Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition
Abstract
1. Introduction
2. Results
2.1. Metabolite Library Construction
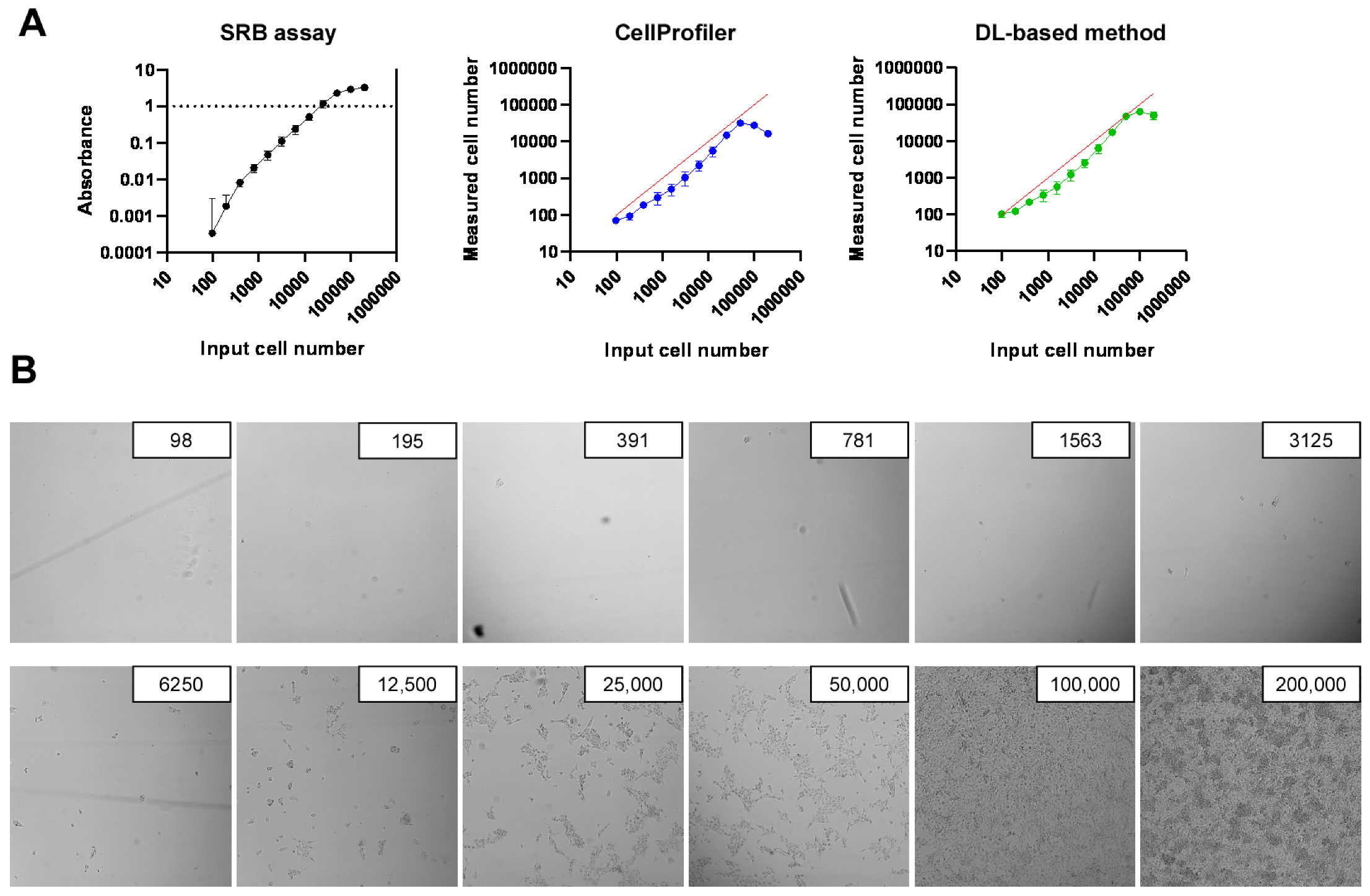
2.2. Development of High-Throughput Methods for Assessing Cell Proliferation
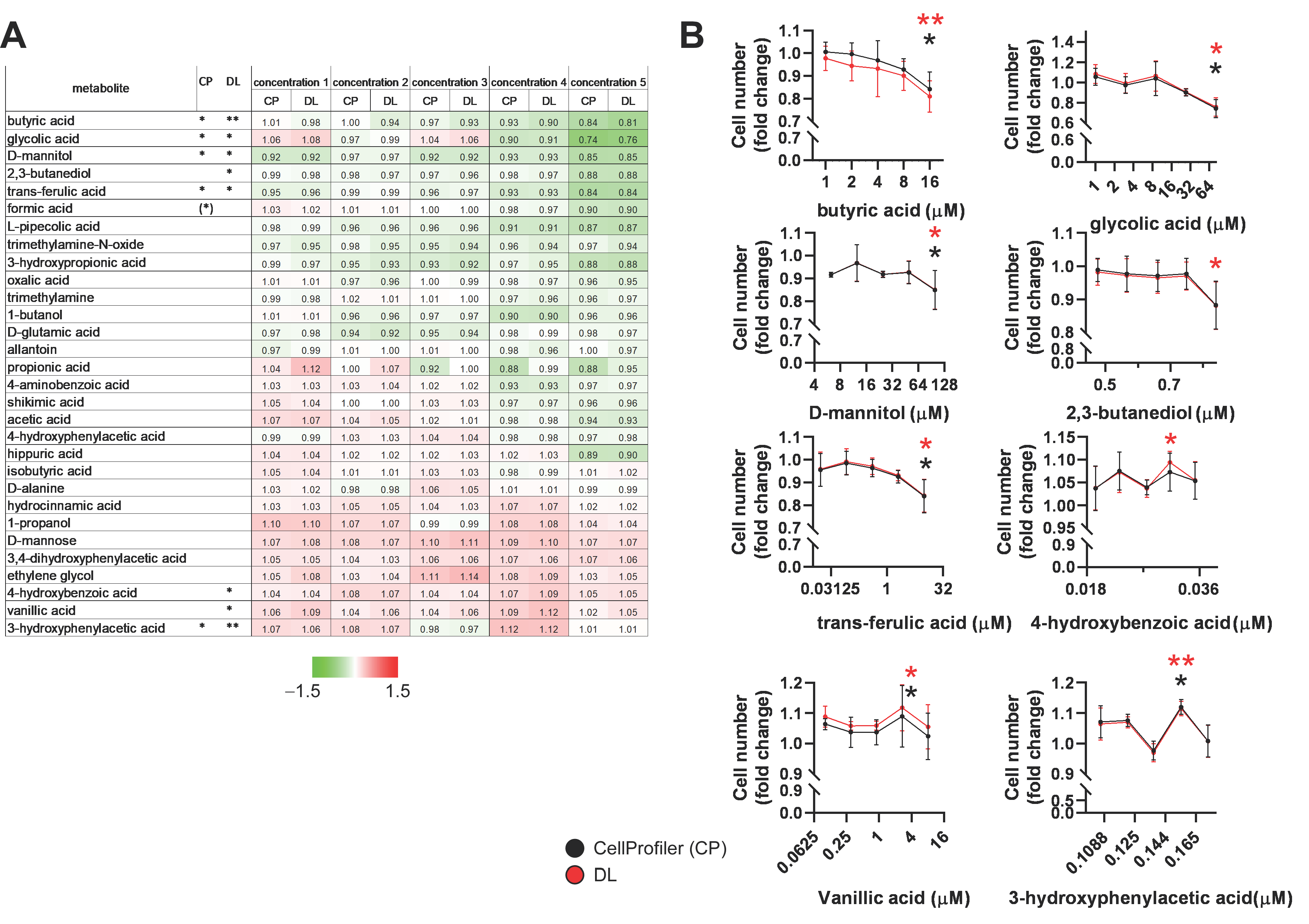
2.3. Identification of Bacterial Metabolites with Pro and Antiproliferative Properties
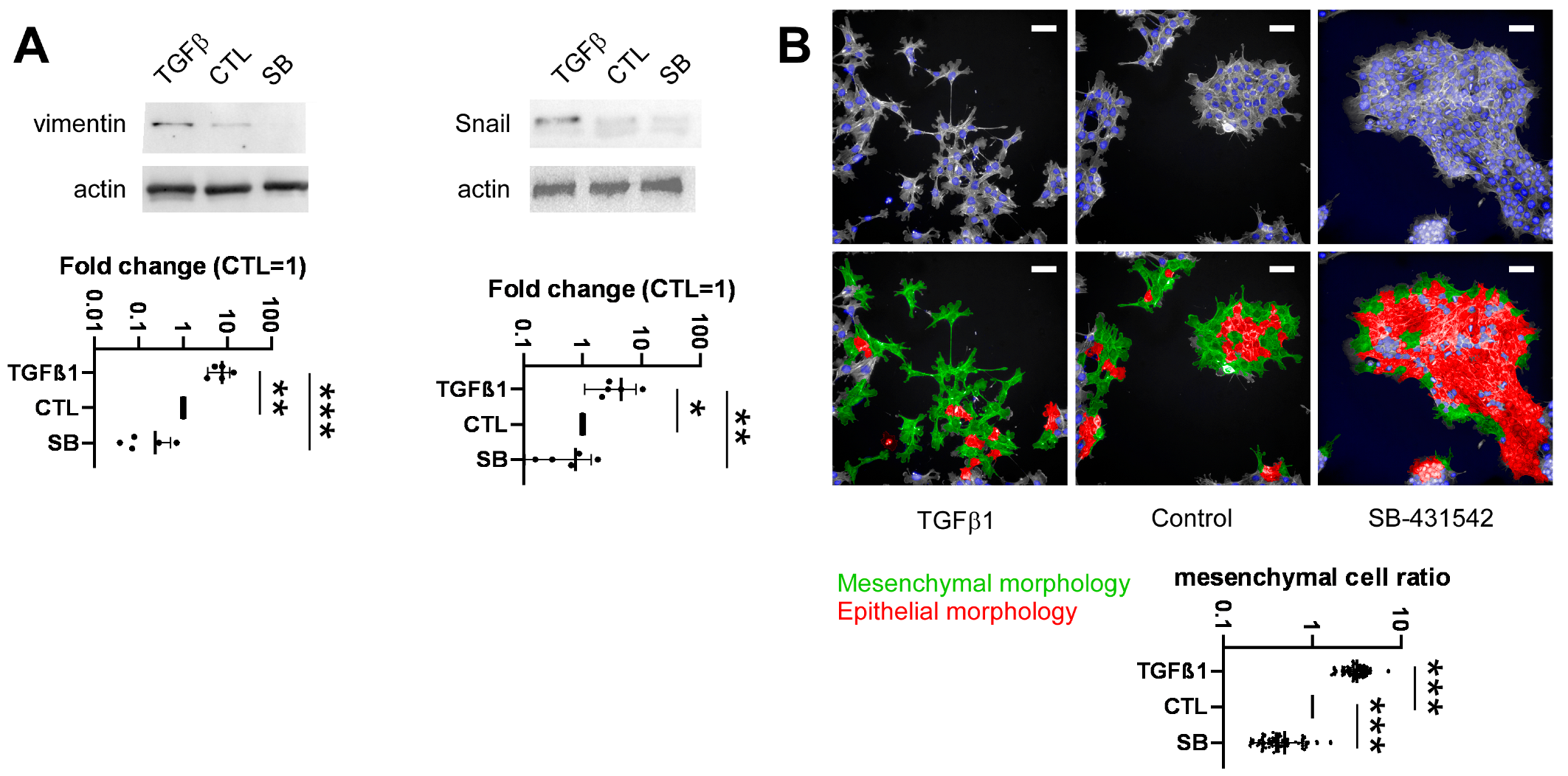
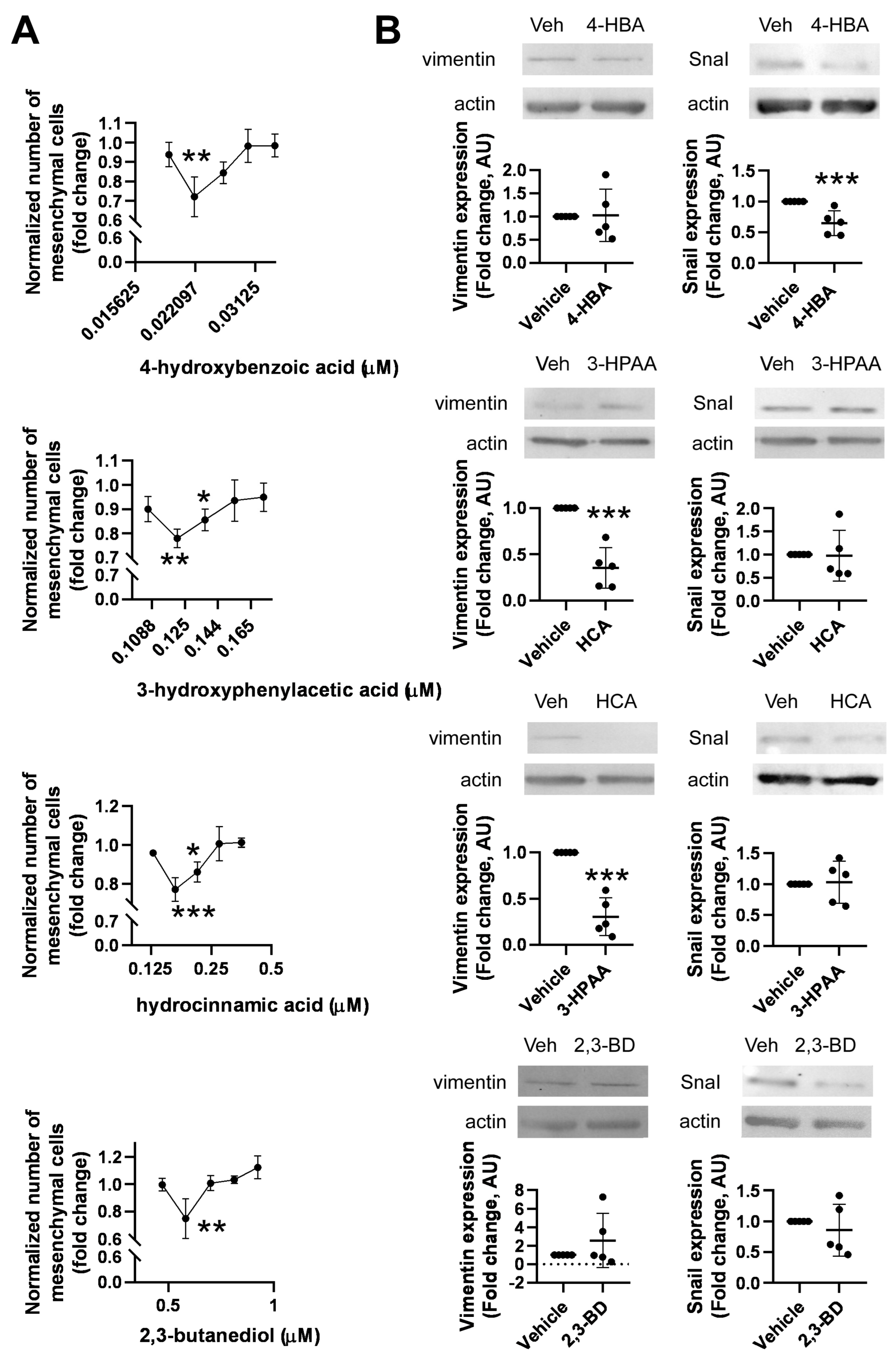
2.4. Development of a High-Throughput Method for Assessing Epithelial-to-Mesenchymal Transition
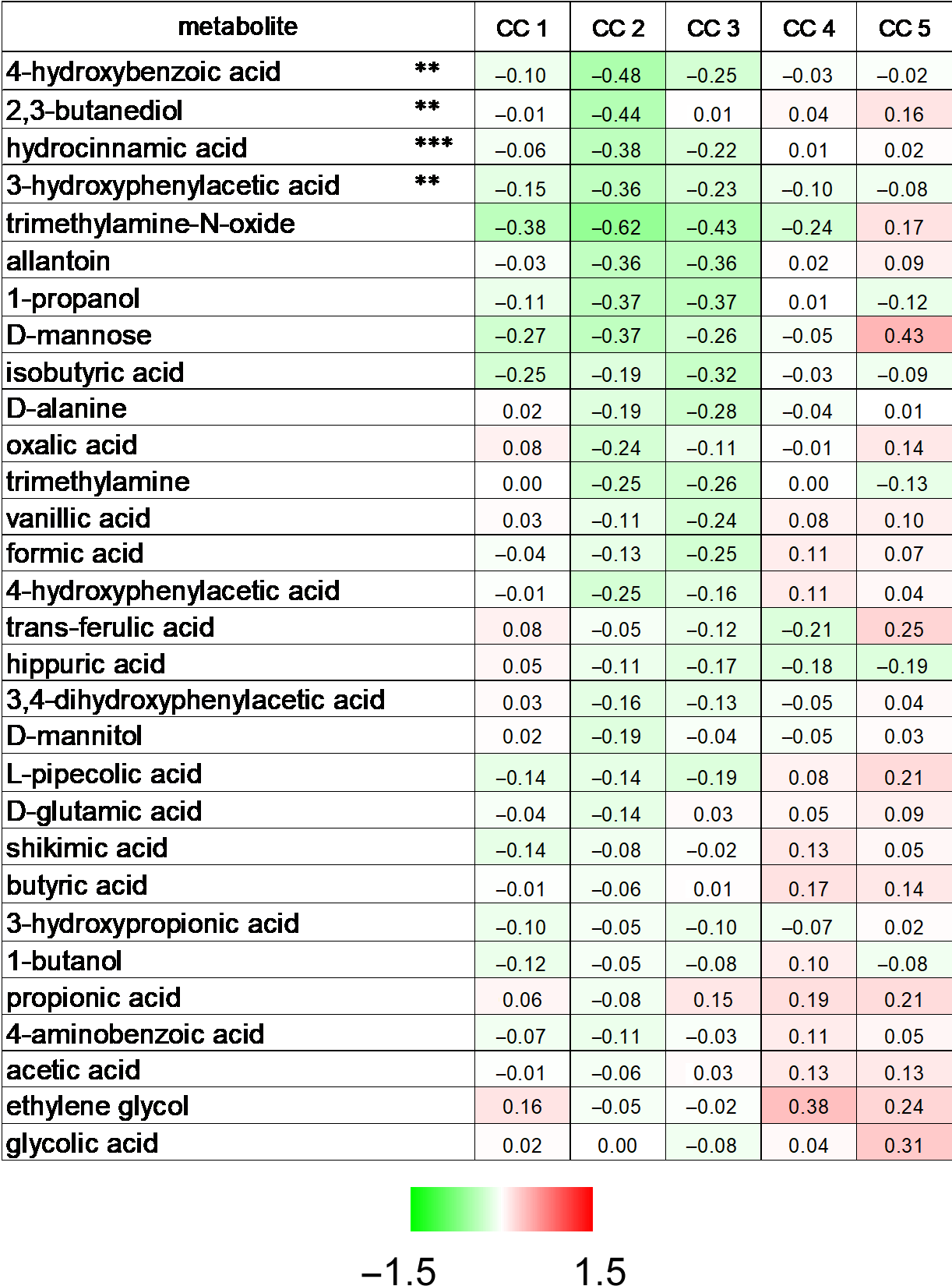
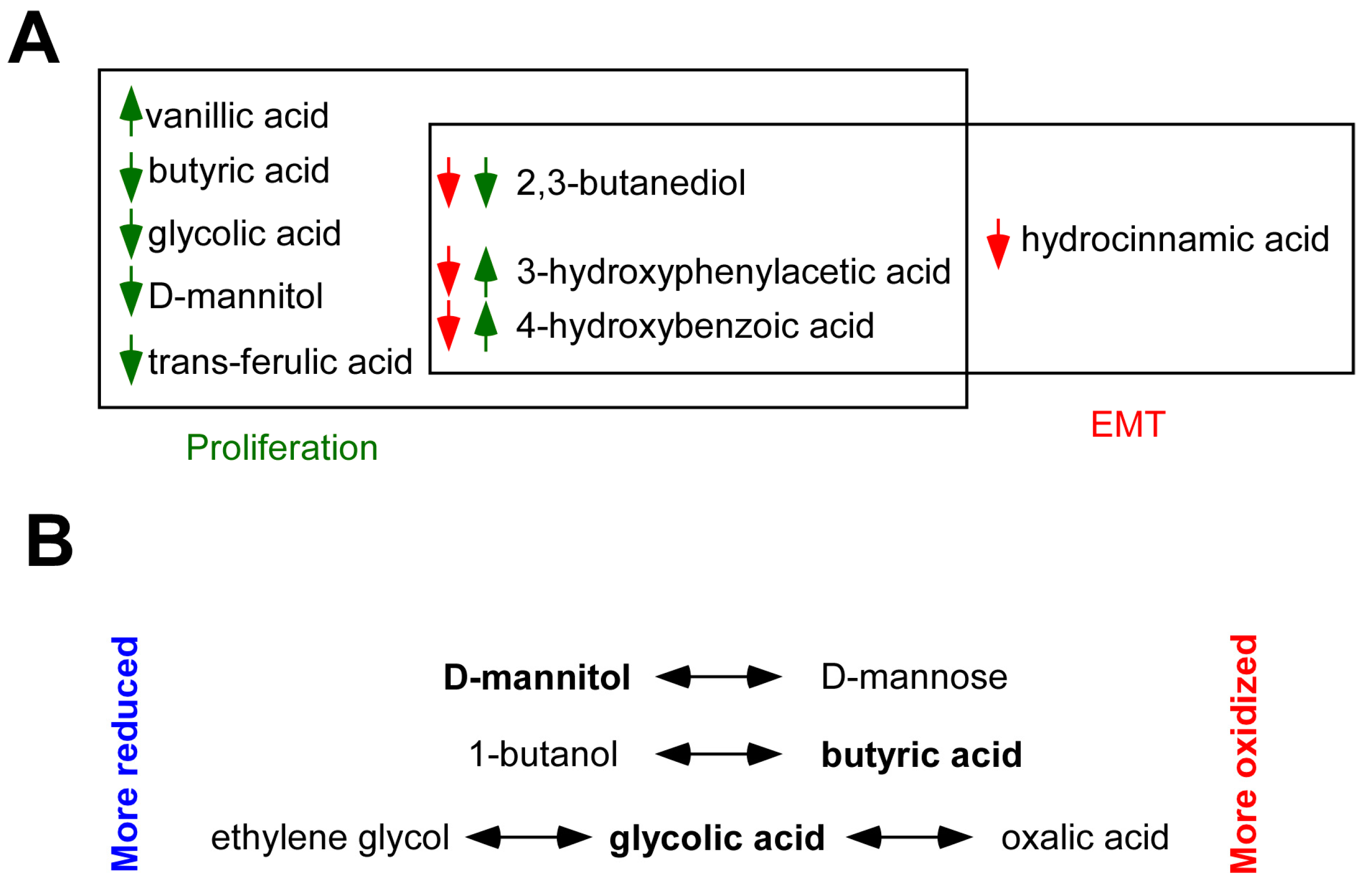
2.5. Identification of Bacterial Metabolites Modulating EMT
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Cytochemistry and Fluorescent Microscopy
4.4. Cell Morphology Analysis
4.5. Training Dataset—DAPI
4.6. Training Parameters
4.7. Nuclei Counting
4.8. Sulforhodamine B Cell Proliferation Assay
4.9. Western Blot
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Dafni, U.; Tsourti, Z.; Alatsathianos, I. Breast Cancer Statistics in the European Union: Incidence and Survival across European Countries. Breast Care 2019, 14, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef]
- Thomas, R.M.; Jobin, C. The Microbiome and Cancer: Is the ‘Oncobiome’ Mirage Real? Trends Cancer 2015, 1, 24–35. [Google Scholar] [CrossRef]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [PubMed]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Chen, B.; Yang, J.; Wang, J.; Zhu, D.; Meng, Q.; Zhang, L. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of in situ Tissue Microbiomes for Promoting Malignancy. Front. Oncol. 2018, 8, 318. [Google Scholar] [CrossRef]
- Smith, A.; Pierre, J.F.; Makowski, L.; Tolley, E.; Lyn-Cook, B.; Lu, L.; Vidal, G.; Starlard-Davenport, A. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 2019, 9, 11940. [Google Scholar] [CrossRef]
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.-C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 2021, 13, 60. [Google Scholar] [CrossRef]
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 2018, 8, 16893. [Google Scholar] [CrossRef]
- Klann, E.; Williamson, J.M.; Tagliamonte, M.S.; Ukhanova, M.; Asirvatham, J.R.; Chim, H.; Yaghjyan, L.; Mai, V. Microbiota composition in bilateral healthy breast tissue and breast tumors. Cancer Causes Control 2020, 31, 1027–1038. [Google Scholar] [CrossRef]
- Thyagarajan, S.; Zhang, Y.; Thapa, S.; Allen, M.S.; Phillips, N.; Chaudhary, P.; Kashyap, M.V.; Vishwanatha, J.K. Comparative analysis of racial differences in breast tumor microbiome. Sci. Rep. 2020, 10, 14116. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Wei, Z.; Tan, F.; Peck, K.N.; Shih, N.; Feldman, M.; Rebbeck, T.R.; Alwine, J.C.; Robertson, E.S. Distinct microbiological signatures associated with triple negative breast cancer. Sci. Rep. 2015, 5, 15162. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.J.; Ingle, J.N.; Tang, X.; Chia, N.; Jeraldo, P.R.; Walther-Antonio, M.R.; Kandimalla, K.K.; Johnson, S.; Yao, J.Z.; Harrington, S.C.; et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS ONE 2017, 12, e0188873. [Google Scholar] [CrossRef] [PubMed]
- Hogan, G.; Eckenberger, J.; Narayanen, N.; Walker, S.P.; Claesson, M.J.; Corrigan, M.; O’Hanlon, D.; Tangney, M. Biopsy bacterial signature can predict patient tissue malignancy. Sci. Rep. 2021, 11, 18535. [Google Scholar] [CrossRef]
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.C.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021, 12, 831. [Google Scholar] [CrossRef]
