Palladium and Platinum Complexes of the Antimetabolite Fludarabine with Vastly Enhanced Selectivity for Tumour over Non-Malignant Cells
Abstract
1. Introduction
2. Results and Discussion
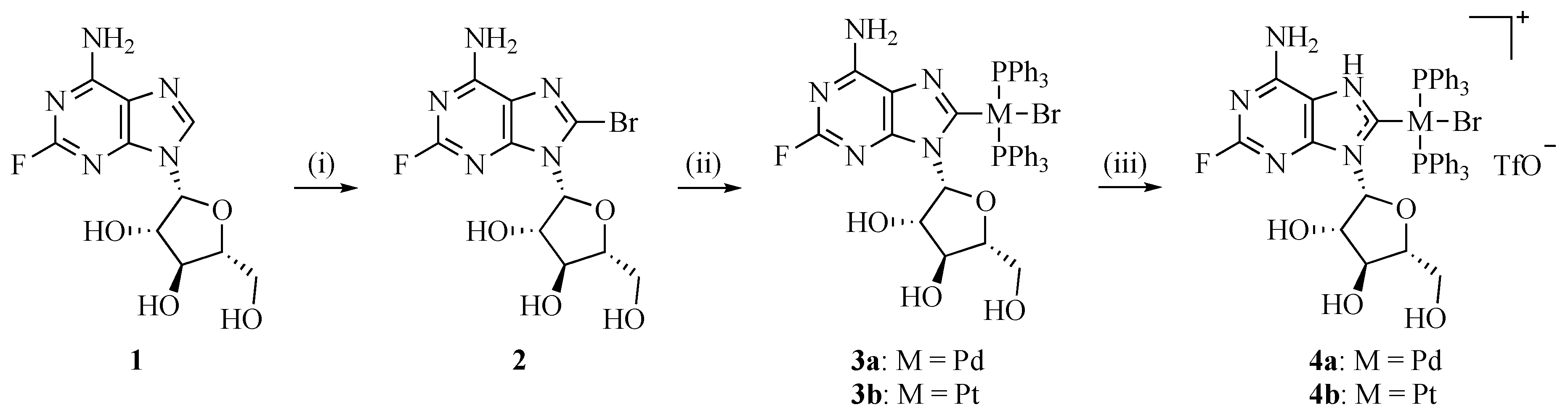
2.1. Complex Syntheses and Characterisation
2.2. Cytotoxicity
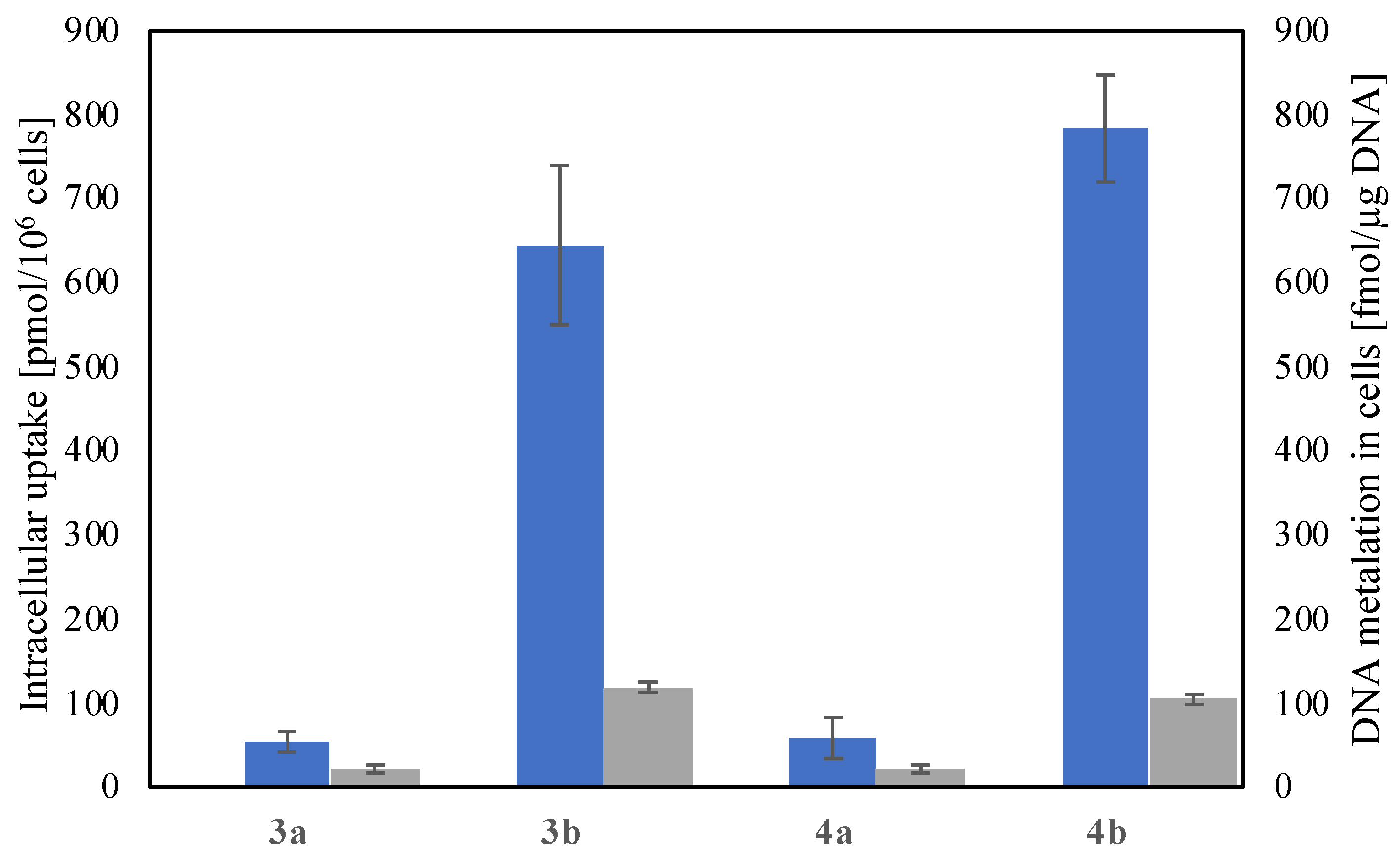
2.3. Cellular Uptake and DNA Metalation
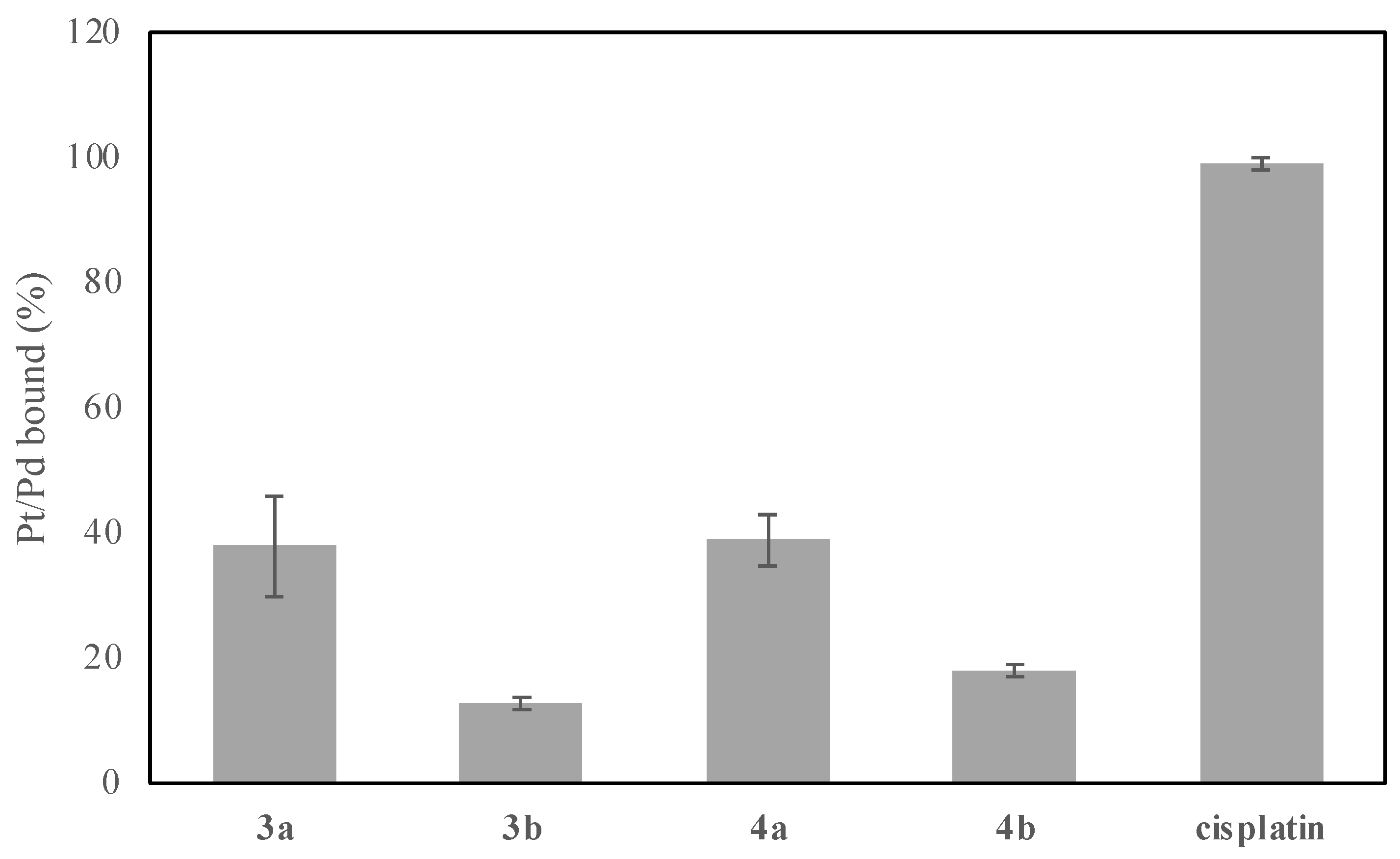
2.4. DNA Binding in Cell-Free Medium
3. Materials and Methods
3.1. General
3.2. Chemistry
3.2.1. 8-Bromofludarabine (2)
3.2.2. General Procedure for the Oxidative Addition of 2
3.2.3. trans-[bromido-(fludarabin-8-yl)-bis(triphenylphosphane)]palladium(II) (3a)
3.2.4. trans-[bromido-(fludarabin-8-yl)-bis(triphenylphosphane)]platinum(II) (3b)
3.2.5. General Procedure for N7-Protonation of Complexes 3
3.2.6. trans-[bromido-(7-H-fludarabin-8-ylidene)-bis(triphenylphosphane)]palladium(II) triflate (4a)
3.2.7. trans-[bromido-(7-H-fludarabin-8-ylidene)-bis(triphenylphosphane)]platinum(II) triflate (4b)
3.3. Bioevaluation
3.3.1. Cell Culture
3.3.2. Resazurin Cell Viability Assay (Suspension Culture)
3.3.3. SRB Assay (Monolayer Culture)
3.3.4. Intracellular Accumulation
3.3.5. Determination of the Amount of Metal Associated with DNA
3.3.6. DNA Binding in Cell-Free Media
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- El-Zine, M.A.Y.; Alhadi, A.M.; Ishak, A.A.; Al-Shamahy, H.A. Prevalence of different types of leukemia and associated factors among children with leukemia in children’s cancer units at Al-Kuwait Hospital, Sana’a City: A cross-sectional study. Glob. J. Ped. Neonatol. Car. 2021, 3, 000569. [Google Scholar] [CrossRef]
