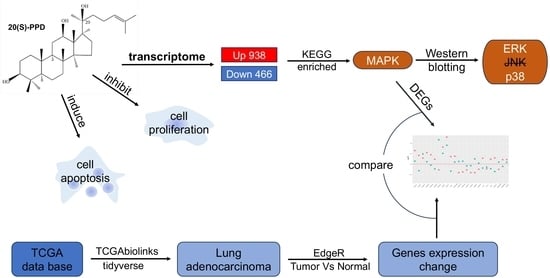
Transcriptome Analysis of the Inhibitory Effects of 20(S)-Protopanaxadiol on NCI-H1299 Non-Small Cell Lung Cancer Cells
Abstract
1. Introduction
2. Results
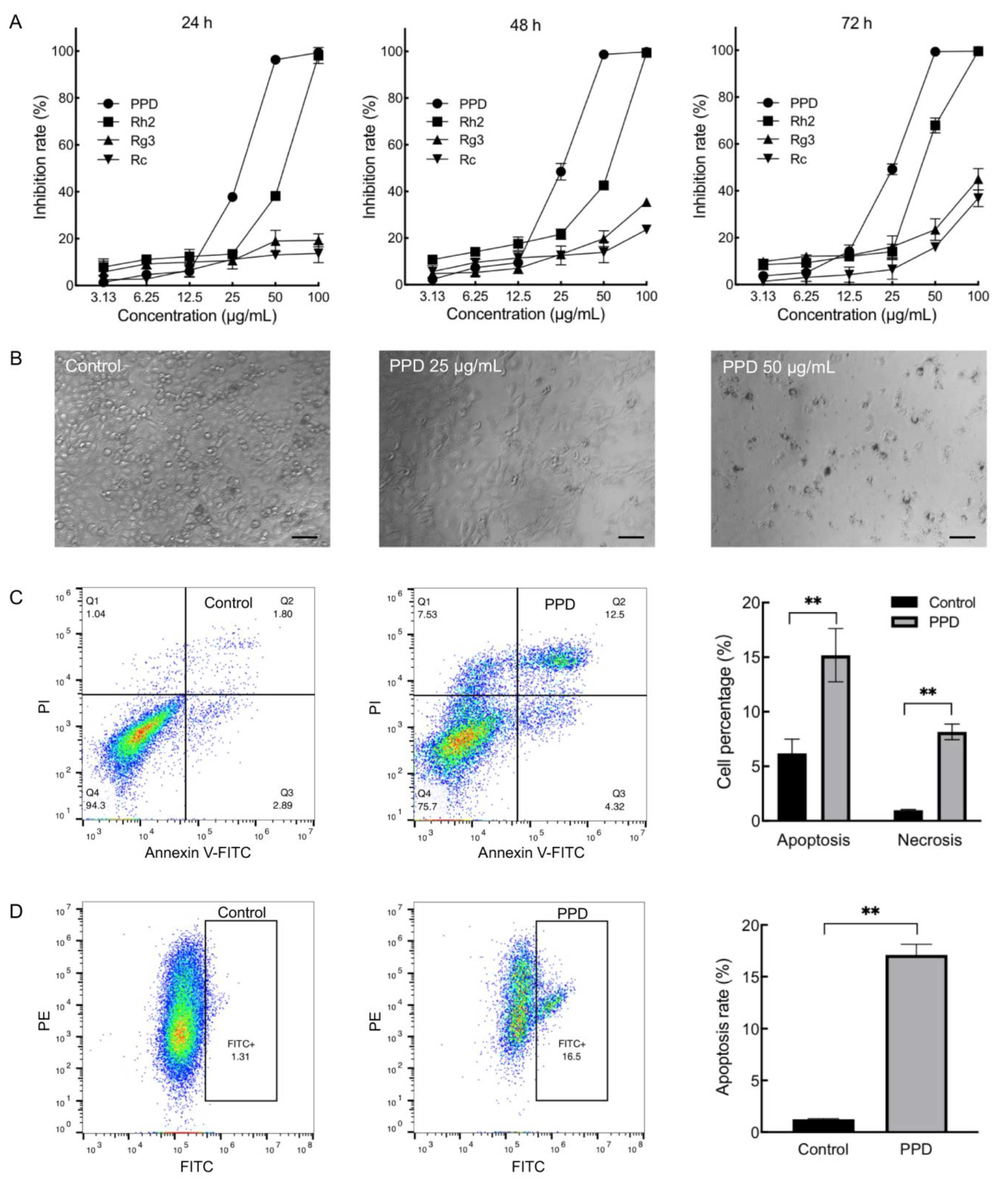
2.1. Inhibition Effects of PPD on NCI-H1299 Cells
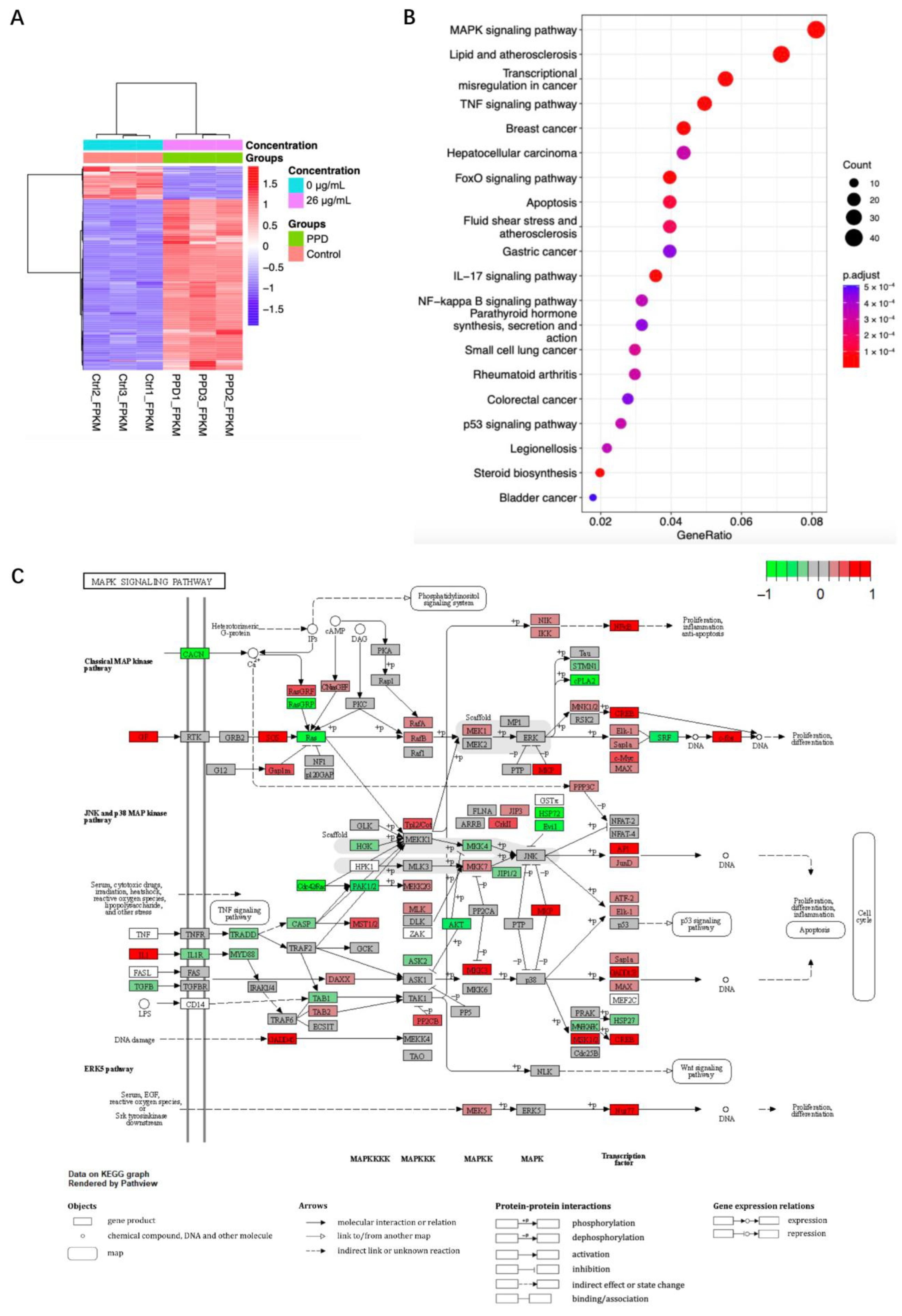
2.2. Transcriptome Sequencing Analysis of PPD Treated NCI-H1299 Cells
2.3. Regulation of MAPK in PPD-Treated NCI-H1299 Cells
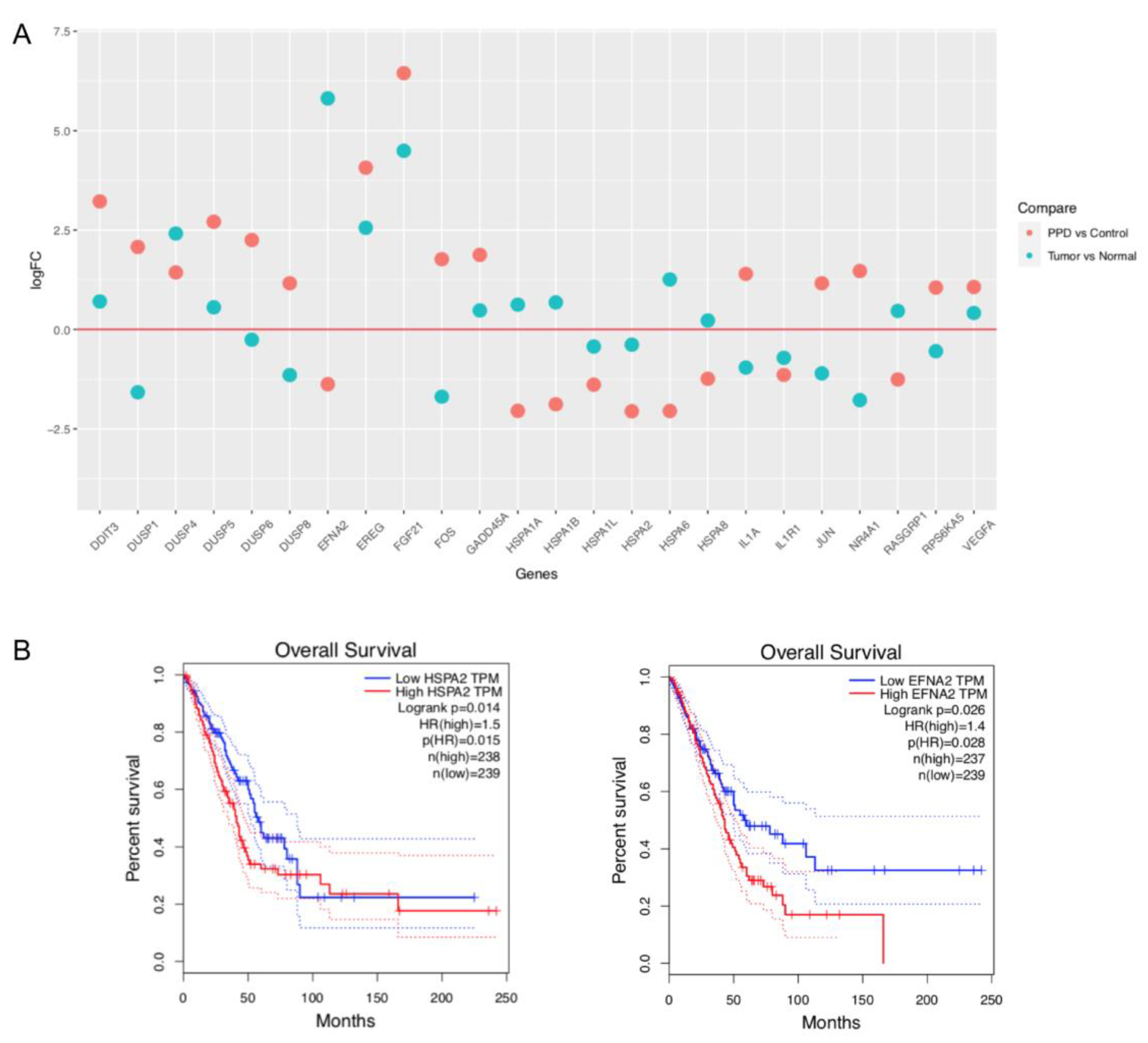
2.4. Comparison of PPD-Regulated DEGs with Clinical Data of Lung Adenocarcinoma Patients
3. Discussion
4. Materials and Methods
4.1. Cell Line and Reagents
4.2. Cell Viability Detection
4.3. Cell Apoptosis and Cell Cycle Analyses
4.4. RNA Sequencing
4.5. Analysis of RNA Sequencing Data
4.6. Western Blotting
4.7. Lung Adenocarcinoma Data Acquisition and Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| A | absorbance |
| CC50 | 50% cytotoxic concentration |
| CCK-8 | cell counting kit-8 |
| DEG | differentially expressed gene |
| DMSO | dimethyl sulfoxide |
| EFNA2 | Ephrin-A2 |
| EGFR-TKI | epidermal growth factor receptor tyrosine kinase inhibitors |
| ELISA | enzyme-linked immunosorbent assay |
| ERK | extracellular signal-regulated kinase |
| FGFR | fibroblast growth factor receptor |
| FITC | fluorescein isothiocyanate |
| FPKM | fragments per kilobase million |
| GEPIA | Gene Expression Profiling Interactive Analysis |
| HSPA | heat shock protein A |
| IC50 | half maximal inhibitory concentration |
| JNK | c-Jun NH2-terminal kinase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | mitogen-activated protein kinases |
| NSCLC | non-small cell lung cancer |
| PI | propidium iodide |
| PPD | 20(S)-Protopanaxadiol |
| RIN | RNA integrity |
| SD | standard deviation |
| TCGA | The Cancer Genome Atlas |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.G.; Haines, C.; Perkel, R.; Enck, R.E. Lung cancer: Diagnosis and management. Am. Fam. Physician 2007, 75, 56–63. [Google Scholar]