- Giallourou, N.; Urbaniak, C.; Puebla-Barragan, S.; Vorkas, P.A.; Swann, J.R.; Reid, G. Characterizing the breast cancer lipidome and its interaction with the tissue microbiota. Commun. Biol. 2021, 4, 1229. [Google Scholar] [CrossRef] [PubMed]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.A.; Vogtmann, E.; Wu, Z.; Han, Y.; Wan, Y.; Clegg-Lamptey, J.-N.; Yarney, J.; Wiafe-Addai, B.; Wiafe, S.; Awuah, B.; et al. Associations of fecal microbial profiles with breast cancer and non-malignant breast disease in the Ghana Breast Health Study. Int. J. Cancer 2021, 148, 2712–2723. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liao, M.; Yao, Z.; Liang, W.; Li, Q.; Liu, J.; Yang, H.; Ji, Y.; Wei, W.; Tan, A.; et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 2018, 6, 136. [Google Scholar] [CrossRef]
- Howe, C.; Kim, S.J.; Mitchell, J.; Im, E.; Kim, Y.S.; Kim, Y.S.; Rhee, S.H. Differential expression of tumor-associated genes and altered gut microbiome with decreased Akkermansia muciniphila confer a tumor-preventive microenvironment in intestinal epithelial Pten-deficient mice. Biochim. Biophys. Acta-Mol. Basis Dis. 2018, 1864, 3746–3758. [Google Scholar] [CrossRef]
- Fruge, A.D.; Van der Pol, W.; Rogers, L.Q.; Morrow, C.D.; Tsuruta, Y.; Demark-Wahnefried, W. Fecal Akkermansia muciniphila Is Associated with Body Composition and Microbiota Diversity in Overweight and Obese Women with Breast Cancer Participating in a Presurgical Weight Loss Trial. J. Acad. Nutr. Diet. 2018, 9, 164. [Google Scholar] [CrossRef]
- Wu, A.H.; Tseng, C.; Vigen, C.; Yu, Y.; Cozen, W.; Garcia, A.A.; Spicer, D. Gut microbiome associations with breast cancer risk factors and tumor characteristics: A pilot study. Breast Cancer Res. Treat. 2020, 182, 451–463. [Google Scholar] [CrossRef]
- Luu, T.H.; Michel, C.; Bard, J.M.; Dravet, F.; Nazih, H.; Bobin-Dubigeon, C. Intestinal Proportion of Blautia sp. is Associated with Clinical Stage and Histoprognostic Grade in Patients with Early-Stage Breast Cancer. Nutr. Cancer 2017, 69, 267–275. [Google Scholar] [CrossRef]
- Miko, E.; Vida, A.; Kovacs, T.; Ujlaki, G.; Trencsenyi, G.; Marton, J.; Sari, Z.; Kovacs, P.; Boratko, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef]
- Sári, Z.; Mikó, E.; Kovács, T.; Jankó, L.; Csonka, T.; Sebő, E.; Toth, J.; Tóth, D.; Árkosy, P.; Boratkó, A.; et al. Indolepropionic acid, a metabolite of the microbiome, has cytostatic properties in breast cancer by activating AHR and PXR receptors and inducing oxidative stress. Cancers 2020, 12, 2411. [Google Scholar] [CrossRef]
- Sári, Z.; Mikó, E.; Kovács, T.; Boratkó, A.; Ujlaki, G.; Jankó, L.; Kiss, B.; Uray, K.; Bai, P. Indoxylsulfate, a Metabolite of the Microbiome, Has Cytostatic Effects in Breast Cancer via Activation of AHR and PXR Receptors and Induction of Oxidative Stress. Cancers 2020, 12, 2915. [Google Scholar] [CrossRef] [PubMed]
- Sári, Z.; Kovács, T.; Csonka, T.; Török, M.; Sebő, É.; Toth, J.; Tóth, D.; Mikó, E.; Kiss, B.; Szeőcs, D.; et al. Fecal expression of E. coli lysine decarboxylase (LdcC) is downregulated in E-cadherin negative lobular breast carcinoma. Physiol. Int. 2020, 107, 349–358. [Google Scholar] [CrossRef]
- Murray, W.R.; Blackwood, A.; Calman, K.C.; MacKay, C. Faecal bile acids and clostridia in patients with breast cancer. Br. J. Cancer 1980, 42, 856–860. [Google Scholar] [CrossRef]
- Goedert, J.J.; Hua, X.; Bielecka, A.; Okayasu, I.; Milne, G.L.; Jones, G.S.; Fujiwara, M.; Sinha, R.; Wan, Y.; Xu, X.; et al. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br. J. Cancer 2018, 23, 435. [Google Scholar] [CrossRef]
