Exploring the Mechanism of Hepatotoxicity Induced by Dictamnus dasycarpus Based on Network Pharmacology, Molecular Docking and Experimental Pharmacology
Abstract
1. Introduction
2. Results
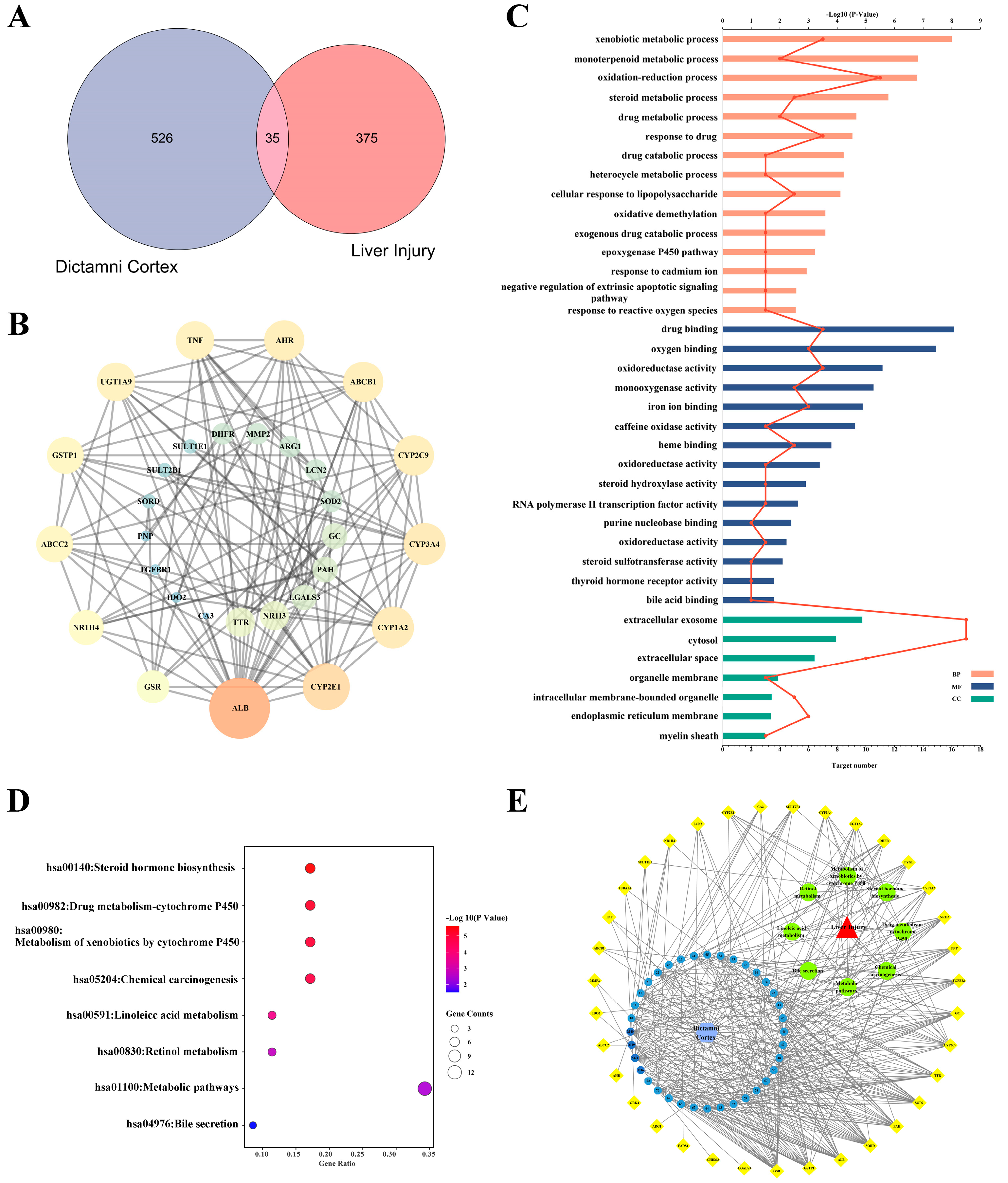
2.1. Construction of “Candidate Compounds-Targets” Network
2.2. Construction of a Protein-Protein Interaction (PPI) Network of DC Hepatotoxicity
2.3. Analysis of GO Pathway and KEGG Pathway
2.4. The Network of “Components-Targets-Pathways” of DC Hepatotoxicity