- Testa, U.; Castelli, G.; Pelosi, E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers 2018, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.G.; Johnson, D.H.; Azzoli, C.G.; Sause, W.; Smith, T.J.; Baker, S., Jr.; Olak, J.; Stover, D.; Strawn, J.R.; Turrisi, A.T.; et al. American Society of Clinical Oncology treatment of unresectable non-small-cell lung cancer guideline: Update 2003. J. Clin. Oncol. 2004, 22, 330–353. [Google Scholar] [CrossRef]
- Lemjabbar-Alaoui, H.; Hassan, O.U.; Yang, Y.W.; Buchanan, P. Lung cancer: Biology and treatment options. Biochim. Biophys. Acta 2015, 1856, 189–210. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Wu, Y.L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.W.; Kato, T.; et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- Zhong, W.Z.; Wang, Q.; Mao, W.M.; Xu, S.T.; Wu, L.; Wei, Y.C.; Liu, Y.Y.; Chen, C.; Cheng, Y.; Yin, R.; et al. Gefitinib Versus Vinorelbine Plus Cisplatin as Adjuvant Treatment for Stage II-IIIA (N1-N2) EGFR-Mutant NSCLC: Final Overall Survival Analysis of CTONG1104 Phase III Trial. J. Clin. Oncol. 2021, 39, 713–722. [Google Scholar] [CrossRef]
- He, J.; Su, C.; Liang, W.; Xu, S.; Wu, L.; Fu, X.; Zhang, X.; Ge, D.; Chen, Q.; Mao, W.; et al. Icotinib versus chemotherapy as adjuvant treatment for stage II-IIIA EGFR-mutant non-small-cell lung cancer (EVIDENCE): A randomised, open-label, phase 3 trial. Lancet Respir. Med. 2021, 9, 1021–1029. [Google Scholar] [CrossRef]
- Lampridis, S.; Scarci, M. Perioperative systemic therapies for non-small-cell lung cancer: Recent advances and future perspectives. Front. Surg. 2022, 9, 1126486. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Hanna, N. Advances in systemic therapy for non-small cell lung cancer. BMJ 2021, 375, n2363. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, D.; Pantuso, T. Panax ginseng. Am. Fam. Physician 2003, 68, 1539–1542. [Google Scholar] [PubMed]
- Ratan, Z.A.; Haidere, M.F.; Hong, Y.H.; Park, S.H.; Lee, J.O.; Lee, J.; Cho, J.Y. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng Res. 2021, 45, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef]