- Bobin-Dubigeon, C.; Luu, H.T.; Leuillet, S.; Lavergne, S.N.; Carton, T.; Le Vacon, F.; Michel, C.; Nazih, H.; Bard, J.M. Faecal Microbiota Composition Varies between Patients with Breast Cancer and Healthy Women: A Comparative Case-Control Study. Nutrients 2021, 13, 2705. [Google Scholar] [CrossRef]
- Horigome, A.; Okubo, R.; Hamazaki, K.; Kinoshita, T.; Katsumata, N.; Uezono, Y.; Xiao, J.Z.; Matsuoka, Y.J. Association between blood omega-3 polyunsaturated fatty acids and the gut microbiota among breast cancer survivors. Benef. Microbes 2019, 10, 751–758. [Google Scholar] [CrossRef]
- Terrisse, S.; Derosa, L.; Iebba, V.; Ghiringhelli, F.; Vaz-Luis, I.; Kroemer, G.; Fidelle, M.; Christodoulidis, S.; Segata, N.; Thomas, A.M.; et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ. 2021, 28, 2778–2796. [Google Scholar] [CrossRef]
- Hou, M.F.; Ou-Yang, F.; Li, C.L.; Chen, F.M.; Chuang, C.H.; Kan, J.Y.; Wu, C.C.; Shih, S.L.; Shiau, J.P.; Kao, L.C.; et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp. Mol. Med. 2021, 53, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Vigen, C.; Tseng, C.; Garcia, A.A.; Spicer, D. Effect of chemotherapy and weight change on the gut microbiome of breast cancer patients during the first year of treatment. Res. Sq. 2022, 14, 433–451. Available online: https://www.researchsquare.com/article/rs-970564/v970561 (accessed on 30 June 2023). [CrossRef] [PubMed]
- Zhu, Q.; Zai, H.; Zhang, K.; Zhang, X.; Luo, N.; Li, X.; Hu, Y.; Wu, Y. L-norvaline affects the proliferation of breast cancer cells based on the microbiome and metabolome analysis. J. Appl. Microbiol. 2022, 133, 1014–1026. [Google Scholar] [CrossRef]
- He, C.; Liu, Y.; Ye, S.; Yin, S.; Gu, J. Changes of intestinal microflora of breast cancer in premenopausal women. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 503–513. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Kim, J.B.; Yang, E.Y.; Kim, H.O.; Lee, W.-H.; Yang, J.; Kwon, H.; Paik, N.S.; Lim, W.; Kim, Y.-K.; et al. Bacterial extracellular vesicles affect endocrine therapy in MCF7 cells. Medicine 2021, 100, e25835. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Ujlaki, G.; Yousef, H.; Csontos, V.; Uray, K.; Bai, P. The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer. Cancer Metastasis Rev. 2021, 40, 1223–1249. [Google Scholar] [CrossRef]
- Wilmanski, T.; Rappaport, N.; Earls, J.C.; Magis, A.T.; Manor, O.; Lovejoy, J.; Omenn, G.S.; Hood, L.; Gibbons, S.M.; Price, N.D. Blood metabolome predicts gut microbiome alpha-diversity in humans. Nat. Biotechnol. 2019, 37, 1217–1228. [Google Scholar] [CrossRef]
- Miko, E.; Kovacs, T.; Sebo, E.; Toth, J.; Csonka, T.; Ujlaki, G.; Sipos, A.; Szabo, J.; Mehes, G.; Bai, P. Microbiome-Microbial Metabolome-Cancer Cell Interactions in Breast Cancer-Familiar, but Unexplored. Cells 2019, 8, E293. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Malla, R.R. Recent advances in metabolomics of triple negative breast cancer. Breast J. 2020, 26, 498–501. [Google Scholar] [CrossRef]
- Hill, M.J.; Goddard, P.; Williams, R.E. Gut bacteria and aetiology of cancer of the breast. Lancet 1971, 2, 472–473. [Google Scholar] [CrossRef]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K.; et al. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef]
- Yonezawa, T.; Kobayashi, Y.; Obara, Y. Short-chain fatty acids induce acute phosphorylation of the p38 mitogen-activated protein kinase/heat shock protein 27 pathway via GPR43 in the MCF-7 human breast cancer cell line. Cell. Signal. 2007, 19, 185–193. [Google Scholar] [CrossRef]
- Thirunavukkarasan, M.; Wang, C.; Rao, A.; Hind, T.; Teo, Y.R.; Siddiquee, A.A.; Goghari, M.A.I.; Kumar, A.P.; Herr, D.R. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS ONE 2017, 12, e0186334. [Google Scholar] [CrossRef]