- Burger, J.A. Treatment of chronic lymphocytic leukemia. N. Engl. J. Med. 2020, 383, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Niemann, C.U.; Kater, A.P.; Gregor, M.; Cymbalista, F.; Buske, C.; on behalf of the ESMO Guidelines Committee; et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Tu, Y.; Freter, C.E. Fludarabine-resistance associates with ceramide metabolism and leukemia stem cell development in chronic lymphocytic leukemia. Oncotarget 2018, 9, 33124–33137. [Google Scholar] [CrossRef]
- Hallek, M.; Fischer, K.; Fingerle-Rowson, G.; Fink, A.M.; Busch, R.; Mayer, J.; Hensel, M.; Hopfinger, G.; Hess, G.; Von Grünhagen, U.; et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukemia: A randomised, open-label, phase 3 trial. Lancet 2010, 376, 1164–1174. [Google Scholar] [CrossRef]
- Hanauske, A.-R.; von Hoff, D.D. Clinical development of fludarabine. In Nucleoside Analogs in Cancer Therapy; Cheson, B.D., Keating, M.J., Plunkett, W., Eds.; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Gandhi, V.; Plunkett, W. Cellular and clinical pharmacology of fludarabine. Clin. Pharmacokinet. 2002, 41, 93–103. [Google Scholar] [CrossRef]
- Huang, P.; Chubb, S.; Plunkett, W. Termination of DNA synthesis by 9-beta-d-arabinosyl-2-fluoroadenine. A mechanism for cytotoxicity. J. Biol. Chem. 1990, 265, 16617–16625. [Google Scholar] [CrossRef]
- Plunkett, W.; Huang, P.; Gandhi, V. Metabolism and action of fludarabine phosphate. Semin. Oncol. 1990, 17, 3–17. [Google Scholar]
- Huang, P.; Sandoval, A.; Van Den Neste, E.; Keating, M.J.; Plunkett, W. Inhibition of RNA transcription: A biochemical mechanism of action against chrocic lymphocytic leukemia cells by fludarabine. Leukemia 2000, 14, 1405–1413. [Google Scholar] [CrossRef]