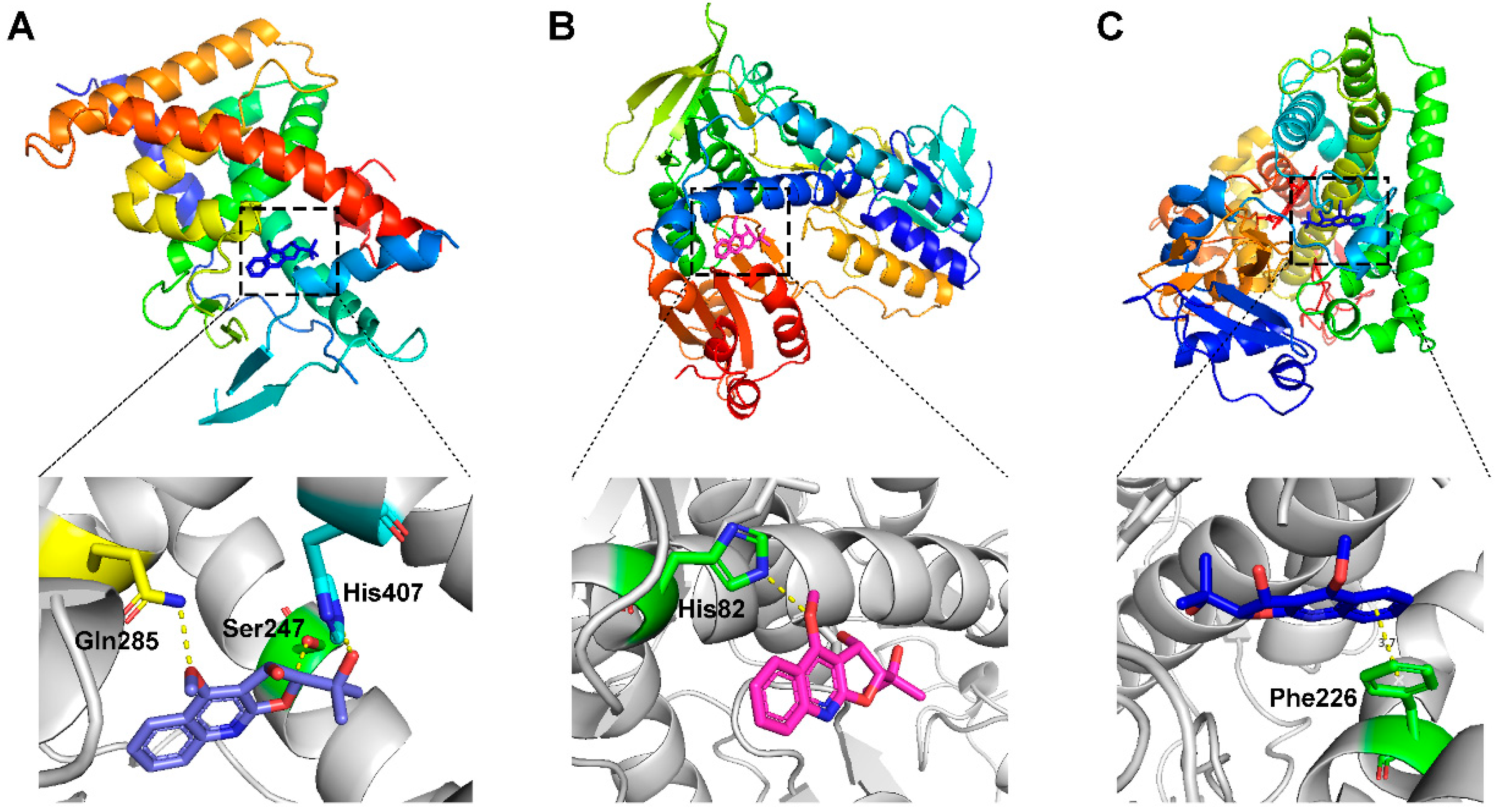
2.5. The Docking Simulation of Protein and Constituent
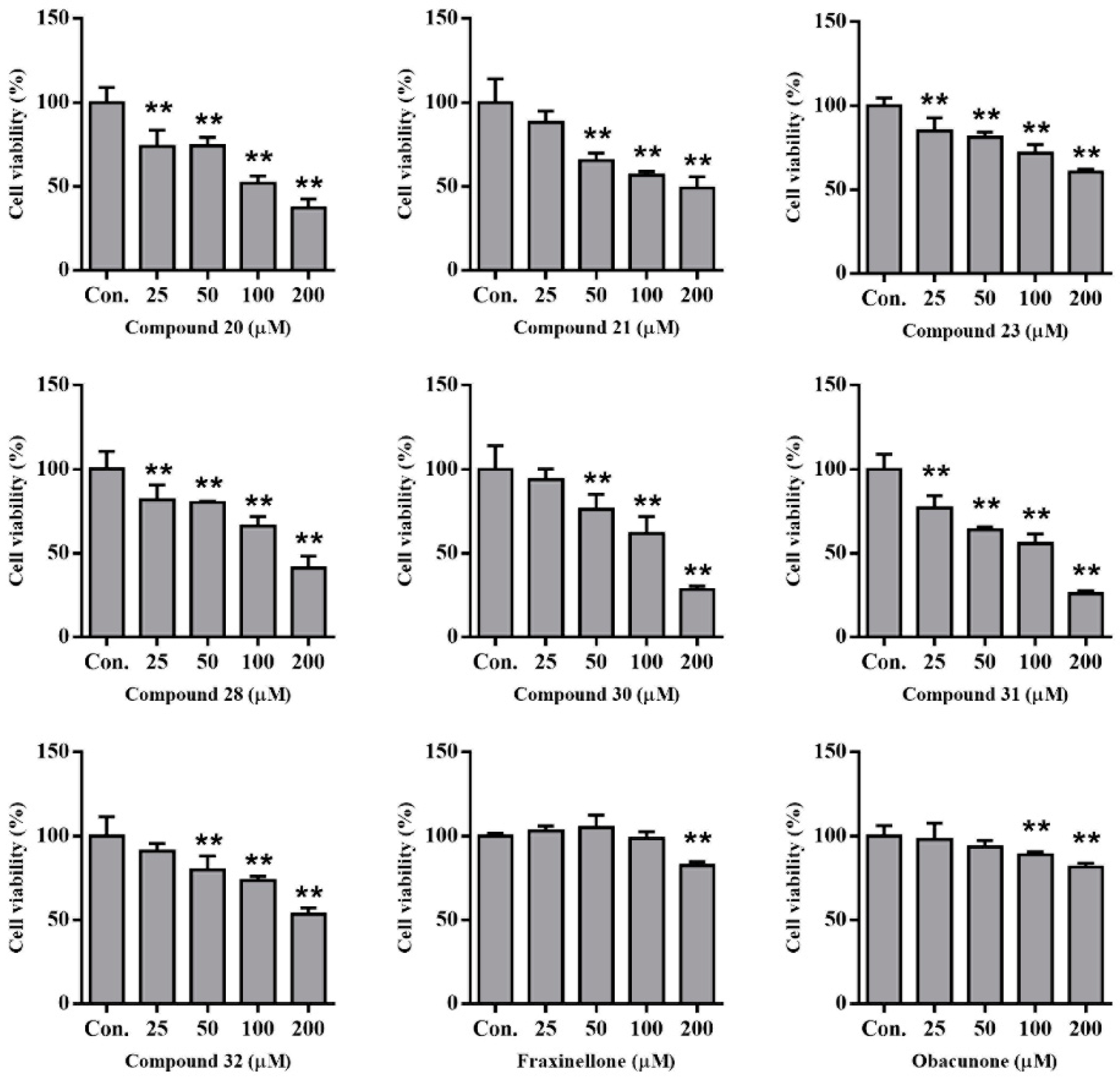
2.6. Compounds from DC-Induced Cytotoxicity of HepG2 Cells
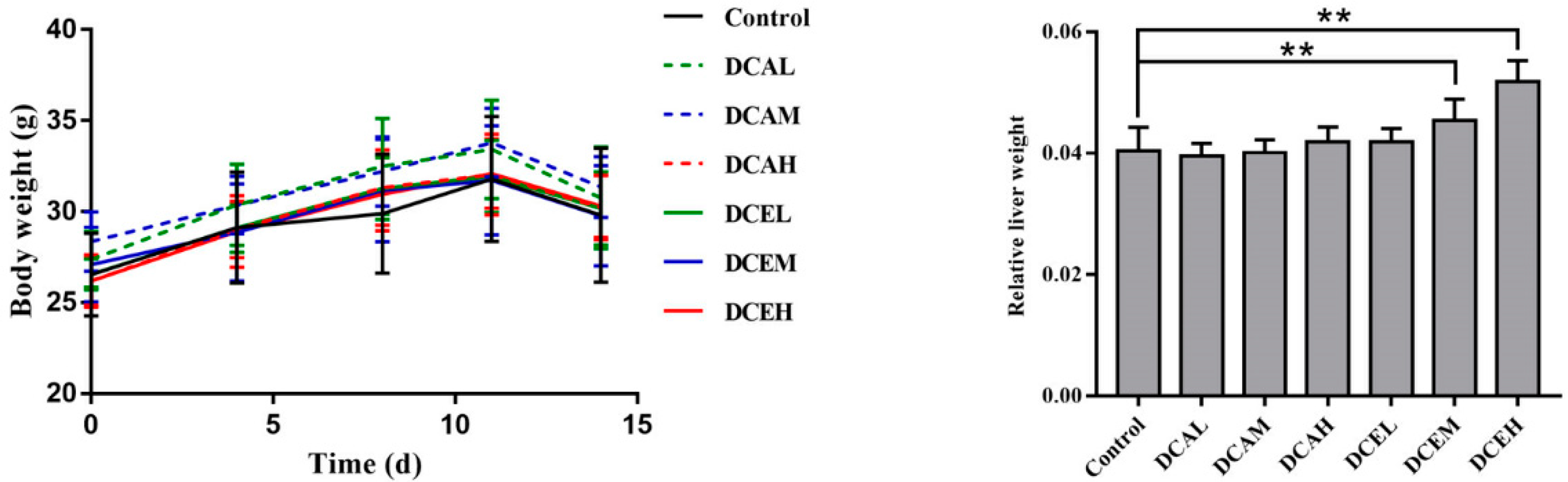
2.7. DC Induced the Increase in Relative Liver Weight
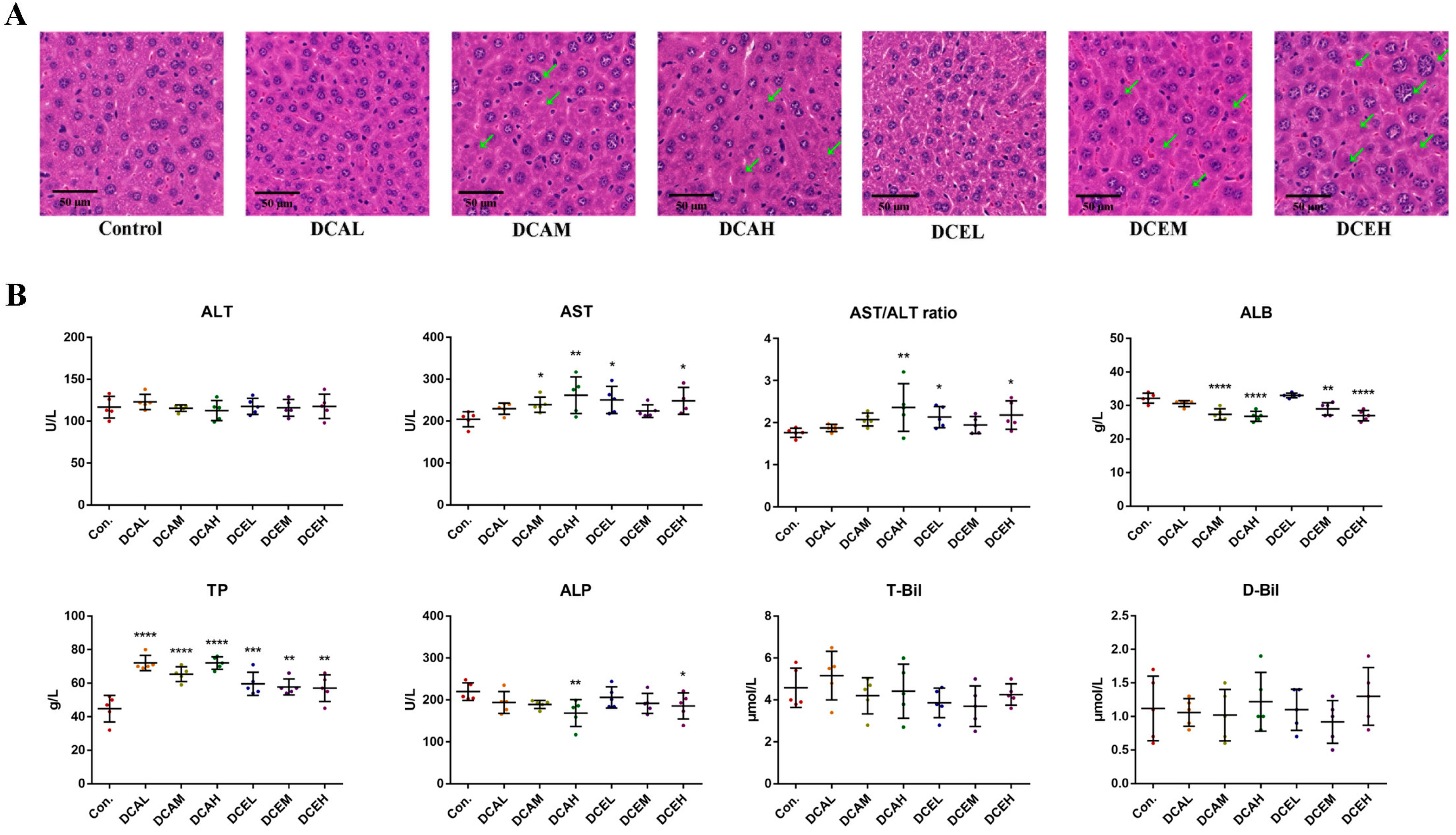
2.8. H&E Staining of Liver Tissues Revealed DC-Induced Injury of Hepatocytes
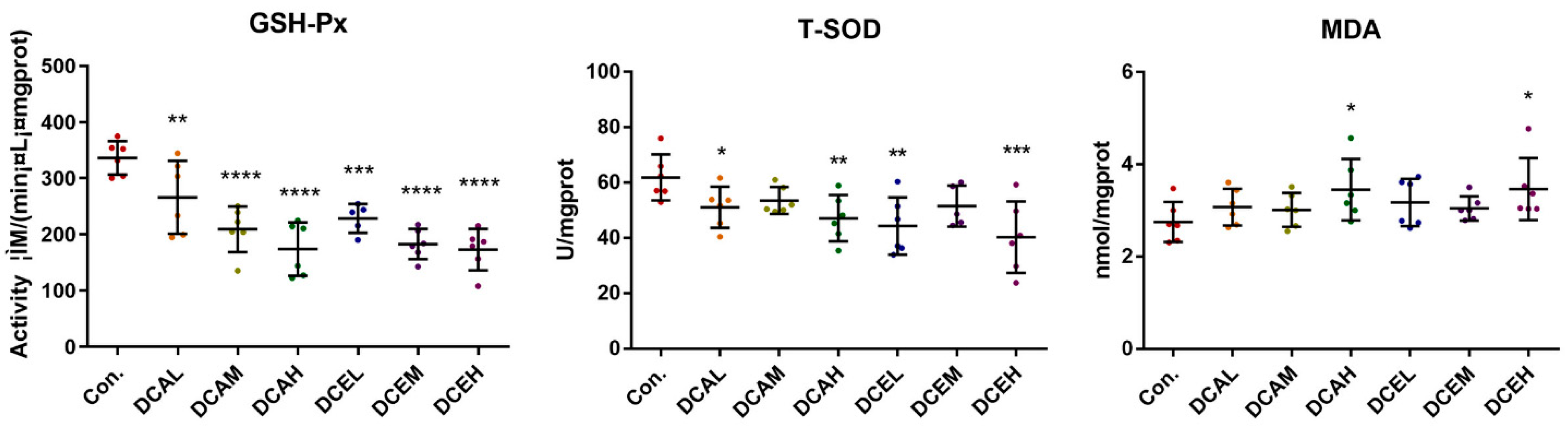
2.9. DC Induced Oxidative Stress, Increased Serum AST and TP, and Decreased ALP and ALB
2.10. DC Downregulated the Expression of CYP1A2
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of DC Extract
4.3. Network Pharmacology Analysis and Computer Docking Simulation
4.3.1. Targets Collecting
4.3.2. Establishment of Protein–Protein Interaction
4.3.3. Docking
4.4. Cell Experiments
4.4.1. Cell Culture
4.4.2. MTT Analysis
4.5. Animal Experiments
4.5.1. Animals
4.5.2. Mice and Drug Administration for the Toxicity Test
4.5.3. Sample Collection
4.5.4. Histology Assay
4.5.5. Biochemical Analysis
4.5.6. Western Blot and Real-Time q-PCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Li, B.; Wang, Z.; Fang, J.J.; Xu, C.Y.; Chen, W.X. Evaluation of Prognostic Markers in Severe Drug-induced Liver Disease. World J. Gastroenterol. 2007, 13, 628–632. Available online: http://www.wjgnet.com/1007-9327/13/628.asp (accessed on 8 June 2020).