- Im, D.S. Pro-Resolving Effect of Ginsenosides as an Anti-Inflammatory Mechanism of Panax ginseng. Biomolecules 2020, 10, 444. [Google Scholar] [CrossRef]
- Fan, W.; Huang, Y.; Zheng, H.; Li, S.; Li, Z.; Yuan, L.; Cheng, X.; He, C.; Sun, J. Ginsenosides for the treatment of metabolic syndrome and cardiovascular diseases: Pharmacology and mechanisms. Biomed. Pharmacother. 2020, 132, 110915. [Google Scholar] [CrossRef]
- Bai, L.; Gao, J.; Wei, F.; Zhao, J.; Wang, D.; Wei, J. Therapeutic Potential of Ginsenosides as an Adjuvant Treatment for Diabetes. Front. Pharmacol. 2018, 9, 423. [Google Scholar] [CrossRef]
- Popovich, D.G.; Kitts, D.D. Structure-function relationship exists for ginsenosides in reducing cell proliferation and inducing apoptosis in the human leukemia (THP-1) cell line. Arch. Biochem. Biophys. 2002, 406, 1–8. [Google Scholar] [CrossRef]
- Bae, E.A.; Han, M.J.; Kim, E.J.; Kim, D.H. Transformation of ginseng saponins to ginsenoside Rh2 by acids and human intestinal bacteria and biological activities of their transformants. Arch. Pharm. Res. 2004, 27, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J.W.; Li, W.; Ma, H.; Sun, J.; Deng, M.C.; Yang, L. Ginsenoside metabolites, rather than naturally occurring ginsenosides, lead to inhibition of human cytochrome P450 enzymes. Toxicol. Sci. 2006, 91, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Shen, T.; Kim, H.G.; Hu, W.; Cho, J.Y. 20(S)-Protopanaxadiol from Panax ginseng Induces Apoptosis and Autophagy in Gastric Cancer Cells by Inhibiting Src. Am. J. Chin. Med. 2023, 51, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, F.F.; Guo, Q.; Zheng, Y.Z.; Liu, T.T.; Yang, Z.; Xu, Q.H.; Jiang, Y.; Liu, D.; Zeng, K.W.; Tu, P.F. Photoaffinity Labeling-Based Chemoproteomic Strategy Reveals RBBP4 as a Cellular Target of Protopanaxadiol against Colorectal Cancer Cells. Chembiochem 2022, 23, e202200038. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Jang, D.; Park, S.K.; Lee, M.G.; Cha, B.; Park, C.; Shin, Y.S.; Park, H.; Baek, J.M.; Heo, H.; et al. Ginsenoside 20(S)-protopanaxadiol induces cell death in human endometrial cancer cells via apoptosis. J. Ginseng Res. 2021, 45, 126–133. [Google Scholar] [CrossRef]