- Liebich, H.M.; Buelow, H.J.; Kallmayer, R. Quantification of endogenous aliphatic alcohols in serum and urine. J. Chromatogr. 1982, 239, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, J.A.; Jetté, M.; Brunengraber, H. Assay of physiological levels of 2,3-butanediol diastereomers in blood and urine by gas chromatography-mass spectrometry. Anal. Biochem. 1990, 185, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, R.P.; Istas, G.; Heiss, C.; Rodriguez-Mateos, A. Plasma and Urinary Phenolic Profiles after Acute and Repetitive Intake of Wild Blueberry. Molecules 2016, 21, 1120. [Google Scholar] [CrossRef] [PubMed]
- Izzo, J.L., Jr.; Greulich, D. Radioenzymatic assay for plasma dihydroxyphenylglycol (DHPG), dihydroxymandelic acid (DOMA) and dihydroxyphenylacetic acid (DOPAC). Life Sci. 1983, 33, 483–488. [Google Scholar] [CrossRef]
- Novarino, G.; El-Fishawy, P.; Kayserili, H.; Meguid, N.A.; Scott, E.M.; Schroth, J.; Silhavy, J.L.; Kara, M.; Khalil, R.O.; Ben-Omran, T.; et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 2012, 338, 394–397. [Google Scholar] [CrossRef]
- Lianidou, E.S.; Ioannou, P.C. Simple spectrofluorometric determination of p-aminobenzoic and p-aminosalicylic acids in biological fluids by use of terbium-sensitized luminescence. Clin. Chem. 1996, 42, 1659–1665. [Google Scholar] [CrossRef]
- Richards, R.H.; Dowling, J.A.; Vreman, H.J.; Feldman, C.; Weiner, M.W. Acetate levels in human plasma. Proc. Clin. Dial. Transpl. Forum 1976, 6, 73–79. [Google Scholar]
- Jakobsdottir, G.; Bjerregaard, J.H.; Skovbjerg, H.; Nyman, M. Fasting serum concentration of short-chain fatty acids in subjects with microscopic colitis and celiac disease: No difference compared with controls, but between genders. Scand. J. Gastroenterol. 2013, 48, 696–701. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Shafaei, A.; Vamathevan, V.; Pandohee, J.; Lawler, N.G.; Broadhurst, D.; Boyce, M.C. Sensitive and quantitative determination of short-chain fatty acids in human serum using liquid chromatography mass spectrometry. Anal. Bioanal. Chem. 2021, 413, 6333–6342. [Google Scholar] [CrossRef] [PubMed]
- Kand’ár, R.; Záková, P. Allantoin as a marker of oxidative stress in human erythrocytes. Clin. Chem. Lab. Med. 2008, 46, 1270–1274. [Google Scholar] [CrossRef]
- Pavitt, D.V.; de Fonseka, S.; Al-Khalaf, N.; Cam, J.M.; Reaveley, D.A. Assay of serum allantoin in humans by gas chromatography-mass spectrometry. Clin. Chim. Acta Int. J. Clin. Chem. 2002, 318, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Gruber, J.; Tang, S.Y.; Jenner, A.M.; Mudway, I.; Blomberg, A.; Behndig, A.; Kasiman, K.; Lee, C.Y.; Seet, R.C.; Zhang, W.; et al. Allantoin in human plasma, serum, and nasal-lining fluids as a biomarker of oxidative stress: Avoiding artifacts and establishing real in vivo concentrations. Antioxid. Redox Signal. 2009, 11, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Yang, H.-T.; Chiu, C.-C.; Lane, H.-Y. Blood levels of D-amino acid oxidase vs. D-amino acids in reflecting cognitive aging. Sci. Rep. 2017, 7, 14849. [Google Scholar] [CrossRef]
- Pitkänen, E.; Pitkänen, O.; Uotila, L. Enzymatic determination of unbound D-mannose in serum. Eur. J. Clin. Chem. Clin. Biochem. J. Forum Eur. Clin. Chem. Soc. 1997, 35, 761–766. [Google Scholar] [CrossRef]
- Campi, B.; Codini, S.; Bisoli, N.; Baldi, S.; Zucchi, R.; Ferrannini, E.; Saba, A. Quantification of d-mannose in plasma: Development and validation of a reliable and accurate HPLC-MS-MS method. Clin. Chim. Acta 2019, 493, 31–35. [Google Scholar] [CrossRef]
- Taguchi, T.; Miwa, I.; Mizutani, T.; Nakajima, H.; Fukumura, Y.; Kobayashi, I.; Yabuuchi, M.; Miwa, I. Determination of d-Mannose in Plasma by HPLC. Clin. Chem. 2003, 49, 181–183. [Google Scholar] [CrossRef]
