Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS
Abstract
1. Introduction
2. Results
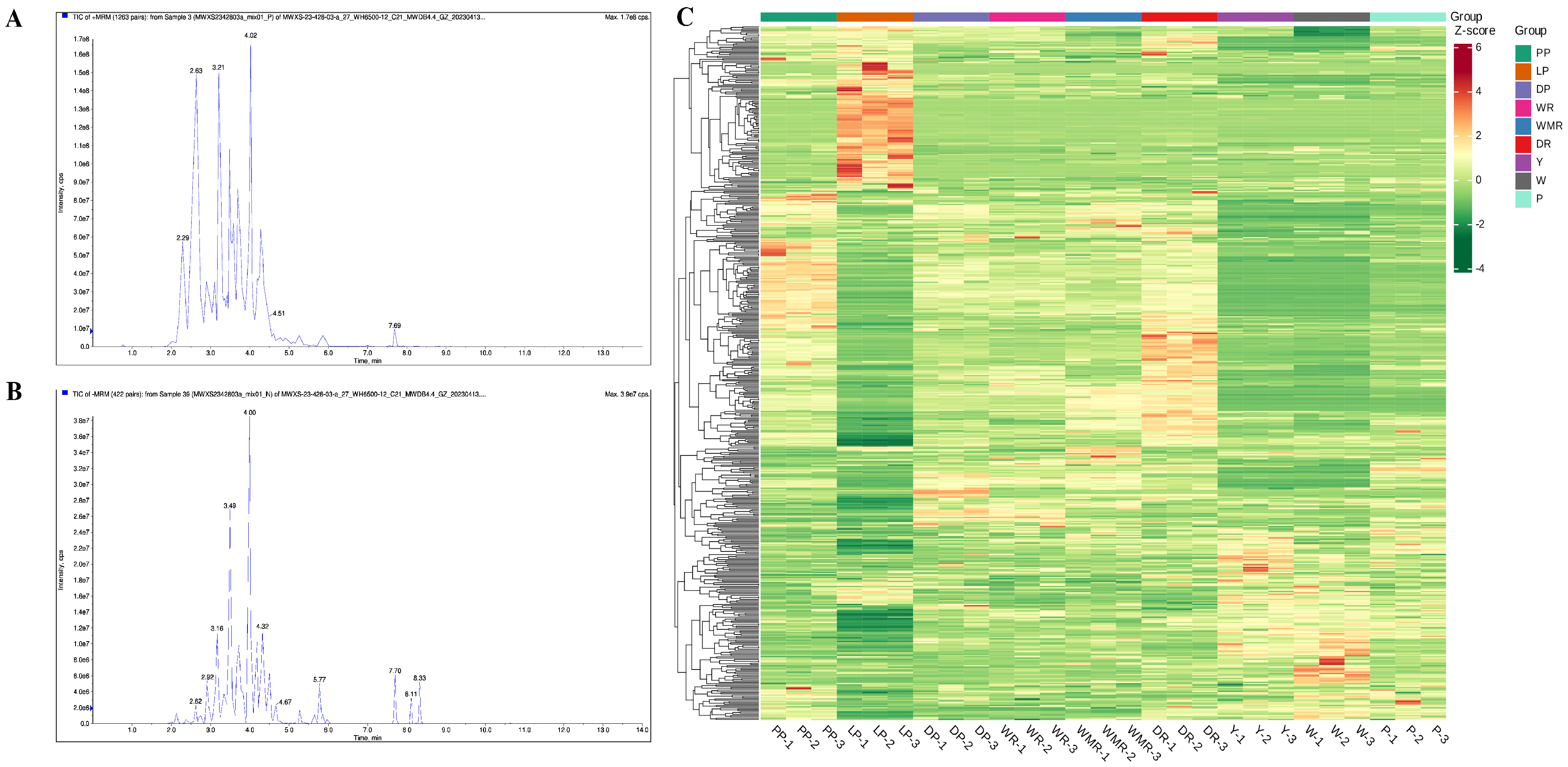
2.1. UPLC-HESI-MS/MS Analysis of Metabolites Profiling in Rapeseed Petals with Different Colors
2.2. Differential Accumulated Metabolites between Yellow and White Petals in Rapeseed
2.3. Differential Accumulated Metabolites in Rapeseed Pink Petals
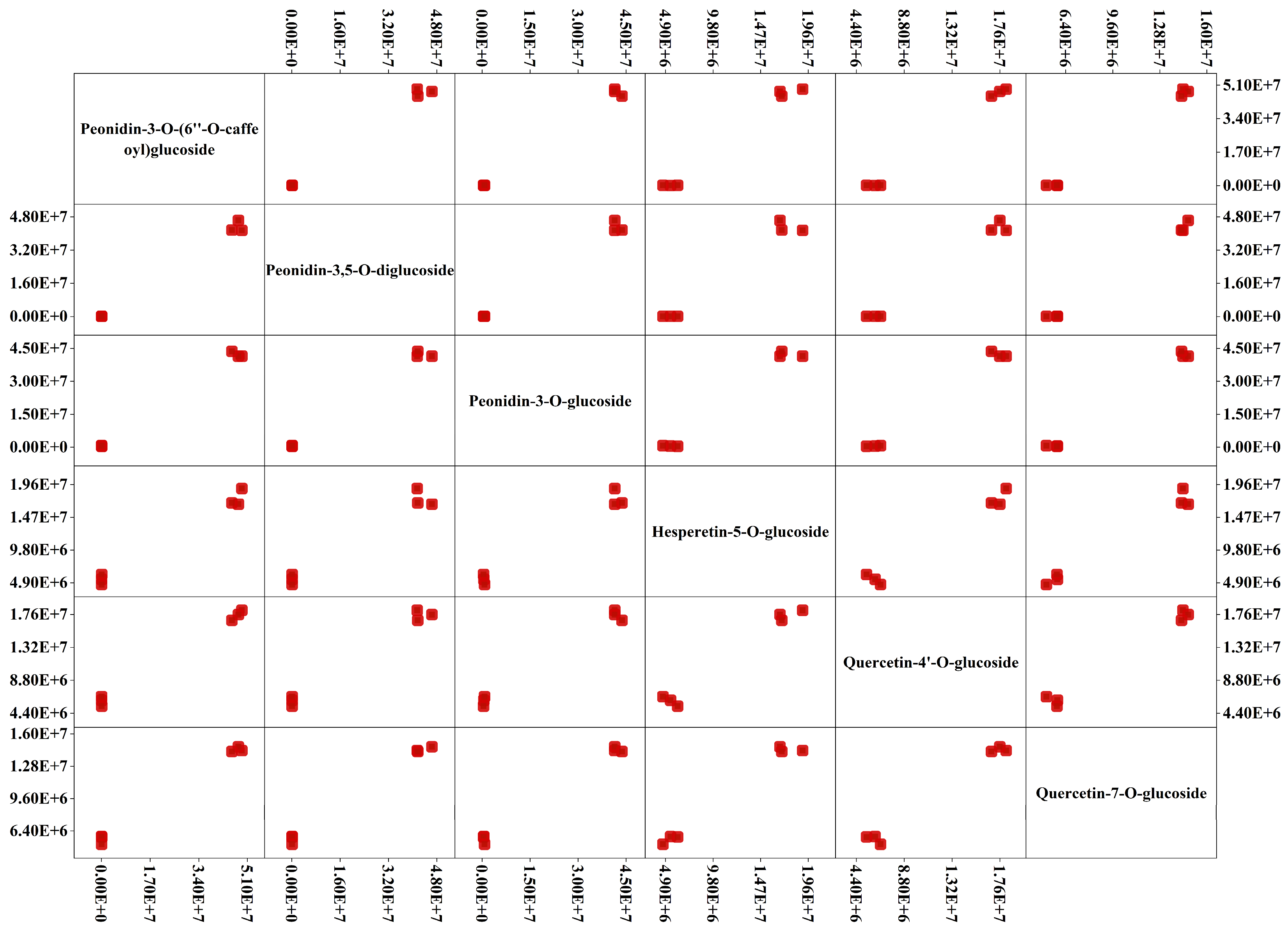
2.4. Differential Accumulated Metabolites between Light Purple, Purple, and Dark Purple Petals in Rapeseed
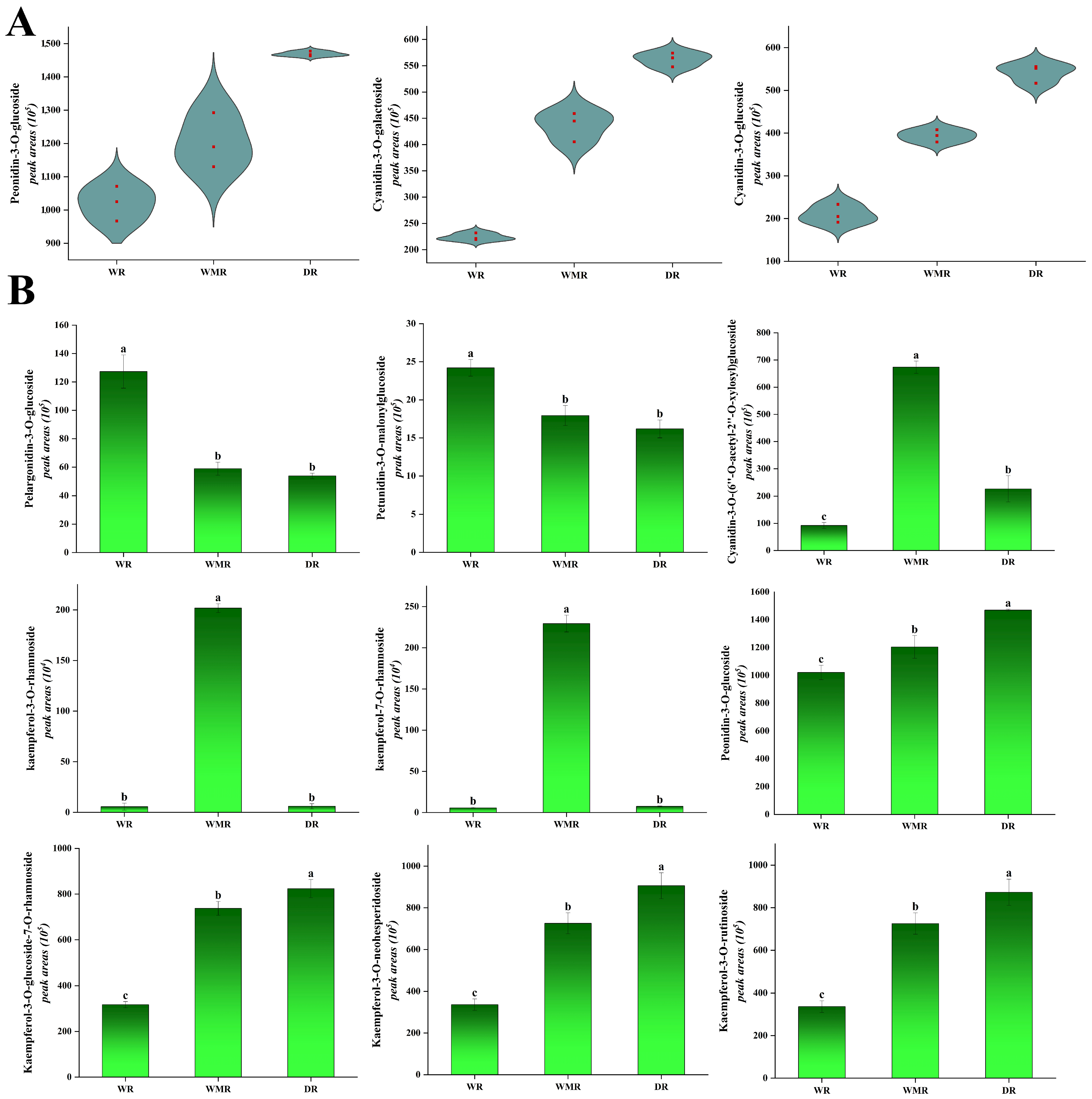
2.5. Differential Accumulated Metabolites between Wine Red, Watermelon Red, and Dark Red Petals in Rapeseed
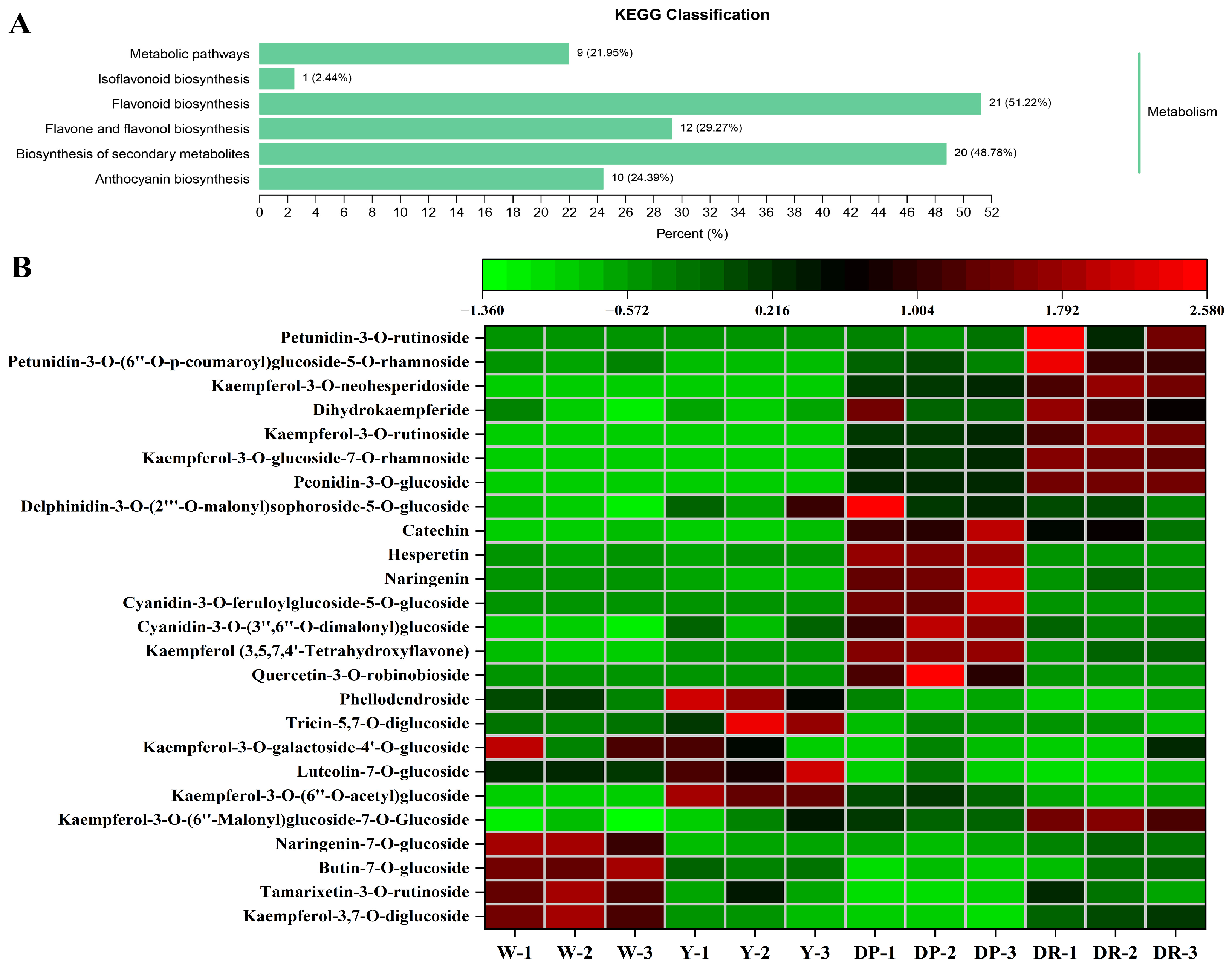
2.6. Differential Accumulated Metabolites between White, Yellow, Purple, and Red Petals in Rapeseed
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Petal Samples Preparation and Extraction
4.3. UPLC-HESI-MS/MS Conditions
4.4. Quantitative Analysis of Metabolites
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Yin, N.-W.; Wang, S.-X.; Jia, L.-D.; Zhu, M.-C.; Yang, J.; Zhou, B.-J.; Yin, J.-M.; Lu, K.; Wang, R.; Li, J.-N.; et al. Identification and characterization of major constituents in different-colored rapeseed petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 2019, 67, 11053–11065. [Google Scholar] [CrossRef]
- Fu, W.; Chen, D.; Pan, Q.; Li, F.; Zhao, Z.; Ge, X.; Li, Z. Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 2018, 16, 367–380. [Google Scholar] [CrossRef]