- Cao, B.; Qi, Y.; Yang, Y.; Liu, X.; Xu, D.; Guo, W.; Zhan, Y.; Xiong, Z.; Zhang, A.; Wang, A.R.; et al. 20(S)-protopanaxadiol inhibition of progression and growth of castration-resistant prostate cancer. PLoS ONE 2014, 9, e111201. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhang, R.; Xu, H.L.; Yu, X.F.; Qu, S.C.; Sui, D.Y. 20(S)-protopanaxadiol triggers mitochondrial-mediated apoptosis in human lung adenocarcinoma A549 cells via inhibiting the PI3K/Akt signaling pathway. Am. J. Chin. Med. 2013, 41, 1137–1152. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Grodzka, A.; Knopik-Skrocka, A.; Kowalska, K.; Kurzawa, P.; Krzyzaniak, M.; Stencel, K.; Bryl, M. Molecular alterations of driver genes in non-small cell lung cancer: From diagnostics to targeted therapy. Excli J. 2023, 22, 415–432. [Google Scholar]
- de Jong, D.; Das, J.P.; Ma, H.; Pailey Valiplackal, J.; Prendergast, C.; Roa, T.; Braumuller, B.; Deng, A.; Dercle, L.; Yeh, R.; et al. Novel Targets, Novel Treatments: The Changing Landscape of Non-Small Cell Lung Cancer. Cancers 2023, 15, 2855. [Google Scholar] [CrossRef]
- Chang, L.; Karin, M. Mammalian MAP kinase signalling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Liu, Y.; Yang, S.; Wu, X.; Li, H.; Wang, Q. MEK inhibitors for the treatment of non-small cell lung cancer. J. Hematol. Oncol. 2021, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.K.; Basu, S.; Hu, J.; Yie, T.A.; Tchou-Wong, K.M.; Rom, W.N.; Lee, T.C. Selective p38 activation in human non-small cell lung cancer. Am. J. Respir. Cell Mol. Biol. 2002, 26, 558–564. [Google Scholar] [CrossRef]
- Pan, L.; Tan, Y.; Wang, B.; Qiu, W.; Yin, Y.; Ge, H.; Zhu, H. Caspase Recruitment Domain Containing Protein 9 Suppresses Non-Small Cell Lung Cancer Proliferation and Invasion via Inhibiting MAPK/p38 Pathway. Cancer Res. Treat. 2020, 52, 867–885. [Google Scholar] [CrossRef]
