Statistical Modelling Investigation of MALDI-MSI-Based Approaches for Document Examination
Abstract
1. Introduction
2. Results and Discussion
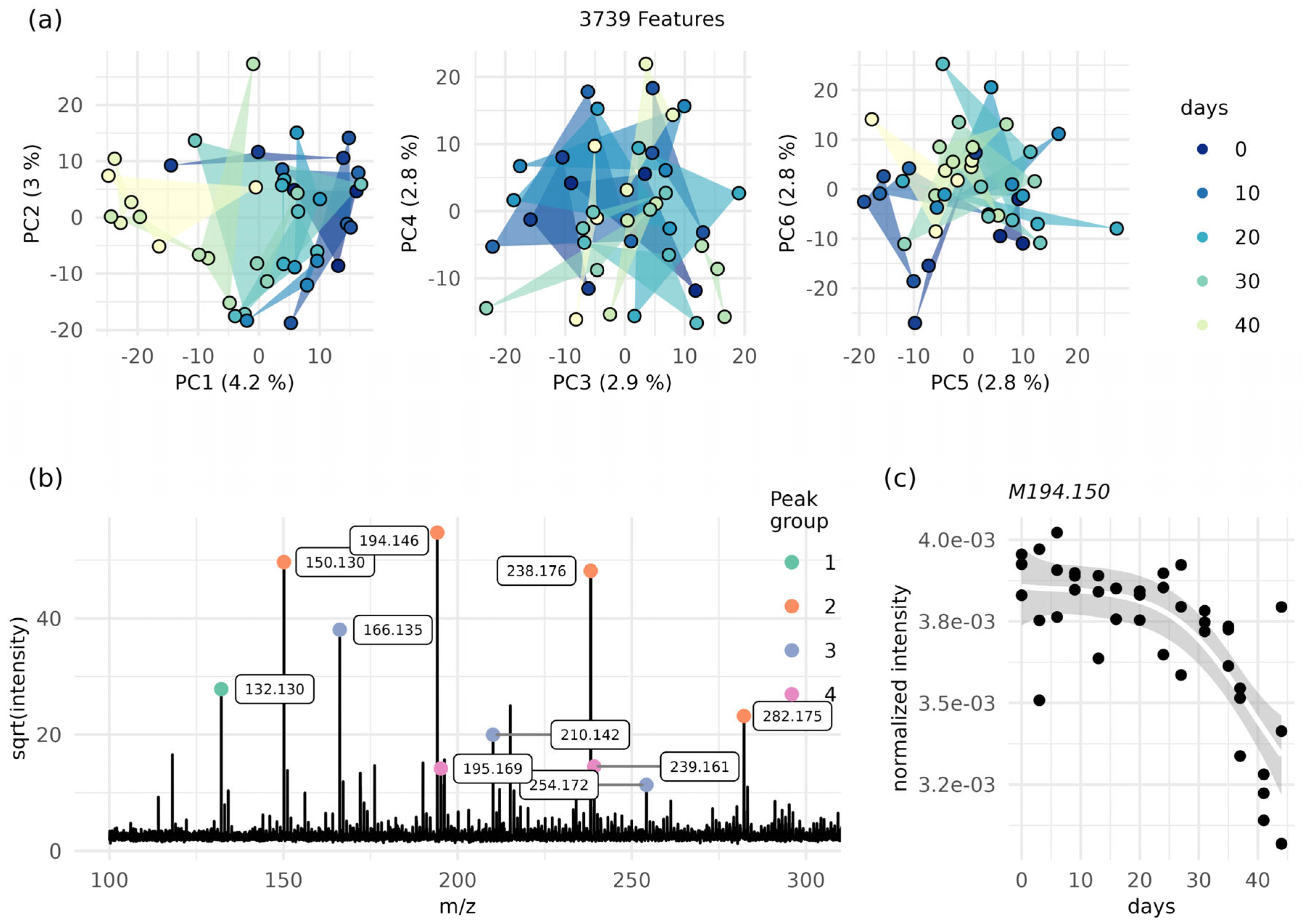
2.1. Determination of Ink Age (Time since Ink Deposition)
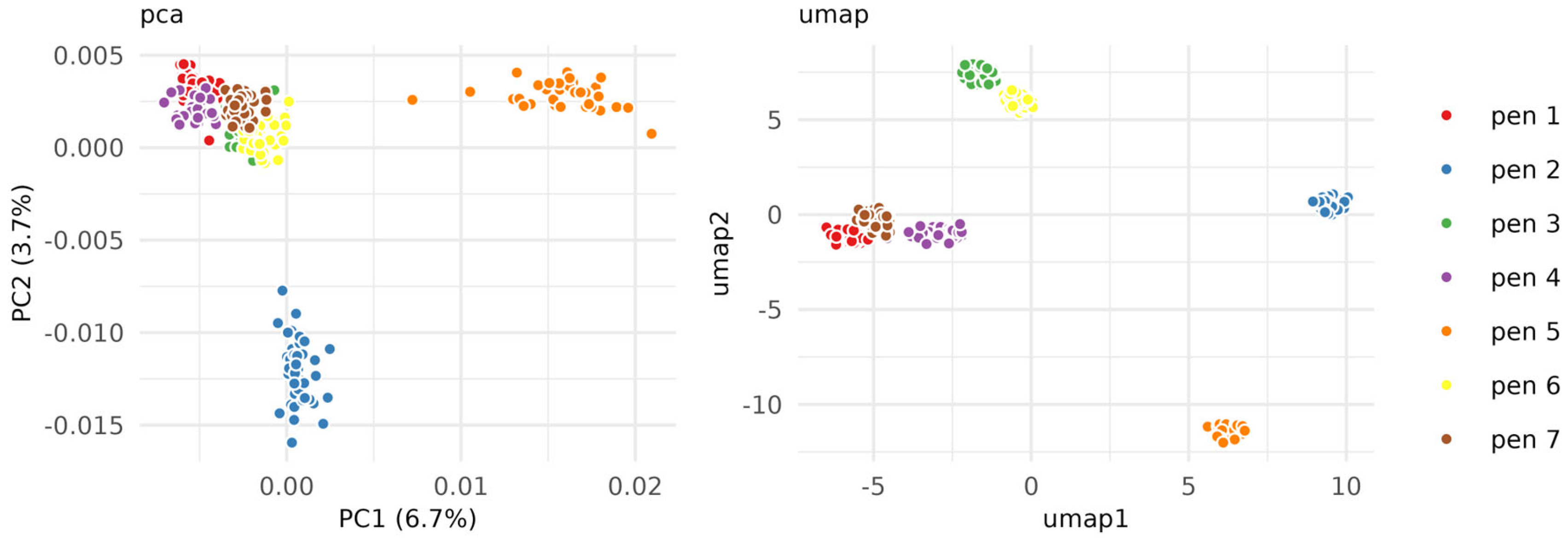
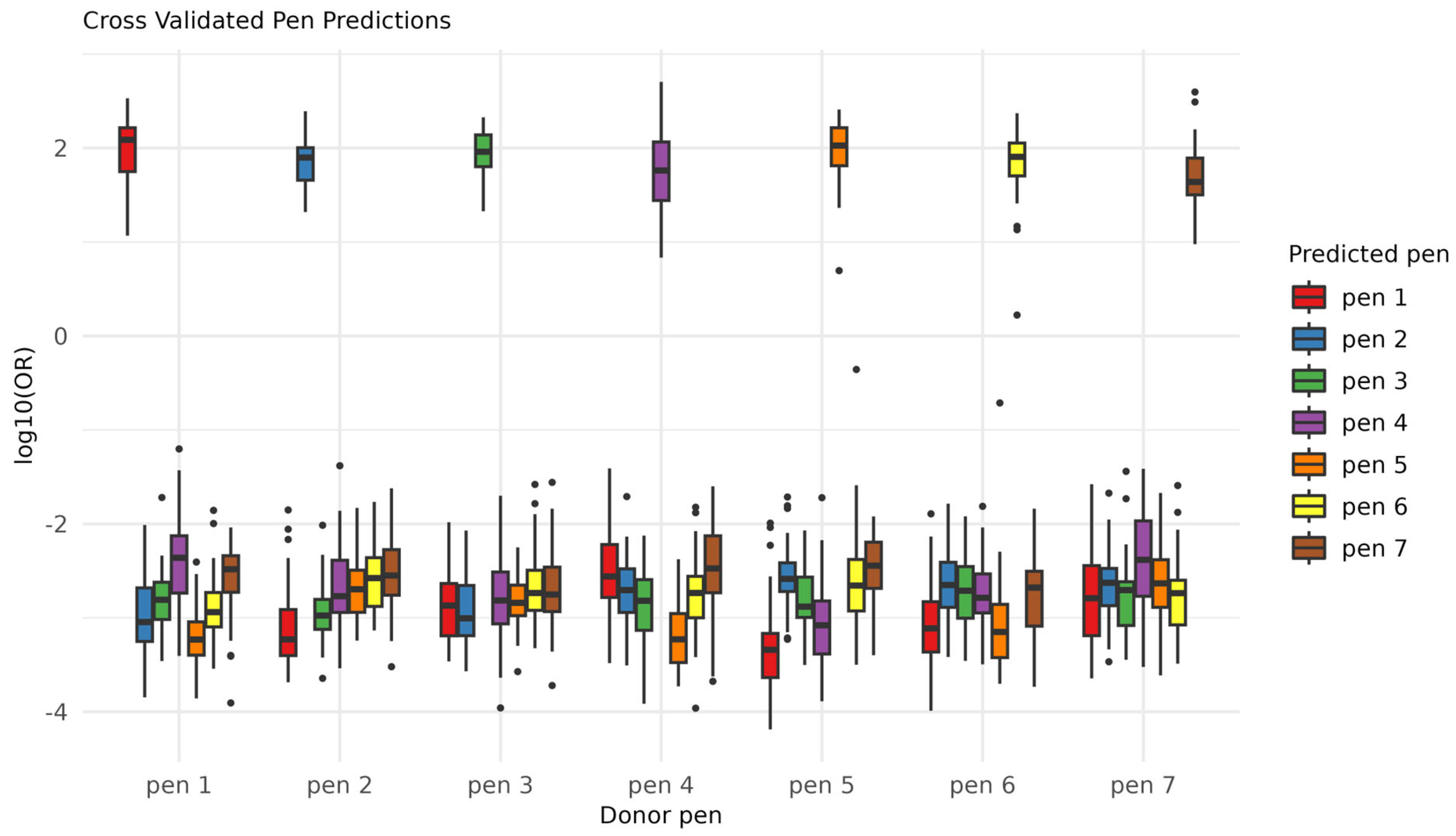
2.2. Ink Classification
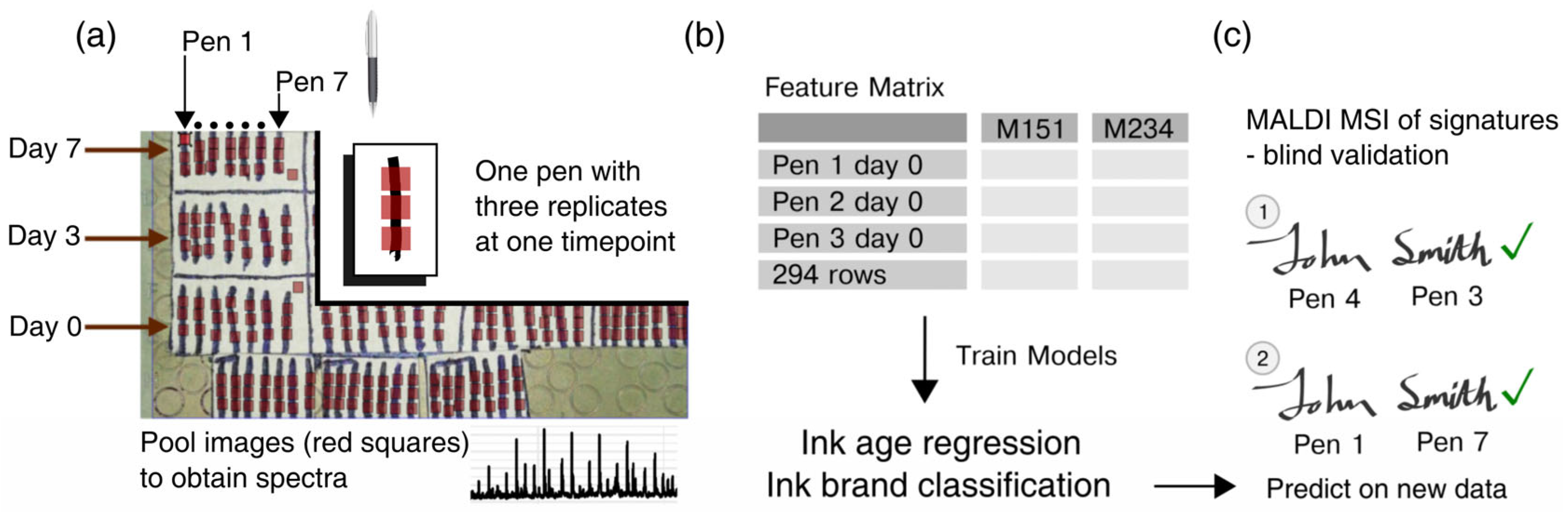
2.3. Method Validation
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Generation of Sample Datasets
3.2.2. MALDI Matrix Deposition
3.2.3. MALDI-MSI of Ink from Black Gel Pen Strokes and Signatures
3.3. Statistical Analysis and Software
3.3.1. Data Preprocessing
3.3.2. Statistical Machine Learning
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.-Z.; Yu, J.; Xie, M.-X.; Liu, Y.; Han, J.; Jing, T.-T. Classification and dating of black gel pen ink by ion-pairing high-performance liquid chromatography. J. Chromatogr. A 2009, 1135, 57–64. [Google Scholar] [CrossRef]
- Erdoğan, A. Analysis and chemical imaging of blue inks for the investigation of document forgery by XPS. Microchem. J. 2022, 183, 108062. [Google Scholar] [CrossRef]
- Silva, C.S.; Pimentel, M.F.; Honorato, R.S.; Pasquini, C.; Prats-Montalbán, J.M.; Ferrer, A. Near infrared hyperspectral imaging for forensic analysis of document forgery. Analyst 2014, 139, 5176–5184. [Google Scholar] [CrossRef]
- de Souza Lins Borba, F.; Saldanha Honorato, R.; de Juan, A. Use of Raman spectroscopy and chemometrics to distinguish blue ballpoint pen inks. Forensic Sci. Int. 2015, 249, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Mohamad Asri, M.N.; Verma, R.; Mahat, N.A.; Nor, N.A.M.; Mat Desa, W.N.S.; Ismail, D. Discrimination and source correspondence of black gel inks using Raman spectroscopy and chemometric analysis with UMAP and PLS-DA. Chemom. Intell. Lab. Syst. 2022, 225, 104557. [Google Scholar] [CrossRef]