- Mushtaq, M.-A.; Pan, Q.; Chen, D.; Zhang, Q.; Ge, X.; Li, Z. Comparative leaves transcriptome analysis emphasizing on accumulation of anthocyanins in Brassica: Molecular regulation and potential interaction with photosynthesis. Front. Plant Sci. 2016, 7, 311. [Google Scholar] [CrossRef]
- Jia, L.; Wang, J.; Wang, R.; Duan, M.; Qiao, C.; Chen, X.; Ma, G.; Zhou, X.; Zhu, M.; Jing, F.; et al. Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus. Planta 2021, 253, 8. [Google Scholar] [CrossRef]
- Zheng, T.; Sun, J.-Q.; Shi, X.-J.; Liu, D.-L.; Sun, B.-Y.; Deng, Y.-J.; Zhang, D.-L.; Liu, S.-M. Evaluation of climate factors affecting the quality of red huajiao (Zanthoxylum bungeanum maxim.) based on UPLC-MS/MS and MaxEnt model. Food Chem. X 2022, 16, 100522. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Long, T.; Wang, S.; Yang, J. Regulation mechanism of plant pigments biosynthesis: Anthocyanins, carotenoids, and betalains. Metabolites 2022, 12, 871. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Quattrocchio flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Chen, S.-M.; Li, C.-H.; Zhu, X.-R.; Deng, Y.-M.; Sun, W.; Wang, L.-S.; Chen, F.-D.; Zhang, Z. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 2012, 56, 458–464. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, W.; Xiang, W.; Wang, D.; Xue, B.; Liu, X.; Xing, L.; Wu, D.; Wang, S.; Guo, Q.; et al. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in Lonicera japonica flower petals during colour-transition. BMC Plant Biol. 2021, 21, 98. [Google Scholar] [CrossRef]