- Liu, C.L.; Chen, S.F.; Wu, M.Z.; Jao, S.W.; Lin, Y.S.; Yang, C.Y.; Lee, T.Y.; Wen, L.W.; Lan, G.L.; Nieh, S. The molecular and clinical verification of therapeutic resistance via the p38 MAPK-Hsp27 axis in lung cancer. Oncotarget 2016, 7, 14279–14290. [Google Scholar] [CrossRef]
- Zarczynska, I.; Gorska-Arcisz, M.; Cortez, A.J.; Kujawa, K.A.; Wilk, A.M.; Skladanowski, A.C.; Stanczak, A.; Skupinska, M.; Wieczorek, M.; Lisowska, K.M.; et al. p38 Mediates Resistance to FGFR Inhibition in Non-Small Cell Lung Cancer. Cells 2021, 10, 3363. [Google Scholar] [CrossRef]
- Ciocca, D.R.; Arrigo, A.P.; Calderwood, S.K. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: An update. Arch. Toxicol. 2013, 87, 19–48. [Google Scholar] [CrossRef] [PubMed]
- Scieglinska, D.; Gogler-Piglowska, A.; Butkiewicz, D.; Chekan, M.; Malusecka, E.; Harasim, J.; Habryka, A.; Krawczyk, Z. HSPA2 is expressed in human tumors and correlates with clinical features in non-small cell lung carcinoma patients. Anticancer. Res. 2014, 34, 2833–2840. [Google Scholar] [PubMed]
- Sojka, D.R.; Gogler-Pigłowska, A.; Vydra, N.; Cortez, A.J.; Filipczak, P.T.; Krawczyk, Z.; Scieglinska, D. Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non-small cell lung carcinoma (NSCLC); an implication for NSCLC treatment. Sci. Rep. 2019, 9, 14394. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Katoh, M. Comparative integromics on Ephrin family. Oncol. Rep. 2006, 15, 1391–1395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, S.; Dong, C.; Zhang, J.; Fu, S.; Lv, Y.; Wu, J. A comprehensive prognostic and immunological analysis of ephrin family genes in hepatocellular carcinoma. Front. Mol. Biosci. 2022, 9, 943384. [Google Scholar] [CrossRef]