- Adam, C.D.; Sherratt, S.L.; Zholobenko, V.L. Classification and individualisation of black ballpoint pen inks using principal component analysis of UV–vis absorption spectra. Forensic Sci. Int. 2008, 174, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sharma, V. A novel combined approach of diffuse reflectance UV–Vis-NIR spectroscopy and multivariate analysis for non-destructive examination of blue ballpoint pen inks in forensic application. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 175, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Sauzier, G.; McGann, J.; Lewis, S.W.; van Bronswijk, W. A study into the ageing and dating of blue ball tip inks on paper using in situ visible spectroscopy with chemometrics. Anal. Methods 2018, 10, 5613–5621. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Yao, L. Dating the writing age of black roller and gel inks by gas chromatography and UV–vis spectrophotometer. Forensic Sci. Int. 2006, 162, 140–143. [Google Scholar] [CrossRef]
- Li, B.; Xie, P.; Guo, Y.-M.; Fei, Q. GC Analysis of Black Gel Pen Ink Stored under Different Conditions. J. Forensic Sci. 2014, 59, 543–549. [Google Scholar] [CrossRef]
- Ifa, D.R.; Gumaelius, L.M.; Eberlin, L.S.; Manicke, N.E.; Cooks, R.G. Forensic analysis of inks by imaging desorption electrospray ionization (DESI) mass spectrometry. Analyst 2007, 132, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Khatami, A.; Prova, S.S.; Bagga, A.K.; Yan Chi Ting, M.; Brar, G.; Ifa, D.R. Detection and imaging of thermochromic ink compounds in erasable pens using desorption electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2017, 31, 983–990. [Google Scholar] [CrossRef]
- Sun, Q.; Luo, Y.; Wang, Y.; Zhang, Q.; Yang, X. Comparative analysis of aged documents by desorption electrospray ionization–Mass spectrometry (DESI-MS) imaging. J. Forensic Sci. 2022, 67, 2062–2072. [Google Scholar] [CrossRef]