- Pan, L.; Li, J.; Yin, H.; Fan, Z.; Li, X. Integrated physiological and transcriptomic analyses reveal a regulatory network of anthocyanin metabolism contributing to the ornamental value in a novel hybrid cultivar of Camellia japonica. Plants 2020, 9, 1724. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; et al. Unraveling the regulatory mechanism of color diversity in Camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Wang, S.-Z.; Huang, S.-Y.; Yang, J.; Li, Z.-L.; Zhang, M.-J.; Fang, Y.-P.; Yang, Q.-F.; Jin, W.-B. Metabolite profiling of violet, white and pink flowers revealing flavonoids composition patterns in Rhododendron pulchrum Sweet. J. Biosci. 2021, 46, 3. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of novel flower colors in floricultural plants: Recent advances via transgenic approaches. Methods Mol. Biol. 2010, 589, 325–347. [Google Scholar]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef]
- Gouvea, A.-C.; Melo, A.; Santiago, M.-C.; Peixoto, F.-M.; Freitas, V.; Godoy, R.-L.; Ferreira, I.-M. Identification and quantification of anthocyanins in fruits from Neomitranthes obscura (DC.) N. Silveira an endemic specie from Brazil by comparison of chromatographic methodologies. Food Chem. 2015, 185, 277–283. [Google Scholar] [CrossRef]
- Khoo, H.-E.; Azlan, A.; Tang, S.-T.; Lim, S.-M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Williams, C.-A.; Grayer, R.-J. Grayer Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef]
- Cartea, M.-E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2010, 16, 251–280. [Google Scholar] [CrossRef]