- Gao, P.; Wang, L.; Zhao, L.; Lu, Y.Y.; Zeng, K.W.; Zhao, M.B.; Jiang, Y.; Tu, P.F.; Guo, X.Y. Rapid Identification, Isolation, and Evaluation on Anti-neuroinflammatory Activity of Limonoids Derivatives from the Root Bark of Dictamnus dasycarpus. J. Pharma. Biomed. Anal. 2021, 200, 114079. [Google Scholar] [CrossRef]
- Kane, J.A.; Kane, S.P.; Jain, S. Hepatitis Induced by Traditional Chinese Herbs; Possible Toxic Components. Gut 1995, 36, 146. [Google Scholar] [CrossRef]
- Lee, W.J.; Kim, H.W.; Lee, H.Y.; Son, C.G. Systematic Review on Herb-induced Liver Injury in Korea. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2015, 84, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Zhao, B.; Wang, C.; Zhang, J.; Wu, J.; Wang, T.; Xu, A. Subchronic Toxicity Studies of Cortex Dictamni Extracts in Mice and Its Potential Hepatotoxicity Mechanisms in vitro. Molecules 2018, 23, 2486. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Li, L.; Li, Y.; Yu, M.; Zhou, Y.; Lv, X.; Arai, H.; Xu, Y. Acute and Sub-chronic Oral Toxicity Profiles of the Aqueous Extract of Cortex Dictamni in Mice and Rats. J. Ethnopharmacol. 2014, 158, 207–215. [Google Scholar] [CrossRef]
- Gao, P.; Wang, L.; Zhao, L.; Zhang, Q.-Y.; Zeng, K.-W.; Zhao, M.-B.; Jiang, Y.; Tu, P.-F.; Guo, X.-Y. Anti-inflammatory Quinoline Alkaloids from the Root Bark of Dictamnus dasycarpus. Phytochemistry 2020, 172, 112260. [Google Scholar] [CrossRef]
- Shi, F.G.; Pan, H.; Cui, B.D.; Huang, L.Y.; Lu, Y.F. Dictamnine-induced Hepatotoxicity in Mice: The Role of Metabolic Activation of Furan. Toxicol. Appl. Pharm. 2018, 364, 68–76. [Google Scholar] [CrossRef]
- Li, Z.Q.; Jiang, L.L.; Zhao, D.S.; Zhou, J.; Wang, L.L.; Wu, Z.T.; Zheng, X.; Shi, Z.Q.; Li, P.; Li, H.J. The Modulatory Role of CYP3A4 in Dictamnine-induced Hepatotoxicity. Front. Pharmacol. 2018, 9, 1033. [Google Scholar] [CrossRef]
- Chang, K.; Gao, P.; Lu, Y.Y.; Tu, P.F.; Jiang, Y.; Guo, X.Y. Identification and Characterization of Quinoline Alkaloids from the Root Bark of Dictamnus dasycarpus and Their Metabolites in Rat Plasma, Urine and Feces by UPLC/Qtrap-MS and UPLC/Q-TOF-MS. J. Pharma. Biomed. Anal. 2021, 204, 114229. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Wang, Y.; Li, Y.; Yang, F.; Zhang, P.; Zhang, Y.; Liu, C. A Strategy for the Discovery and Validation of Toxicity Quality Marker of Chinese Medicine Based on Network Toxicology. Phytomedicine 2019, 54, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, X.Y.; Li, Y.Z.; Liu, C.X. Network Toxicology and Its application in Predicting the Toxicity of Traditional Chinese Medicine. Drug Eval. Res. 2018, 41, 709–715. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, C.; Wang, W.; Yuan, F.; Huang, J. Integrated Metabolomics and Network Toxicology to Reveal Molecular Mechanism of Celastrol Induced Cardiotoxicity. Toxicol. Appl. Pharm. 2019, 383, 114785. [Google Scholar] [CrossRef]
- Liu, C.X.; Zhang, C.N.; He, T.; Sun, L.; Wang, Q.; Han, S.; Wang, W.X.; Kong, J.; Yuan, F.L.; Huang, J.M. Study on Potential Toxic Material Base and Mechanisms of Hepatotoxicity Induced by Dysosma versipellis Based on Toxicological Evidence Chain (TEC) Concept. Ecotox. Environ. Saf. 2020, 190, 110073. [Google Scholar] [CrossRef] [PubMed]