- Siegel, J.; Allison, J.; Mohr, D.; Dunn, J. The use of laser desorption/ionization mass spectrometry in the analysis of inks in questioned documents. Talanta 2005, 67, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Weyermann, C.; Bucher, L.; Majcherczyk, P.; Mazzella, W.; Roux, C.; Esseiva, P. Statistical discrimination of black gel pen inks analysed by laser desorption/ionization mass spectrometry. Forensic Sci. Int. 2012, 217, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Weyermann, C.; Bucher, L.; Majcherczyk, P. A statistical methodology for the comparison of blue gel pen inks analyzed by laser desorption/ionization mass spectrometry. Sci. Justice 2011, 51, 122–130. [Google Scholar] [CrossRef]
- Kumar, R.; Sharma, V. Chemometrics in forensic science. TrAC Trends Anal. Chem. 2018, 105, 191–201. [Google Scholar] [CrossRef]
- Calcerrada, M.; García-Ruiz, C. Analysis of questioned documents: A review. Anal. Chim. Acta 2015, 853, 143–166. [Google Scholar] [CrossRef]
- Khan, M.J.; Khan, H.S.; Yousaf, A.; Khurshid, K.; Abbas, A. Modern Trends in Hyperspectral Image Analysis: A Review. IEEE Access 2018, 6, 14118–14129. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, C.-X.; Yu, J.; Liu, H.-L.; Xie, M.-X. Differentiation and dating of gel pen ink entries on paper by laser desorption ionization- and quadruple-time of flight mass spectrometry. Dye. Pigment. 2012, 94, 525–532. [Google Scholar] [CrossRef]
- Weyermann, C.; Kirsch, D.; Vera, C.C.; Spengler, B. A GC/MS study of the drying of ballpoint pen ink on paper. Forensic Sci. Int. 2007, 168, 119–127. [Google Scholar] [CrossRef]
- Fischer, T.; Marchetti-Deschmann, M.; Assis, C.A.; Levin Elad, M.; Algarra, M.; Barac, M.; Radovic, I.B.; Cicconi, F.; Claes, B.; Frascione, N.; et al. Profiling and imaging of forensic evidence—A pan-European forensic round robin study part 1: Document forgery. Sci. Justice 2022, 62, 433–447. [Google Scholar] [CrossRef]