- Velasco, P.; Francisco, M.; Moreno, D.-A.; Ferreres, F.; Garcia-Viguera, C.; Cartea, M.-E. Phytochemical fingerprinting of vegetable Brassica oleracea and Brassica napus by simultaneous identification of glucosinolates and phenolics. Phytochem. Anal. 2011, 22, 144–152. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Sun, J.; Chen, P.; Harnly, J. UHPLC-PDA-ESI/HRMS/MS(n) analysis of anthocyanins, flavonol glycosides, and hydroxycinnamic acid derivatives in red mustard greens (Brassica juncea Coss variety). J. Agric. Food Chem. 2011, 59, 12059–12072. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Li, X.; Zhang, H.; Zhang, H.; Zhou, J.; Ma, X.; Sun, W.; Yang, W.; He, R.; Wahab, A.T.; et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem. X 2023, 17, 100594. [Google Scholar] [CrossRef] [PubMed]
- Auger, B.; Marnet, N.; Gautier, V.; Maia-Grondard, A.; Leprince, F.; Renard, M.; Guyot, S.; Nesi, N.; Routaboul, J.-M. A detailed survey of seed coat flavonoids in developing seeds of Brassica napus L. J. Agric. Food Chem. 2010, 58, 6246–6256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Q.; Liu, G.-M.; Hu, L.-P.; Zhao, X.-Z.; Zhang, D.-S.; He, H.-J. Prediction of anthocyanidins content in purple Chinese cabbage based on visible/near infrared spectroscopy. Foods 2023, 12, 1922. [Google Scholar] [CrossRef]
- Geng, F.; Nie, R.; Yang, N.; Cai, L.; Hu, Y.; Chen, S.; Cheng, X.; Wang, Z.; Chen, L. Integrated transcriptome and metabolome profiling of Camellia reticulata reveal mechanisms of flower color differentiation. Front. Genet. 2022, 13, 1059717. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, L.-Y.; Wang, S.-L.; Niu, X.-Y. Analysis of anthocyanins and flavonols in petals of 10 Rhododendron species from the Sygera Mountains in Southeast Tibet. Plant Physiol. Biochem. 2016, 104, 250–256. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef]