- Carino, A.; Graziosi, L.; Marchianò, S.; Biagioli, M.; Marino, E.; Sepe, V.; Zampella, A.; Distrutti, E.; Donini, A.; Fiorucci, S. Analysis of Gastric Cancer Transcriptome Allows the Identification of Histotype Specific Molecular Signatures with Prognostic Potential. Front. Oncol. 2021, 11, 663771. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, C.; Zhang, M.; Shi, L.; Wang, J.; Zhang, H.; Ma, P.; Li, S. Ephrin-A2 promotes prostate cancer metastasis by enhancing angiogenesis and promoting EMT. J. Cancer Res. Clin. Oncol. 2021, 147, 2013–2023. [Google Scholar] [CrossRef]
- Huang, Z.R.; Tipparaju, S.K.; Kirpotin, D.B.; Pien, C.; Kornaga, T.; Noble, C.O.; Koshkaryev, A.; Tran, J.; Kamoun, W.S.; Drummond, D.C. Formulation optimization of an ephrin A2 targeted immunoliposome encapsulating reversibly modified taxane prodrugs. J. Control Release 2019, 310, 47–57. [Google Scholar] [CrossRef]
- Lu, J.; Shi, W.; Liang, B.; Chen, C.; Wu, R.; Lin, H.; Zhang, Y.; Han, J. Efficient engulfment of necroptotic and pyroptotic cells by nonprofessional and professional phagocytes. Cell Discov. 2019, 5, 39. [Google Scholar] [CrossRef]
- Xin, H.; Jiang, X.; Gu, J.; Sha, X.; Chen, L.; Law, K.; Chen, Y.; Wang, X.; Jiang, Y.; Fang, X. Angiopep-conjugated poly(ethylene glycol)-co-poly(ε-caprolactone) nanoparticles as dual-targeting drug delivery system for brain glioma. Biomaterials 2011, 32, 4293–4305. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Z.; Hu, J.; Feng, J.; Zhu, Z.; Fan, Y.; Lin, Q.; Ding, G. Mfn2 Regulates High Glucose-Induced MAMs Dysfunction and Apoptosis in Podocytes via PERK Pathway. Front. Cell Dev. Biol. 2021, 9, 769213. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sha, X.; Xin, H.; Chen, L.; Gao, X.; Wang, X.; Law, K.; Gu, J.; Chen, Y.; Jiang, Y.; et al. Self-aggregated pegylated poly (trimethylene carbonate) nanoparticles decorated with c(RGDyK) peptide for targeted paclitaxel delivery to integrin-rich tumors. Biomaterials 2011, 32, 9457–9469. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, Y.; Lou, D.; Gao, Y.; Yu, J.; Kong, D.; Zhang, Q.; Jia, Y.; Zhang, H.; Wang, Z. Sodium arsenite exposure inhibits histone acetyltransferase p300 for attenuating H3K27ac at enhancers in mouse embryonic fibroblast cells. Toxicol. Appl. Pharmacol. 