- He, L.N.; Yang, A.H.; Cui, T.Y.; Zhai, Y.R.; Zhang, F.L.; Chen, J.X.; Jin, C.H.; Fan, Y.W.; Wu, Z.J.; Wang, L.L. Reactive Metabolite Activation by CYP2C19-mediated Rhein Hepatotoxicity. Xenobiotica 2015, 45, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.; Nabila, J.; Adelusi Temitope, I.; Wang, S. Current Etiological Comprehension and Therapeutic Targets of Acetaminophen-induced Hepatotoxicity. Pharmacol. Res. 2020, 161, 105102. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H.; McGill, M.R.; Ramachandran, A. Oxidant Stress, Mitochondria, and Cell Death Mechanisms in Drug-induced Liver Injury: Lessons Learned from Acetaminophen Hepatotoxicity. Drug Metab. Rev. 2012, 44, 88–106. [Google Scholar] [CrossRef] [PubMed]
- Luedde, T.; Kaplowitz, N.; Schwabe, R.F. Cell Death and Cell Death Responses in Liver Disease: Mechanisms and Clinical Relevance. Gastroenterology 2014, 147, 765–783.e4. [Google Scholar] [CrossRef]
- Li, C.Y.; Li, X.F.; Tu, C.; Li, N.; Ma, Z.J.; Pang, J.Y.; Jia, G.L.C.; Cui, H.R.; You, Y. The Idiosyncratic Hepatotoxicity of Polygonum Multiflorum Based on Endotoxin Model. Acta Pharm. Sin. 2015, 50, 28–33. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, C.; Yang, T.; Li, X.; Wei, L.; Chen, D.; Zhou, J.; Yin, Y.; Yu, X.; Li, F. Systematically Explore the Potential Hepatotoxic Material Basis and Molecular Mechanism of Radix Aconiti Lateralis Based on the Concept of Toxicological Evidence Chain (TEC). Ecotoxicol Environ. Saf. 2020, 205, 111342. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Gao, H.Y.; Li, J.; Zhou, T.Y.; Wang, S.T.; Yang, J.B.; Hao, R.R.; Pang, F.; Wei, F.; Liu, Z.G.; et al. Integrated Spatially Resolved Metabolomics and Network Toxicology to Investigate the Hepatotoxicity Mechanisms of Component D of Polygonum multiflorum Thunb. J. Ethnopharmacol. 2022, 298, 115630. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhu, X.; Badawy, S.; Ihsan, A.; Liu, Z.; Xie, C.; Wang, X. Metabolism and Mechanism of Human Cytochrome P450 Enzyme 1A2. Curr. Drug Metab. 2021, 22, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ma, H.; Xu, Y.; Yang, F.; Li, Y.; Shi, F.; Lu, Y. Potentiation of Flutamide-Induced Hepatotoxicity in Mice by Xian-Ling-Gu-Bao Through Induction of CYP1A2. J. Ethnopharmacol. 2021, 278, 114299. [Google Scholar] [CrossRef] [PubMed]
- Danek, P.J.; Wójcikowski, J.; Daniel, W.A. Asenapine and Iloperidone Decrease the Expression of Major Cytochrome P450 Enzymes CYP1A2 and CYP3A4 in Human Hepatocytes. A Significance for Drug-drug Interactions During Combined Therapy. Toxicol. Appl. Pharm. 2020, 406, 115239. [Google Scholar] [CrossRef] [PubMed]
- Shertzer, H.G.; Clay, C.D.; Genter, M.B.; Schneider, S.N.; Nebert, D.W.; Dalton, T.P. CYP1A2 Protects Against Reactive Oxygen Production in Mouse Liver Microsomes. Free Radic. Biol. Med. 2004, 36, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Wood, J.; Barber, J. The Role of Glutathione Reductase and Related Enzymes on Cellular Redox Homoeostasis Network. Free Radic. Bio. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dong, H.; Thompson, D.C.; Shertzer, H.G.; Nebert, D.W.; Vasiliou, V. Glutathione Defense Mechanism in Liver Injury: Insights from Animal Models. Food Chem. Toxicol. 2013, 60, 38–44. [Google Scholar] [CrossRef]
- Pei, J.; Xiao, W.; Zhu, D.; Ji, X.; Deng, X.J. Cytochrome P450 Enzyme-mediated Bioactivation as an Underlying Mechanism of Columbin-induced Hepatotoxicity. Chem. Res. Toxicol 2020, 33, 940–947. [Google Scholar] [CrossRef]
- Lin, D.; Li, W.; Peng, Y.; Jiang, C.; Xu, Y.; Gao, H.Y.; Zheng, J.J. Role of Metabolic Activation in 8-Epidiosbulbin E Acetate-induced Liver Injury: Mechanism of Action of the Hepatotoxic Furanoid. Chem. Res. Toxicol. 2016, 29, 359–366. [Google Scholar] [CrossRef]