- Christopherson, R.I.; Mactier, S.; Almazi, J.G.; Kohnke, P.L.; Best, O.G.; Mulligan, S.P. Mechanism of action of fludarabine nucleoside against human Raji lymphoma cells. Nucleos. Nucleot Nucl. 2014, 33, 375–383. [Google Scholar] [CrossRef]
- Takagi, K.; Kawai, Y.; Yamauchi, T.; Iwasaki, H.; Ueda, T. Synergistic effects of combination with fludarabine and carboplatin depend on fludarabine-mediated inhibition of enhanced nucleotide excision repair in leukemia. Int. J. Hematol. 2011, 94, 378–389. [Google Scholar] [CrossRef]
- Zecevic, A.; Sampath, D.; Ewald, B.; Chen, R.; Wierda, W.; Plunkett, W. Killing of chronic lymphocytic leukemia by the combination of fludarabine and oxaliplatin is dependent on the activity of XPF endonuclease. Clin. Cancer Res. 2011, 17, 4731–4741. [Google Scholar] [CrossRef]
- Moufarij, M.A.; Sampath, D.; Keating, M.J.; Plunkett, W. Fludarabine increases oxaliplatin cytotoxicity in normal and chronic lymphocytic leukemia lymphocytes by suppressing interstrand DNA crosslink removal. Blood 2006, 108, 4187–4193. [Google Scholar] [CrossRef]
- Lohmann, G.; Vasyutina, E.; Bloehdorn, J.; Reinart, N.; Schneider, J.; Babu, V.; Knittel, G.; Mayer, P.; Prinz, C.; Biersack, B.; et al. Targeting transcription-coupled nucleotide excision repair overcomes resistance in chronic lymphocytic leukemia. Leukemia 2017, 31, 1177–1186. [Google Scholar] [CrossRef]
- Gianferrara, T.; Bratsos, I.; Alessio, E. A categorization of metal anticancer compounds based on their mode of action. Dalton Trans. 2009, 37, 7588–7598. [Google Scholar] [CrossRef]
- Bär, S.I.; Gold, M.; Schleser, S.W.; Rehm, T.; Bär, A.; Köhler, L.; Carnell, L.R.; Biersack, B.; Schobert, R. Guided antitumoral drugs: (imidazol-2-ylidene)(L)gold(I) complexes seeking cellular targets controlled by the nature of ligand L. Chem. Eur. J. 2021, 27, 5003–5010. [Google Scholar] [CrossRef]
