study: A Comparative Interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef]
- Tang, H.; Huang, L.; Sun, C.; Zhao, D. Exploring the structure-activity relationship and interaction mechanism of flavonoids and α-glucosidase based on experimental analysis and molecular docking studies. Food Funct. 2020, 11, 3332–3350. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, Y.; Si, Y.; Wang, W.; Zhang, S.; Sun, S.; Liu, X.; Wang, R.; Wang, W. Isolation, characterization, and xanthine oxidase inhibitory activities of flavonoids from the leaves of Perilla frutescens. Nat. Prod. Res. 2020, 34, 2566–2572. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, B.X.; Guo, M. Current advances in screening for bioactive components from medicinal plants by affinity ultrafiltration mass spectrometry. Phytochem. Anal. 2018, 29, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, B.; Cao, J.; Zheng, H.; Ye, L.H. Ligand fishing based on bioaffinity ultrafiltration for screening xanthine oxidase inhibitors from citrus plants. J. Sep. Sci. 2021, 44, 1353–1360. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, G.; Zhang, Y.; Yang, M.; Chen, J.; Guo, M. Potential hypoglycemic, hypolipidemic, and anti-inflammatory bioactive components in Nelumbo nucifera leaves explored by bioaffinity ultrafiltration with multiple targets. Food Chem. 2022, 375, 131856. [Google Scholar] [CrossRef]
- Fan, M.X.; Chen, G.L.; Guo, M.Q. Potential Antioxidative Components in Azadirachta indica Revealed by Bio-Affinity Ultrafiltration with SOD and XOD. Antioxidants 2022, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Sawikowska, A.; Piasecka, A.; Kachlicki, P.; Krajewski, P. Separation of Chromatographic Co-Eluted Compounds by Clustering and by Functional Data Analysis. Metabolites 2021, 11, 214. [Google Scholar] [CrossRef]
- Grewal, H.K.; Martinez, J.R.; Espinoza, L.R. Febuxostat: Drug review and update. Expert Opin. Drug Metab. Toxicol. 2014, 10, 747–758. [Google Scholar] [CrossRef]
- Zhong, H.; Abdullah; Zhang, Y.P.; Deng, L.L.; Zhao, M.J.; Tang, J.; Zhang, H.; Feng, F.Q.; Wang, J. Exploring the potential of novel xanthine oxidase inhibitory peptide (ACECD) derived from Skipjack tuna hydrolysates using affinity-ultrafiltration coupled with HPLC-MALDI-TOF/TOF-MS. Food Chem. 2021, 347, 129068. [Google Scholar] [CrossRef]