- Sugawara, S. Comparison of preprocessing and machine learning methods for identifying writing inks using mid-infrared hyperspectral imaging and machine learning. Infrared Phys. Technol. 2022, 127, 104357. [Google Scholar] [CrossRef]
- Bradshaw, R.; Denison, N.; Francese, S. Implementation of MALDI MS profiling and imaging methods for the analysis of real crime scene fingermarks. Analyst 2017, 142, 1581–1590. [Google Scholar] [CrossRef]
- Bradshaw, R.; Wilson, G.; Denison, N.; Francese, S. Application of MALDI MS imaging after sequential processing of latent fingermarks. Forensic Sci. Int. 2021, 319, 110643. [Google Scholar] [CrossRef]
- Bandey, H. Fingermark Visualisation Manual, 2nd ed.; Dstl: Salisbury, UK, 2023; ISBN 978-1-3999-0976-1. [Google Scholar]
- Kennedy, K.; Heaton, C.; Langenburg, G.; Cole, L.; Clark, T.; Clench, M.R.; Sears, V.; Sealey, M.; McColm, R.; Francese, S. Pre-validation of a MALDI MS proteomics-based method for the reliable detection of blood and blood provenance. Sci. Rep. 2020, 10, 17087. [Google Scholar] [CrossRef]
- Kennedy, K.; Gannicliffe, C.; Cole, L.M.; Sealey, M.; Francese, S. In depth investigation of the capabilities and limitations of combined proteomic-MALDI MS based approach for the forensic detection of blood. Sci. Justice 2022, 62, 602–609. [Google Scholar] [CrossRef]
- Kennedy, K.; Bengiat, R.; Heaton, C.; Herman, Y.; Oz, C.; Elad, M.L.; Cole, L.; Francese, S. “MALDI-CSI”: A proposed method for the tandem detection of human blood and DNA typing from enhanced fingermarks. Forensic Sci. Int. 2021, 323, 110774. [Google Scholar] [CrossRef]
- Flinders, B.; Bassindale, T.; Heeren, R.M.A. Recent Technological Developments in MALDI-MSI Based Hair Analysis. In Emerging Technologies for the Analysis of Forensic Traces; Francese, S., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 133–149. [Google Scholar]