- Nguyen, A.; Vessieres, A.; Hillard, E.A.; Top, S.; Pigeon, P.; Jaouen, G. Ferrocifens and ferrocifenols as new potential weapons against breast cancer. Chimia 2007, 61, 716–724. [Google Scholar] [CrossRef]
- Knauer, S.; Biersack, B.; Zoldakova, M.; Effenberger, K.; Milius, W.; Schobert, R. Melanoma specific ferrocene esters of the fungal cytotoxin illudin M. Anticancer Drugs 2009, 20, 676–681. [Google Scholar] [CrossRef]
- Kober, L.; Schleser, S.W.; Bär, S.I.; Schobert, R. Revisiting the anticancer properties of phosphane(9-ribosylpurine-6-thiolato)gold(I) complexes and their 9H-purine precursors. J. Biol. Inorg. Chem. 2022, 27, 731–745. [Google Scholar] [CrossRef]
- Leitão, M.I.P.S.; Herrera, F.; Petronilho, A. N-heterocyclic carbenes derived from guanosine: Synthesis and evidences of their antiproliferative activity. ACS Omega 2018, 3, 15653–15656. [Google Scholar] [CrossRef] [PubMed]
- Kampert, F.; Brackemeyer, D.; Tan, T.T.Y.; Hahn, F.E. Selective C8-metalation of purine nucleosides via oxidative addition. Organometallics 2018, 37, 4181–4185. [Google Scholar] [CrossRef]
- Goossen, L.J.; Koley, D.; Hermann, H.L.; Thiel, W. Mechanistic pathways for oxidative addition of aryl halides to palladium(0) complexes: A DFT study. Organometallics 2005, 24, 2398–2410. [Google Scholar] [CrossRef]
- Casado, A.L.; Espinet, P. A novel reversible aryl exchange involving two organometallics: mechanism of the gold(I)-catalyzed isomerization of trans-[PdR2L2] complexes (R = aryl, L = SC4H8). Organometallics 1998, 17, 3677–3683. [Google Scholar] [CrossRef]