- Li, W.; Lin, D.; Gao, H.; Xu, Y.; Meng, D.; Smith, C.V.; Peng, Y.; Zheng, J.J. Metabolic Activation of Furan Moiety Makes Diosbulbin B Hepatotoxic. Arch. Toxicol. 2016, 90, 863–872. [Google Scholar] [CrossRef]
- Lin, Q.; Guan, H.; Ma, C.; Chen, L.; Wang, C. Pharmacology. Biotransformation Patterns of Dictamnine in Vitro/in Vivo and Its Relative Molecular Mechanism of Dictamnine-induced Acute Liver Injury in Mice. Environ. Toxicol. Pharm. 2021, 85, 103628. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Zhu, Y.; Sun, J.; Yerke, A.; Sang, S.; Yu, Z. Metabolism of Dictamnine in Liver Microsomes from Mouse, Rat, Dog, Monkey, and Human. J. Pharm. Biomed. Anal. 2016, 119, 166–174. [Google Scholar] [CrossRef] [PubMed]

| Compound ID | Structure | FXR | TNF | PXR | ALB | CDK2 | IDH2 | GSR | P53 | CYP3A4 | CYP1A2 | CYP2C9 | CYP2E1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 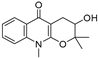 | −8.046 | −5.288 | −8.328 | −7.729 | −6.825 | −9.542 | −4.485 | −7.169 | −6.608 | −7.161 | −4.703 | — |
| 2 | 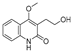 | −6.727 | −5.762 | −7.918 | −8.4 | −7.972 | −9.713 | −3.551 | −7.924 | −5.827 | −7.163 | −4.029 | — |
| 3 | 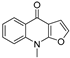 | −7.409 | −4.142 | −7.445 | −7.908 | −5.996 | −7.734 | −3.854 | −6.462 | −1.06 | −6.816 | −4.68 | — |
| 4 |  | −9.303 | −6.333 | −8.841 | −10.145 | −7.922 | −10.494 | −4.549 | −8.136 | −6.948 | −9.582 | −5.299 | — |
| 5 |  | −6.047 | −3.995 | −7.58 | −6.798 | −5.826 | −5.878 | −2.933 | −6.069 | −4.201 | −6.071 | −4.637 | −5.08 |
| 6 | 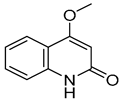 | −5.888 | −4.087 | −7.094 | −5.954 | −6.581 | −8.858 | −2.504 | −7.434 | −4.508 | −6.489 | −3.953 | −5.053 |
| 7 | 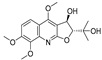 | −7.845 | −6.365 | −8.852 | −8.813 | −7.474 | −9.584 | −4.281 | −6.978 | −7.041 | −9.526 | −6.167 | — |
| 8 |  | −6.641 | −4.495 | −7.13 | −6.695 | −7.396 | −5.76 | −3.461 | −5.554 | −0.951 | −6.982 | −3.89 | — |
| 9 | 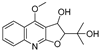 | −9.512 | −6.213 | −9.92 | −9.254 | −7.648 | −10.26 | −5.467 | −8.249 | −7.687 | −9.073 | −5.834 | — |
| 10 | 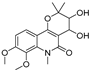 | −9.821 | −6.217 | −9.357 | −9.505 | −6.283 | −10.52 | −4.948 | −8.345 | −6.562 | −9.218 | −5.132 | — |
| 11 | 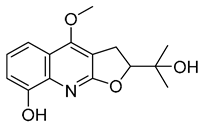 | −8.83 | −5.672 | −9.025 | −9.052 | −7.662 | −9.652 | −4.207 | −8.042 | −6.212 | −8.858 | −5.403 | — |
| 12 | 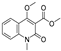 | −7.32 | −4.448 | −7.041 | −8.101 | −5.494 | −8.236 | — | −5.944 | −4.084 | −7.276 | −4.318 | — |
| 14 |  | −6.635 | −4.315 | −6.949 | −6.927 | −6.177 | −5.952 | −3.215 | −6.383 | −3.126 | −6.683 | −4.446 | — |
| 13 | 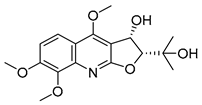 | −8.042 | −5.915 | −9.189 | −7.942 | −7.040 | −10.471 | −4.931 | −7.477 | −6.799 | −9.261 | −6.167 | — |
| 15 |  | −8.853 | −5.738 | −8.561 | −9.658 | −6.392 | −9.573 | −4.075 | −7.54 | −6.542 | −8.801 | −3.841 | — |
| 16 | 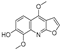 | −7.221 | −4.811 | −7.186 | −7.859 | −8.648 | −8.578 | −3.901 | −7.09 | −5.673 | −7.112 | — | — |
| 17 | 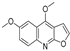 | −7.041 | −4.449 | −6.692 | −7.241 | −7.073 | −5.56 | −3.884 | −6.106 | −5.164 | −7.29 | — | — |
| 18 | 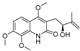 | −8.798 | −6.439 | −8.404 | −8.157 | −8.706 | −8.803 | −3.133 | −6.597 | −6.532 | −7.991 | — | — |
| 19 | 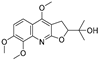 | −7.439 | −5.675 | −8.039 | −8.574 | −6.163 | −9.865 | −4.202 | −7.072 | −5.907 | −9.236 | — | — |