- Grasso, G.; Calcagno, M.; Rapisarda, A.; D’Agata, R.; Spoto, G. Atmospheric pressure MALDI for the noninvasive characterization of carbonaceous ink from Renaissance documents. Anal. Bioanal. Chem. 2017, 409, 3943–3950. [Google Scholar] [CrossRef]
- Dunn, J.D.; Allison, J. The detection of multiply charged dyes using matrix-assisted laser desorption/ionization mass spectrometry for the forensic examination of pen ink dyes directly from paper. J. Forensic Sci. 2007, 52, 1205–1211. [Google Scholar] [CrossRef]
- Weyermann, C.; Kirsch, D.; Costa-Vera, C.; Spengler, B. Photofading of ballpoint dyes studied on paper by LDI and MALDI MS. J. Am. Soc. Mass Spectrom. 2006, 17, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Gibb, S.; Strimmer, K. MALDIquant: A versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012, 28, 2270–2271. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Luo, Y.; Xiang, P.; Yang, X.; Shen, M. Analysis of PEG oligomers in black gel inks: Discrimination and ink dating. Forensic Sci. Int. 2017, 277, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and Variable Selection via the Elastic Net. J. R. Stat. Society. Ser. B (Stat. Methodol.) 2005, 67, 301–320. Available online: http://www.jstor.org/stable/3647580 (accessed on 1 April 2023). [CrossRef]
- Ortiz-Herrero, L.; Bartolomé, L.; Durán, I.; Velasco, I.; Alonso, M.L.; Maguregui, M.I.; Ezcurra, M. DATUVINK pilot study: A potential non-invasive methodology for dating ballpoint pen inks using multivariate chemometrics based on their UV–vis-NIR reflectance spectra. Microchem. J. 2018, 140, 158–166. [Google Scholar] [CrossRef]
- Weyermann, C.; Spengler, B. The potential of artificial aging for modelling of natural aging processes of ballpoint ink. Forensic Sci. Int. 2008, 180, 23–31. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Lassen, J.; Nielsen, K.L.; Johannsen, M.; Villesen, P. Assessment of XCMS Optimization Methods with Machine-Learning Performance. Anal. Chem. 2021, 93, 13459–13466. [Google Scholar] [CrossRef]