- Leitão, M.I.P.S.; Francescato, G.; Gomes, C.S.B.; Petronilho, A. Synthesis of platinum(II) N-heterocyclic carbenes based on adenosine. Molecules 2021, 26, 5384. [Google Scholar] [CrossRef]
- Kucukdumlu, A.; Tuncbilek, M.; Guven, E.B.; Atalay, R.C. Design, synthesis and in vitro cytotoxic activity of new 6,9-disubstituted purine analogues. Acta Chim. Slov. 2020, 67, 70–82. [Google Scholar] [CrossRef]
- Kapdi, A.R.; Fairlamb, I.J.S. Anti-cancer palladium complexes: A focus on PdX2L2, palladacycles and related complexes. Chem. Soc. Rev. 2014, 43, 4751–4777. [Google Scholar] [CrossRef]
- Heinrich, A.-K.; Lucas, H.; Schindler, L.; Chytil, P.; Etrych, T.; Mäder, K.; Mueller, T. Improved tumor-specific drug accumulation by polymer therapeutics with pH-sensitive drug release overcomes chemotherapy resistance. Mol. Cancer Ther. 2016, 15, 998–1007. [Google Scholar] [CrossRef]
| cmpd | A2780 | A2780cis | A549 | MCF7 | HT29 | CCD18Co | RI | SI1 | SI2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.17 ± 0.05 | 0.24 ± 0.00 | 0.13 ± 0.04 | 0.24 ± 0.06 | 1.8 ± 0.57 | 0.62 ± 0.44 | 1.4 | 3.6 | 0.3 |
| 3a | 3.18 ± 1.45 | 6.55 ± 2.4 | 7.74 ± 2.90 | 17.64 ± 8.43 | 23.85 ± 9.60 | >30 | 2.1 | 9.4 | 1.3 |
| 3b | 1.06 ± 0.22 | 1.63 ± 0.2 | 1.77 ± 0.4 | 1.10 ± 0.27 | 4.07 ± 0.83 | >30 | 1.5 | 28.3 | 7.4 |
| 4a | 3.99 ± 2.28 | 7.67 ± 1.70 | 9.83 ± 5.42 | 17.23 ± 8.42 | 23.05 ± 8.18 | >30 | 1.9 | 7.5 | 1.3 |
| 4b | 0.97 ± 0.23 | 1.56 ± 0.32 | 1.50 ± 0.3 | 1.17 ± 0.27 | 3.32 ± 1.18 | >30 | 1.6 | 30.9 | 9.0 |
| cmpd | HH | DERL-2 | Oci-Ly1 | HG-3 |
|---|---|---|---|---|
| 1 | 12.2 ± 8.0 | 2.8 ± 0.4 | 2.1 ± 1.6 | 3.6 ± 2.1 |
| 3a | 25.6 ± 4.0 | 30.1 ± 12.9 | 13.9 ± 6.2 | 23.1 ± 4.3 |
| 3b | 23.8 ± 2.3 | 14.8 ± 16.4 | 3.6 ± 3.0 | 6.3 ± 1.9 |
| 4a | 29.7 ± 5.4 | 32.0 ± 2.7 | 18.4 ± 7.7 | 24.6 ± 5.0 |