- Baumann, K.; Angerer, J. Occupational chronic exposure to organic solvents. VI. Formic acid concentration in blood and urine as an indicator of methanol exposure. Int. Arch. Occup. Environ. Health 1979, 42, 241–249. [Google Scholar] [CrossRef]
- Marangella, M.; Petrarulo, M.; Bianco, O.; Vitale, C.; Finocchiaro, P.; Linari, F. Glycolate determination detects type I primary hyperoxaluria in dialysis patients. Kidney Int. 1991, 39, 149–154. [Google Scholar] [CrossRef]
- Nowak, P.J.; Wilk, R.; Prymont-Przyminska, A.; Zwolinska, A.; Sarniak, A.; Wlodarczyk, A.; de Graft-Johnson, J.; Mamelka, B.; Zasowska-Nowak, A.; Bartnicki, P.; et al. Hemodialysis Decreases the Concentration of Accumulated Plant Phenols in the Plasma of Patients on Maintenance Dialysis: Influence of Residual Renal Function. Ther. Apher. Dial. 2017, 21, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Afessa, B.; Mullon, J.; Badley, A.; Gajic, O. The impact of protocolized sepsis order set on the process of care in patients with severe sepsis/septic shock. Crit. Care 2007, 11, P6. [Google Scholar] [CrossRef][Green Version]
- Baas, J.C.; van de Laar, R.; Dorland, L.; Duran, M.; Berger, R.; Poll-The, B.T.; de Koning, T.J. Plasma pipecolic acid is frequently elevated in non-peroxisomal disease. J. Inherit. Metab. Dis. 2002, 25, 699–701. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Zarembski, P.M.; Hodgkinson, A. The fluorimetric determination of oxalic acid in blood and other biological materials. Biochem. J. 1965, 96, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Bain, M.A.; Faull, R.; Fornasini, G.; Milne, R.W.; Evans, A.M. Accumulation of trimethylamine and trimethylamine-N-oxide in end-stage renal disease patients undergoing haemodialysis. Nephrol. Dial. Transplant. 2006, 21, 1300–1304. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Akolekar, R.; Mandal, R.; Dong, E.; Xia, J.; Kruger, M.; Wishart, D.S.; Nicolaides, K. Metabolomics and first-trimester prediction of early-onset preeclampsia. J. Matern. Fetal Neonatal Med. 2012, 25, 1840–1847. [Google Scholar] [CrossRef]
- Garcia, E.; Wolak-Dinsmore, J.; Wang, Z.; Li, X.S.; Bennett, D.W.; Connelly, M.A.; Otvos, J.D.; Hazen, S.L.; Jeyarajah, E.J. NMR quantification of trimethylamine-N-oxide in human serum and plasma in the clinical laboratory setting. Clin. Biochem. 2017, 50, 947–955. [Google Scholar] [CrossRef]
- Nie, J.; Xie, L.; Zhao, B.X.; Li, Y.; Qiu, B.; Zhu, F.; Li, G.F.; He, M.; Wang, Y.; Wang, B.; et al. Serum Trimethylamine N-Oxide Concentration Is Positively Associated With First Stroke in Hypertensive Patients. Stroke 2018, 49, 2021–2028. [Google Scholar] [CrossRef]
- Randrianarisoa, E.; Lehn-Stefan, A.; Wang, X.; Hoene, M.; Peter, A.; Heinzmann, S.S.; Zhao, X.; Königsrainer, I.; Königsrainer, A.; Balletshofer, B.; et al. Relationship of Serum Trimethylamine N-Oxide (TMAO) Levels with early Atherosclerosis in Humans. Sci. Rep. 2016, 6, 26745. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.; Terrisse, S.; Alves Costa Silva, C.; Lafarge, A.; Iebba, V.; Ferrere, G.; Goubet, A.G.; Fahrner, J.E.; Lahmar, I.; Ueda, K.; et al. Cancer induces a stress ileopathy depending on B-adrenergic receptors and promoting dysbiosis that contribute to carcinogenesis. Cancer Discov. 2021, 12, 1128–1151. [Google Scholar] [CrossRef]