- Lassen, J.K.; Wang, T.; Nielsen, K.L.; Hasselstrøm, J.B.; Johannsen, M.; Villesen, P. Large-Scale metabolomics: Predicting biological age using 10,133 routine untargeted LC–MS measurements. Aging Cell 2023, 22, e13813. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]

| Brand | Pen ID | No. of Features with FDRs < 0.01 | RMSE (Days) |
|---|---|---|---|
| ASDA Gel Pen | 1 | 122 | 13.3 |
| Uniball Signo UMN 207 | 4 | 117 | 12.8 |
| Pentel Energel BL77-A | 5 | 68 | 14.4 |
| Pilot G1-100 | 2 | 66 | 12.9 |
| Tesco Gel Pen | 3 | 64 | 14.9 |
| Zebra Sarasa | 6 | 51 | 14.9 |
| WHSmith Rollerball Gel Pen | 7 | 44 | 15.1 |
| Training Data | Validation Data | |
|---|---|---|
| # of Paper Sheets | 1 | 1 |
| # of MALDI-MSI Samples | 294 | 5 |
| # of Pens | 7 | Up to 7 (blinded) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kjeldbjerg Lassen, J.; Bradshaw, R.; Villesen, P.; Francese, S. Statistical Modelling Investigation of MALDI-MSI-Based Approaches for Document Examination. Molecules 2023, 28, 5207. https://doi.org/10.3390/molecules28135207
Kjeldbjerg Lassen J, Bradshaw R, Villesen P, Francese S. Statistical Modelling Investigation of MALDI-MSI-Based Approaches for Document Examination. Molecules. 2023; 28(13):5207. https://doi.org/10.3390/molecules28135207
Chicago/Turabian StyleKjeldbjerg Lassen, Johan, Robert Bradshaw, Palle Villesen, and Simona Francese. 2023. "Statistical Modelling Investigation of MALDI-MSI-Based Approaches for Document Examination" Molecules 28, no. 13: 5207. https://doi.org/10.3390/molecules28135207
APA StyleKjeldbjerg Lassen, J., Bradshaw, R., Villesen, P., & Francese, S. (2023). Statistical Modelling Investigation of MALDI-MSI-Based Approaches for Document Examination. Molecules, 28(13), 5207. https://doi.org/10.3390/molecules28135207