| 4b | 18.8 ± 6.5 | 13.8 ± 11.0 | 3.7 ± 2.5 | 7.5 ± 4.4 |
| 1 | 3a | 3b | 4a | 4b | |
|---|---|---|---|---|---|
| IC50 [µM] | 8.00 ± 3.4 * | 32.2 ± 4.8 | 2.30 ± 0.4 | 25.9 ± 2.4 | 2.50 ± 0.5 |
| Cell Line | HH | DERL-2 | Oci-Ly1 | HG-3 |
|---|---|---|---|---|
| Cell density | 5000 | 75,000 | 20,000 | 10,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schleser, S.W.; Krytovych, O.; Ziegelmeier, T.; Groß, E.; Kasparkova, J.; Brabec, V.; Weber, T.; Schobert, R.; Mueller, T. Palladium and Platinum Complexes of the Antimetabolite Fludarabine with Vastly Enhanced Selectivity for Tumour over Non-Malignant Cells. Molecules 2023, 28, 5173. https://doi.org/10.3390/molecules28135173
Schleser SW, Krytovych O, Ziegelmeier T, Groß E, Kasparkova J, Brabec V, Weber T, Schobert R, Mueller T. Palladium and Platinum Complexes of the Antimetabolite Fludarabine with Vastly Enhanced Selectivity for Tumour over Non-Malignant Cells. Molecules. 2023; 28(13):5173. https://doi.org/10.3390/molecules28135173
Chicago/Turabian StyleSchleser, Sebastian W., Oleksandr Krytovych, Tim Ziegelmeier, Elisabeth Groß, Jana Kasparkova, Viktor Brabec, Thomas Weber, Rainer Schobert, and Thomas Mueller. 2023. "Palladium and Platinum Complexes of the Antimetabolite Fludarabine with Vastly Enhanced Selectivity for Tumour over Non-Malignant Cells" Molecules 28, no. 13: 5173. https://doi.org/10.3390/molecules28135173
APA StyleSchleser, S. W., Krytovych, O., Ziegelmeier, T., Groß, E., Kasparkova, J., Brabec, V., Weber, T., Schobert, R., & Mueller, T. (2023). Palladium and Platinum Complexes of the Antimetabolite Fludarabine with Vastly Enhanced Selectivity for Tumour over Non-Malignant Cells. Molecules, 28(13), 5173. https://doi.org/10.3390/molecules28135173