- Song, H.P.; Zhang, H.; Fu, Y.; Mo, H.Y.; Zhang, M.; Chen, J.; Li, P. Screening for selective inhibitors of xanthine oxidase from Flos Chrysanthemum using ultrafiltration LC-MS combined with enzyme channel blocking. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 961, 56–61. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
| NO | Rt (min) | Identification | MS+ (m/z) | ppm | Element Composition | MS2 Fragment Ion (m/z) | References |
|---|---|---|---|---|---|---|---|
| 1 | 1.89 | Adenosine | 268.1042 | 0.6 | C10H13N5O4 | 136.0620; 119.0350 | * |
| 2 | 2.50 | Fructose-phenylalanine | 328.1390 | −0.2 | C15H21NO7 | 310.1326; 292.1213; 254.1228; 246.1097; 166.0861; 132.0792 | [18] |
| 3 | 2.99 | Pantothenic acid | 220.1176 | −1.6 | C9H17NO5 | 220.1164; 202.1063; 184.0972; 128.0257; 90.0550 | * |
| 4 | 4.38 | Tryptophan | 205.0972 | −0.8 | C11H12N2O2 | 188.0696; 146.0593; 118.0645; 115.0539 | [19] * |
| 5 | 8.35 | Chlorogenic acid | 355.1020 | −1.0 | C16H18O9 | NA | [20] * |
| 6 | 10.62 | p-Hydroxybenzaldehyde | 123.0441 | 0.4 | C7H6O2 | 123.0465; 95.0482; 77.0383 | [21] * |
| 7 | 11.54 | Coumaroylquinic acid | 339.1075 | 0.2 | C16H18O8 | 147.0436; 119.0481; 91.0532 | [20] |
| 8 | 13.54 | Coumaroylquinic acid or isomer | 339.1076 | 0.5 | C16H18O8 | 147.0440; 119.0489; 91.0541 | [20] |
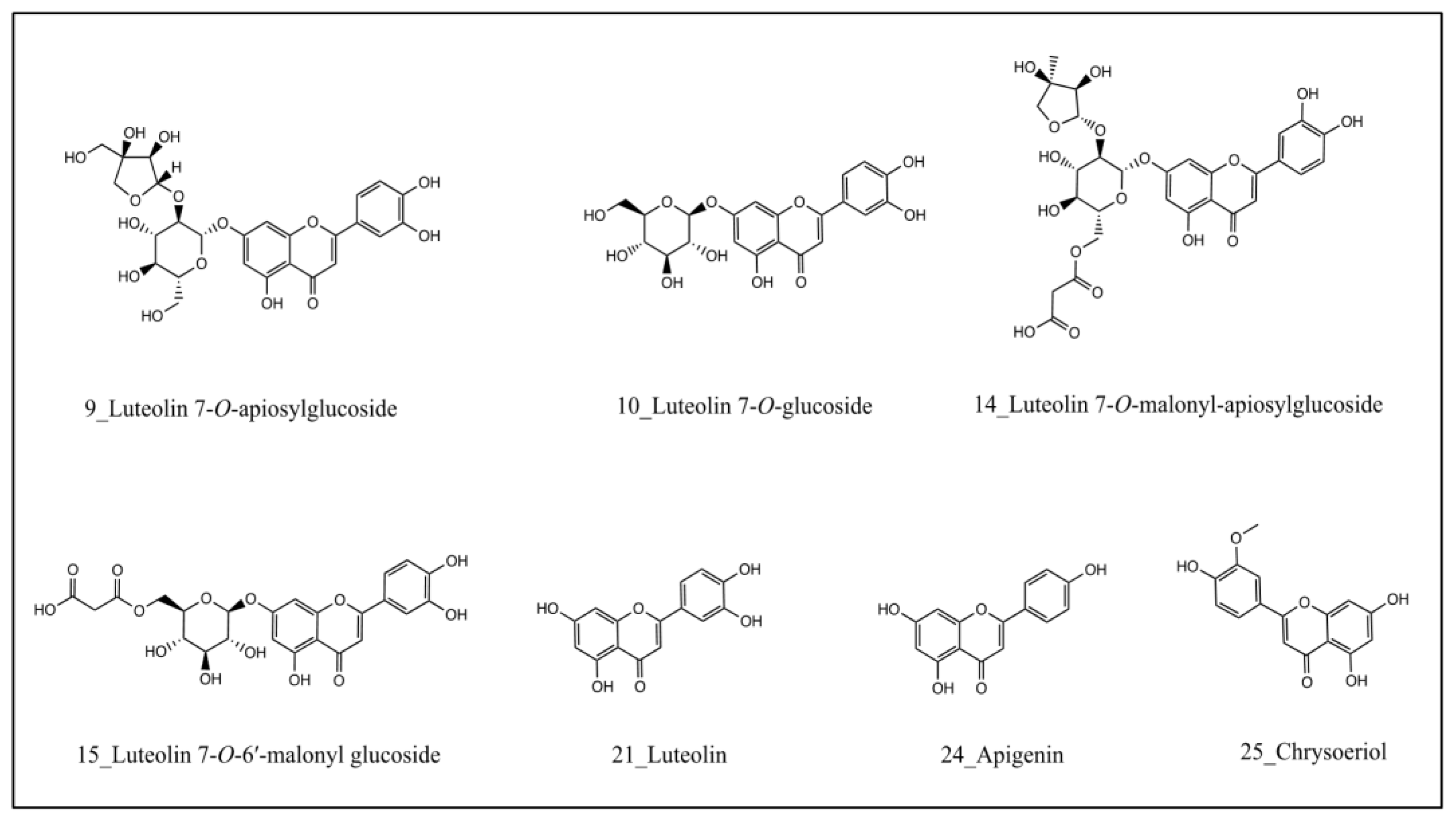
| 9 | 17.11 | Luteolin 7-O-apiosylglucoside | 581.1502 | 0.2 | C26H28O15 | 287.0554 | [20] |
| 10 | 17.38 | Luteolin 7-O-glucoside | 449.1075 | −0.8 | C21H20O11 | 287.0543 | [20] * |
| 11 | 18.91 | Senkyunolide J | 227.1274 | −1.7 | C12H18O4 | 209.1164; 191.1134; 153.0542 | [19,22] |
| 12 | 19.20 | Apiin | 565.1553 | 0.2 | C26H28O14 | 271.0609 | [20] * |
| 13 | 19.65 | Chrysoeriol 7-O-apiosylglucoside | 595.1652 | −0.9 | C27H30O15 | 463.1226; 301.0701; 286.0463; 258.0526 | [20] |
| 14 | 19.85 | Luteolin 7-O-malonyl-apiosylglucoside | 667.1488 | −2.5 | C29H30O18 | 535.1115; 287.0558; | [20] |
| 15 | 20.22 | Luteolin 7-O-6′-malonyl glucoside | 535.1082 | −0.1 | C24H22O14 | 535.1062; 449.1039; 287.0529 | [20] |
| 16 | 20.32 | Chrysoeriol 7-O-glucoside | 463.1234 | −0.2 | C22H22O11 | 301.0694; 286.0507 | [20] |
| 17 | 20.85 | Malonylapiin A | 651.1539 | −2.6 | C29H30O17 | NA | [20] |
| 18 | 21.15 | Malonylapiin B | 651.1547 | −1.3 | C29H30O17 | 519.1109; 271.0594 | [20] |
| 19 | 21.91 | 6″-Malonylapiin | 651.1549 | −1.0 | C29H30O17 | 519.1121; 271.0556 | [20] |
| 20 | 22.22 | Chrysoeriol 7-O-6″-malonyl apiosylglucoside | 681.1661 | −0.1 | C30H32O18 | 549.1281; 301.0717; 286.0474 | [20] |
| 21 | 25.19 | Luteolin | 287.0552 | 0.6 | C15H10O6 | NA | [23] * |