- Mekapogu, M.; Vasamsetti, B.-M.-K.; Kwon, O.-K.; Ahn, M.-S.; Lim, S.-H.; Jung, J.-A. Anthocyanins in floral colors: Biosynthesis and regulation in chrysanthemum flowers. Int. J. Mol. Sci. 2020, 21, 6537. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.-D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Zhao, C.-C.; Shen, J.; Chen, J.; Shao, J.-H.; Li, K.-H.; Gu, W.-Y.; Miao, B.-J. Phenolic glycoside constituents from Brassica rapa flowers and their alpha-glucosidase inhibitory activity. Nat. Prod. Res. 2019, 33, 3398–3403. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Simal-Gandara, I.; Ma, W.-J.; Peng, Q.-H.; Shi, Y.-L.; Xu, Q.; Lin, Z.; Lv, H.-P. Insight into the pigmented anthocyanins and the major potential co-pigmented flavonoids in purple-coloured leaf teas. Food Chem. 2021, 363, 130278. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xia, Q.; Chen, A.J.; Jin, Z. Copigmentation effect of flavonols on anthocyanins in black mulberry juice and their interaction mechanism investigation. Food Chem. 2023, 399, 133927. [Google Scholar] [CrossRef]
- Fossen, T.; Slimestad, R.; Ovstedal, D.-O.; Andersen, O.-M. Covalent anthocyanin-flavonol complexes from flowers of chive, Allium schoenoprasum. Phytochemistry 2000, 54, 317–323. [Google Scholar] [CrossRef]
- Heursel, J. Diversity of flower colours in Rhododendron simsii Planch. and prospects for breeding. Euphytica 1981, 30, 9–14. [Google Scholar] [CrossRef]
- Gao, S.; Liu, J.; Tang, Y.-Q.; Tang, Z.-H.; Zhang, J. Analysis on the key physicochemical properties of the coloration of varietals of Catharanthus roseus. Non-Wood For. Res. 2022, 40, 214–227. [Google Scholar]