- Malik, A.; Morya, R.K.; Bhadada, S.K.; Rana, S. Type 1 diabetes mellitus: Complex interplay of oxidative stress, cytokines, gastrointestinal motility and small intestinal bacterial overgrowth. Eur. J. Clin. Investig. 2018, 48, e13021. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- González-Martínez, S.; Pérez-Mies, B.; Pizarro, D.; Caniego-Casas, T.; Cortés, J.; Palacios, J. Epithelial Mesenchymal Transition and Immune Response in Metaplastic Breast Carcinoma. Int. J. Mol. Sci. 2021, 22, 7398. [Google Scholar] [CrossRef]
- Trelford, C.B.; Dagnino, L.; Di Guglielmo, G.M. Transforming growth factor-β in tumour development. Front. Mol. Biosci. 2022, 9, 991612. [Google Scholar] [CrossRef]
- Inman, G.J.; Nicolás, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef]
- Ng, C.; Jung, M.-y.; Lee, J.; Oh, M.-K. Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb. Cell Factories 2012, 11, 68. [Google Scholar] [CrossRef]
- Garg, S.K.; Jain, A. Fermentative production of 2,3-butanediol: A review. Bioresour. Technol. 1995, 51, 103–109. [Google Scholar] [CrossRef]
- Million, M.; Tidjani Alou, M.; Khelaifia, S.; Bachar, D.; Lagier, J.C.; Dione, N.; Brah, S.; Hugon, P.; Lombard, V.; Armougom, F.; et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci. Rep. 2016, 6, 26051. [Google Scholar] [CrossRef]
- Cha, Y.J.; Kim, E.S.; Koo, J.S. Amino Acid Transporters and Glutamine Metabolism in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 907. [Google Scholar] [CrossRef] [PubMed]
- Geck, R.C.; Toker, A. Nonessential amino acid metabolism in breast cancer. Adv. Biol. Regul. 2016, 62, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, A.S.; Tokuhara, C.K.; Fakhoury, V.S.; Pagnan, A.L.; Oliveira, G.S.N.; Sanches, M.L.R.; Inacio, K.K.; Costa, B.C.; Ximenes, V.F.; Oliveira, R.C. The dimerization of methyl vanillate improves its effect against breast cancer cells via pro-oxidant effect. Chem. Biol. Interact. 2022, 361, 109962. [Google Scholar] [CrossRef]
- Rouxel, M.; Barthe, M.; Marchand, P.; Juin, C.; Mondamert, L.; Berges, T.; Blanc, P.; Verdon, J.; Berjeaud, J.M.; Aucher, W. Characterization of antifungal compounds produced by lactobacilli in cheese-mimicking matrix: Comparison between active and inactive strains. Int. J. Food Microbiol. 2020, 333, 108798. [Google Scholar] [CrossRef] [PubMed]
- Aura, A.M.; O’Leary, K.A.; Williamson, G.; Ojala, M.; Bailey, M.; Puupponen-Pimiä, R.; Nuutila, A.M.; Oksman-Caldentey, K.M.; Poutanen, K. Quercetin Derivatives Are Deconjugated and Converted to Hydroxyphenylacetic Acids but Not Methylated by Human Fecal Flora in Vitro. J. Agric. Food Chem. 2002, 50, 1725–1730. [Google Scholar] [CrossRef]
- Rechner, A.R.; Smith, M.A.; Kuhnle, G.; Gibson, G.R.; Debnam, E.S.; Srai, S.K.S.; Moore, K.P.; Rice-Evans, C.A. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic. Biol. Med. 2004, 36, 212–225. [Google Scholar] [CrossRef]
- Williamson, G.; Clifford, M.N. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochem. Pharmacol. 2017, 139, 24–39. [Google Scholar] [CrossRef]
- Ozdemir, O.O.; Soyer, F. Pseudomonas aeruginosa Presents Multiple Vital Changes in Its Proteome in the Presence of 3-Hydroxyphenylacetic Acid, a Promising Antimicrobial Agent. ACS Omega 2020, 5, 19938–19951. [Google Scholar] [CrossRef]
- Liu, Y.; Myojin, T.; Li, K.; Kurita, A.; Seto, M.; Motoyama, A.; Liu, X.; Satoh, A.; Munemasa, S.; Murata, Y.; et al. A Major Intestinal Catabolite of Quercetin Glycosides, 3-Hydroxyphenylacetic Acid, Protects the Hepatocytes from the Acetaldehyde-Induced Cytotoxicity through the Enhancement of the Total Aldehyde Dehydrogenase Activity. Int. J. Mol. Sci. 2022, 23, 1762. [Google Scholar] [CrossRef]
- Chen, W.; Wu, H.; Hernandez, R.J.; Mehta, A.K.; Ticku, M.K.; France, C.P.; Coop, A. Ethers of 3-hydroxyphenylacetic acid as selective gamma-hydroxybutyric acid receptor ligands. Bioorganic Med. Chem. Lett. 2005, 15, 3201–3202. [Google Scholar] [CrossRef] [PubMed]