| 22 | 26.91 | Sedanolide | 195.1380 | 0.2 | C12H18O2 | 177.1276; 149.1323; 125.0617; 111.0436; 97.0645; 79.0542 | [23,24] * |
| 23 | 27.22 | Isomer of procyanidin B1 | 579.1487 | −1.7 | C30H26O12 | 427.0852; 409.0739; 291.0699; 247.0802; 205.0693 | [25] |
| 24 | 27.50 | Apigenin | 271.0605 | 1.5 | C15H10O5 | 271.0584; 163.0352; 153.0182; 119.0480 | [26] * |
| 25 | 27.75 | Chrysoeriol | 301.0707 | −0.9 | C16H12O6 | 301.0710; 286.0476; 258.0500; 229.0465 | [20,26] |
| 26 | 27.82 | Dihydroxy-3-butylphthalide | 223.0969 | 1.9 | C12H14O4 | 177.0905; 167.0327; 149.0218; 121.0271 | [27] |
| 27 | 28.22 | Lunularic acid | 257.0841 | 8.4 | C15H14O4 | 213.0919; 171.0809; 107.0497; 106.0415 | * |
| 28 | 28.53 | 3-Butylphthalide | 191.1067 | 0.2 | C12H14O2 | 129.0684; 117.0703; 91.0526; 77.0383 | [21,28] |
| 29 | 28.92 | Senkyunolide F | 207.1016 | 0.1 | C12H14O3 | 189.0882; 161.0959; 151.0386; 146.0726; 105.0328; 77.0381 | [11,19] |
| 30 | 29.56 | Cinnamaldehyde | 133.0648 | 0.1 | C9H8O | 133.0644; 105.0700; 79.0536; 77.0739 | * |
| 31 | 30.19 | Sedenenolide | 193.1222 | −0.6 | C12H16O2 | 175.1126; 147.1175; 137.0603; 105.0696; 91.0540; 77.0383 | [29] |
| 32 | 31.91 | Linolenic acid | 279.2317 | −0.6 | C18H30O2 | 131.0828; 95.0853; 67.0531 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, X.; Peng, B.; Chen, L.; Jiang, Y.; Li, T.; Li, B.; Liu, X. Identification of Xanthine Oxidase Inhibitors from Celery Seeds Using Affinity Ultrafiltration–Liquid Chromatography–Mass Spectrometry. Molecules 2023, 28, 6048. https://doi.org/10.3390/molecules28166048
Gan X, Peng B, Chen L, Jiang Y, Li T, Li B, Liu X. Identification of Xanthine Oxidase Inhibitors from Celery Seeds Using Affinity Ultrafiltration–Liquid Chromatography–Mass Spectrometry. Molecules. 2023; 28(16):6048. https://doi.org/10.3390/molecules28166048
Chicago/Turabian StyleGan, Xiaona, Bo Peng, Liang Chen, Yanjun Jiang, Tingzhao Li, Bo Li, and Xiaodong Liu. 2023. "Identification of Xanthine Oxidase Inhibitors from Celery Seeds Using Affinity Ultrafiltration–Liquid Chromatography–Mass Spectrometry" Molecules 28, no. 16: 6048. https://doi.org/10.3390/molecules28166048
APA StyleGan, X., Peng, B., Chen, L., Jiang, Y., Li, T., Li, B., & Liu, X. (2023). Identification of Xanthine Oxidase Inhibitors from Celery Seeds Using Affinity Ultrafiltration–Liquid Chromatography–Mass Spectrometry. Molecules, 28(16), 6048. https://doi.org/10.3390/molecules28166048