- Hao, Y.; Wang, J.; Hu, C.; Zhou, Q.; Mubeen, H.-M.; Hou, X. Regulation of BcMYB44 on anthocyanin synthesis and drought tolerance in non-heading Chinese cabbage (Brassica campestris ssp. chinensis Makino). Horticulturae 2022, 8, 351. [Google Scholar] [CrossRef]
- Portu, J.; Rosa Gutierrez-Viguera, A.; Gonzalez-Arenzana, L.; Santamaria, P. Characterization of the color parameters and monomeric phenolic composition of ‘Tempranillo’ and ‘Graciano’ wines made by carbonic maceration. Food Chem. 2023, 406, 134327. [Google Scholar] [CrossRef]
- Deng, K.; Ouyang, J.; Hu, N.; Dong, Q.; Chen, C.; Wang, H. Improved stability of blue colour of anthocyanins from Lycium ruthenicum Murr. based on copigmentation. Molecules 2022, 27, 6089. [Google Scholar] [CrossRef]
- Labadie, M.; Vallin, G.; Potier, A.; Petit, A.; Ring, L.; Hoffmann, T.; Gaston, A.; Munoz-Blanco, J.; Caballero, J.-L.; Schwab, W.; et al. High resolution quantitative trait locus mapping and whole genome sequencing enable the design of an anthocyanidin reductase-specific homoeo-allelic marker for fruit colour improvement in octoploid strawberry (Fragaria × ananassa). Front. Plant Sci. 2022, 13, 869655. [Google Scholar] [CrossRef]
- Chen, K.; Kortesniemi, M.-K.; Linderborg, M.-K.; Yang, B.-R. Anthocyanins as promising molecules affecting energy homeostasis, inflammation, and gut microbiota in type 2 diabetes with special reference to impact of acylation. J. Agric. Food Chem. 2022, 71, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Matsuba, Y.; Abe, Y.; Okamura, M.; Momose, M.; Umemoto, N.; Nakayama, M.; Itoh, Y.; Ozeki, Y. Recent advances in understanding the anthocyanin modification steps in carnation flowers. Sci. Hortic. 