- Ticku, M.K.; Mehta, A.K. Characterization and Pharmacology of the GHB Receptor. Ann. N. Y. Acad. Sci. 2008, 1139, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, D.; Pope, G.S.; Darbre, P.D. Oestrogenic activity of p-hydroxybenzoic acid (common metabolite of paraben esters) and methylparaben in human breast cancer cell lines. J. Appl. Toxicol. 2005, 25, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Rosiles-Alanis, W.; Zamilpa, A.; García-Macedo, R.; Zavala-Sánchez, M.A.; Hidalgo-Figueroa, S.; Mora-Ramiro, B.; Román-Ramos, R.; Estrada-Soto, S.E.; Almanza-Perez, J.C. 4-Hydroxybenzoic Acid and β-Sitosterol from Cucurbita ficifolia Act as Insulin Secretagogues, Peroxisome Proliferator-Activated Receptor-Gamma Agonists, and Liver Glycogen Storage Promoters: In Vivo, In Vitro, and In Silico Studies. J. Med. Food 2022, 25, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Valle-González, E.R.; Jackman, J.A.; Yoon, B.K.; Mokrzecka, N.; Cho, N.J. pH-Dependent Antibacterial Activity of Glycolic Acid: Implications for Anti-Acne Formulations. Sci. Rep. 2020, 10, 7491. [Google Scholar] [CrossRef]
- Sousa, M.; Afonso, A.C.; Teixeira, L.S.; Borges, A.; Saavedra, M.J.; Simões, L.C.; Simões, M. Hydrocinnamic Acid and Perillyl Alcohol Potentiate the Action of Antibiotics against Escherichia coli. Antibiotics 2023, 12, 360. [Google Scholar] [CrossRef]
- Sharma, S.; Gopu, V.; Sivasankar, C.; Shetty, P.H. Hydrocinnamic acid produced by Enterobacter xiangfangensis impairs AHL-based quorum sensing and biofilm formation in Pseudomonas aeruginosa. RSC Adv. 2019, 9, 28678–28687. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Liu, L.; Zhang, Y.; Yuan, J. Microbial synthesis of 4-hydroxybenzoic acid from renewable feedstocks. Food Chem. Mol. Sci. 2021, 3, 100059. [Google Scholar] [CrossRef]
- Dillon, V.M. Natural Anti-Microbial Systems | Preservative Effects During Storage. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 941–947. [Google Scholar]
- Kacsir, I.; Sipos, A.; Bényei, A.; Janka, E.; Buglyó, P.; Somsak, L.; Bai, P.; Bokor, É. Reactive oxygen species production is responsible for antineoplastic activity of osmium, ruthenium, iridium and rhodium half-sandwich type complexes with bidentate glycosyl hetero-cyclic ligands in various cancer cell models. Int. J. Mol. Sci. 2022, 23, 813. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Marton, J.; Fodor, T.; Nagy, L.; Vida, A.; Kis, G.; Brunyanszki, A.; Antal, M.; Luscher, B.; Bai, P. PARP10 (ARTD10) modulates mitochondrial function. PLoS ONE 2018, 13, e0187789. [Google Scholar] [CrossRef] [PubMed]
- Marton, J.; Peter, M.; Balogh, G.; Bodi, B.; Vida, A.; Szanto, M.; Bojcsuk, D.; Janko, L.; Bhattoa, H.P.; Gombos, I.; et al. Poly(ADP-ribose) polymerase-2 is a lipid-modulated modulator of muscular lipid homeostasis. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2018, 1863, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
| Metabolite Name | Solvent | Catalog Number | Serum Reference Concentrations (µM) | Applied Concentrations (μM) | |
|---|---|---|---|---|---|
| 1 | 1-Butanol | PBS | Sigma B7906-500ML | 0–0.27 [52] | 0.005, 0.015, 0.044, 0.13, 0.4 |
| 2 | 1-Propanol | PBS | Sigma 96566-5ML-F | 0–0.8 [52] | 0.05, 0.1, 0.2, 0.4, 0.8 |
| 3 | 2,3-butanediol | PBS | Sigma B84904 | 0.5–0.9 [53] | 0.48, 0.56, 0.66, 0.77, 0.9 |
| 4 | 3,4-dihydroxyphenyl acetic acid | PBS | Sigma 850217-14 | 0.0102–0.104 [54,55] | 0.01, 0.018, 0.032, 0.058, 0.104 |
| 5 | 3-Hydroxyphenylacetic acid | PBS | Sigma H49901 | 0.11–0.174 [54] | 0.106, 0.120, 0.136, 0.154, 0.174 |
| 6 | 3-Hydroxypropionic acid | PBS | Sigma 792659-1G | 3, 6, 8 (individual values) [56] | 0.5, 1, 2, 4, 8 |