2018, 357, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Meng, Q.; Han, J.; Shen, L.; Sui, X.; Gu, Y.; Wu, G. Regulation of dynamin-related protein 1 (DRP1) levels modulates myoblast atrophy induced by C26 colon cancer-conditioned medium. Transl. Cancer Res. 2021, 10, 3020–3032. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps, R Package, Version 1.0.12; CRAN: Sao Paulo, Brazil, 2019; Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 30 August 2022).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Lin, Y.J.; Flaczyk, A.; Wolfheimer, S.; Goretzki, A.; Jamin, A.; Wangorsch, A.; Vieths, S.; Scheurer, S.; Schülke, S. The Fusion Protein rFlaA:Betv1 Modulates DC Responses by a p38-MAPK and COX2-Dependent Secretion of PGE(2) from Epithelial Cells. Cells 2021, 10, 3415. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, Z.; Zhang, X.; Lv, Z.; Jiang, J.; Wang, L.; Li, J.; Wang, J.; Zhao, J. Transcriptome Analysis of the Inhibitory Effects of 20(S)-Protopanaxadiol on NCI-H1299 Non-Small Cell Lung Cancer Cells. Molecules 2023, 28, 5746. https://doi.org/10.3390/molecules28155746
Cong Z, Zhang X, Lv Z, Jiang J, Wang L, Li J, Wang J, Zhao J. Transcriptome Analysis of the Inhibitory Effects of 20(S)-Protopanaxadiol on NCI-H1299 Non-Small Cell Lung Cancer Cells. Molecules. 2023; 28(15):5746. https://doi.org/10.3390/molecules28155746
Chicago/Turabian StyleCong, Zhongyi, Xinmin Zhang, Zeqi Lv, Jingyuan Jiang, Lei Wang, Jiapeng Li, Jie Wang, and Jianjun Zhao. 2023. "Transcriptome Analysis of the Inhibitory Effects of 20(S)-Protopanaxadiol on NCI-H1299 Non-Small Cell Lung Cancer Cells" Molecules 28, no. 15: 5746. https://doi.org/10.3390/molecules28155746
APA StyleCong, Z., Zhang, X., Lv, Z., Jiang, J., Wang, L., Li, J., Wang, J., & Zhao, J. (2023). Transcriptome Analysis of the Inhibitory Effects of 20(S)-Protopanaxadiol on NCI-H1299 Non-Small Cell Lung Cancer Cells. Molecules, 28(15), 5746. https://doi.org/10.3390/molecules28155746