2013, 163, 37–45. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Hou, H.; Ma, X.; Sun, S.; Wang, H.; Kong, L. Metabolomics and gene expression analysis reveal the accumulation patterns of phenylpropanoids and flavonoids in different colored-grain wheats (Triticum aestivum L.). Food Res. Int. 2020, 138, 109711. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Wang, X.; Chang, T.; Peng, Z.; Guan, C.; Guan, M. Flavonoid synthesis-related genes determine the color of flower petals in Brassica napus L. Int. J. Mol. Sci. 2023, 24, 6472. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, H.; Zheng, T.; Li, Y.; Chen, Q.; Xue, Y.; Tang, Q.; Xu, H.; Chen, M. Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS. Molecules 2023, 28, 5670. https://doi.org/10.3390/molecules28155670
Zeng H, Zheng T, Li Y, Chen Q, Xue Y, Tang Q, Xu H, Chen M. Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS. Molecules. 2023; 28(15):5670. https://doi.org/10.3390/molecules28155670
Chicago/Turabian StyleZeng, Haitao, Tao Zheng, Ying Li, Qiao Chen, Yan Xue, Qi Tang, Hao Xu, and Mengjiao Chen. 2023. "Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS" Molecules 28, no. 15: 5670. https://doi.org/10.3390/molecules28155670
APA StyleZeng, H., Zheng, T., Li, Y., Chen, Q., Xue, Y., Tang, Q., Xu, H., & Chen, M. (2023). Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS. Molecules, 28(15), 5670. https://doi.org/10.3390/molecules28155670