| 7 | 4-aminobenzoic acid | EtOH | Sigma A9878-5G | 5.01–32.0 [57] | 0.3, 1.2, 3.6, 10.8, 32.4 |
| 8 | 4-hydroxybenzoic acid | PBS | Sigma 8218140250 | 0.019–0.035 [54] | 0.019, 0.022, 0.026, 0.03, 0.035 |
| 9 | 4-Hydroxyphenylacetic acid | PBS | Sigma H50004-54 | 0.283–0.61 [54] | 0.28, 0.36, 0.41, 0.5, 0.61 |
| 10 | Acetic acid | PBS | VWR UN2789 | 23–254.4 [52,58,59,60,61] | 15, 30, 60, 120, 240 |
| 11 | Allantoin | DMSO | Sigma 05670 | 1.0–24.0 [62,63,64] | 0.99, 2.2, 4.9, 10.8, 24 |
| 12 | Butyric acid | PBS | Sigma B103500-5ML | 1.39–14.15 [59,60,61] | 1, 2, 4, 8, 16 |
| 13 | d-alanine | PBS | Sigma A7377-5G | 0–0.77 [65] | 0.048, 0.96, 0.193, 0.385, 0.77 |
| 14 | d-glutamic acid | PBS | Sigma G1001-1G | 7.42–14.6 [65] | 7.28, 8.66, 10.31, 12.27, 14.6 |
| 15 | d-mannitol | PBS | Sigma M4125-10MG | no report, same concentrations as for d-mannose | 6.25, 12.5, 25, 50, 100 |
| 16 | d-mannose | DMSO | no data | 13–73.87 [66,67,68] | 6.25, 12.5, 25, 50, 100 |
| 17 | Ethylene glycol | PBS | no data | no data, toxic (1.56 mg/kg) | 1, 3, 9,27, 81 |
| 18 | Formic acid | PBS | Sigma F0507-500ML | 11.84–224.5 [69] | 10, 30, 90, 270, 810 |
| 19 | Glycolic acid | PBS | Sigma 124737 | 6.1–69 [56,70] | 1, 3, 9,27, 81 |
| 20 | Hippuric acid | PBS | Sigma 112003 | 1.5–21.2 [54,71] | 0.024, 0.12, 0.6, 3, 15 |
| 21 | Hydrocinnamic acid | EtOH | Sigma 135232-5G | 0.131–0.354 [72] | 0.128, 0.165, 0.213, 0.274, 0.354 |
| 22 | Isobutyric acid | PBS | Sigma I1754-100ML | 1.02–14.15 [59,61] | 1.02, 1.97, 3.80, 7.33, 14.15 |
| 23 | l-pipecolic acid | PBS | Sigma P2519 | 1.2–3.72 [73] | 0.25, 0.5, 1, 2, 4 |
| 24 | oxalic acid | PBS | Sigma 75688 | 6.5–35.5 [74,75] | 6.5, 9.9, 15.2, 23.2, 35.5 |
| 25 | Propionic acid | PBS | Sigma P1386-1L | 4.86–15.33 [59,60,61] | 1.25, 2.5, 5, 10, 20 |
| 26 | Shikimic acid | DMSO | Sigma S5375-10MG | 0.03–0.23 [76] | 0.01, 0.03, 0.09, 0.27, 0.81 |
| 27 | trans-ferulic acid | DMSO | Sigma 52229 | 0.04–15.7 [54,71] | 0.016, 0.08, 0.4, 2, 10 |
| 28 | Trimethylamine (TMA) | PBS | Sigma 92260 | 0.3–14.44 [77,78] | 0.3, 0.79, 2.1, 5.4, 14.35 |
| 29 | Trimethylamine-N-oxide (TMAO) | PBS | Sigma 317594 | 1.21–21.1 [79,80,81] | 1.22, 2.48, 5.065, 10.33, 21.1 |
| 30 | Vanillic acid | PBS | Sigma H36001 | 0.01–0.338 [54,71] | 0.01, 0.024, 0.058, 0.140,0.338 |
| Antibody | Catalog Number | Company | Dilution |
|---|---|---|---|
| SnaiI | 3879S | Cell Signaling Technology | 1:1000 |
| Vimentin | 5741S | ||
| β-actin | A3854 | Sigma | 1:20,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ujlaki, G.; Kovács, T.; Vida, A.; Kókai, E.; Rauch, B.; Schwarcz, S.; Mikó, E.; Janka, E.; Sipos, A.; Hegedűs, C.; et al. Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition. Molecules 2023, 28, 5898. https://doi.org/10.3390/molecules28155898
Ujlaki G, Kovács T, Vida A, Kókai E, Rauch B, Schwarcz S, Mikó E, Janka E, Sipos A, Hegedűs C, et al. Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition. Molecules. 2023; 28(15):5898. https://doi.org/10.3390/molecules28155898
Chicago/Turabian StyleUjlaki, Gyula, Tünde Kovács, András Vida, Endre Kókai, Boglára Rauch, Szandra Schwarcz, Edit Mikó, Eszter Janka, Adrienn Sipos, Csaba Hegedűs, and et al. 2023. "Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition" Molecules 28, no. 15: 5898. https://doi.org/10.3390/molecules28155898
APA StyleUjlaki, G., Kovács, T., Vida, A., Kókai, E., Rauch, B., Schwarcz, S., Mikó, E., Janka, E., Sipos, A., Hegedűs, C., Uray, K., Nagy, P., & Bai, P. (2023). Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition. Molecules, 28(15), 5898. https://doi.org/10.3390/molecules28155898