| 20 | 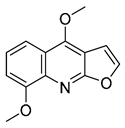 | −6.405 | −4.389 | −6.581 | −7.598 | −5.329 | −6.919 | −3.303 | −6.737 | −4.824 | −6.146 | — | — |
| 21 | 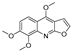 | −5.114 | −4.593 | −5.714 | −5.292 | −4.549 | −6.365 | −3.32 | −5.619 | −4.22 | −7.011 | — | — |
| 22 | 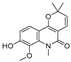 | −8.882 | −5.915 | −8.8 | −8.998 | −9.098 | −8.728 | −3.594 | −7.598 | −6.348 | −8.297 | — | — |
| 23 | 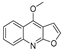 | −6.749 | −4.382 | −6.552 | −10.834 | −5.792 | −6.789 | −3.916 | −9.861 | −4.767 | −7.665 | — | — |
| 24 | 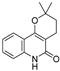 | −8.314 | −6.522 | −7.407 | −7.875 | −8.172 | −10.103 | −3.942 | −8.651 | −5.46 | −7.555 | — | — |
| 25 | 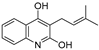 | −6.658 | −6.363 | −7.61 | −6.733 | −7.297 | −7.867 | −2.334 | −7.687 | −5.699 | −7.169 | — | — |
| 26 |  | −7.887 | −6.064 | −7.79 | −7.863 | −6.917 | −10.625 | −3.979 | −8.332 | −5.645 | −6.882 | — | — |
| 27 | 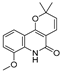 | −8.3 | −5.228 | −8.438 | −6.799 | −8.524 | −8.313 | −3.379 | −7.344 | −5.454 | −7.437 | — | — |
| 28 | 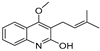 | −8.122 | −5.349 | −7.91 | −6.456 | −8.923 | −10.824 | −4.135 | −8.317 | −6.669 | −8.258 | — | — |
| 29 | 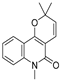 | −8.156 | −5.049 | −7.571 | −7.455 | −5.983 | −8.465 | −3.472 | −7.264 | −5.663 | −7.515 | — | — |
| 30 | 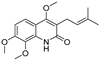 | −8.774 | −5.239 | −7.954 | −7.224 | −9.76 | −10.38 | −3.901 | −5.585 | −5.31 | −7.276 | — | — |
| 31 | 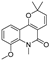 | −8.443 | −4.814 | −6.797 | −7.803 | −7.696 | −8.308 | −3.278 | −7.189 | −5.506 | −8.345 | — | — |
| 32 | 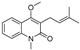 | −8.092 | −4.395 | −7.071 | −7.457 | −5.573 | −8.168 | −3.439 | −6.715 | −6.408 | −7.98 | −4.367 | — |
| 33 | 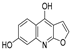 | −7.411 | −4.922 | −6.544 | −7.948 | −9.275 | −8.186 | −3.748 | −6.302 | −5.755 | −7.006 | — | — |
| 34 | 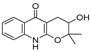 | −10.404 | −5.62 | −8.012 | −7.523 | −7.125 | −9.707 | −4.316 | −8.043 | −6.531 | −7.345 | — | — |
| 35 |  | −7.204 | −5.489 | −7.801 | −6.961 | −6.152 | −8.646 | −4.529 | −5.604 | −6.075 | −7.993 | — | — |
| 36 |  | −8.285 | −5.693 | −7.836 | −7.065 | −8.184 | −7.151 | −3.512 | −7.228 | −5.787 | −7.471 | — | — |
| Obacunone | 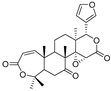 | — | −4.734 | −9.098 | — | −2.579 | — | −1.778 | −3.543 | −3.828 | — | — | — |
| Fraxinellone | 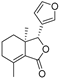 | −8.418 | −4.264 | −9.335 | −7.529 | −5.352 | −8.29 | −3.661 | −6.916 | −0.906 | −6.557 | −3.226 | — |
| Gene Name | Sequences |
|---|---|
| Actb | Forward: CCTAGCACCATGAAGATCAAGAT |
| Reverse: ACTCATCGTACTCCTGCTTGCT | |
| Cyp2e1 | Forward: TCCAGAGACATTTAAACCTGAGC |
| Reverse: GACAAAAGCAGAAACAGTTCCAT | |
| Cyp1a2 | Forward: GCTTCTCCATAGCCTCGGAC |
| Reverse: CTGGCTGACTGGTTCGAAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, P.; Chang, K.; Yuan, S.; Wang, Y.; Zeng, K.; Jiang, Y.; Tu, P.; Lu, Y.; Guo, X. Exploring the Mechanism of Hepatotoxicity Induced by Dictamnus dasycarpus Based on Network Pharmacology, Molecular Docking and Experimental Pharmacology. Molecules 2023, 28, 5045. https://doi.org/10.3390/molecules28135045
Gao P, Chang K, Yuan S, Wang Y, Zeng K, Jiang Y, Tu P, Lu Y, Guo X. Exploring the Mechanism of Hepatotoxicity Induced by Dictamnus dasycarpus Based on Network Pharmacology, Molecular Docking and Experimental Pharmacology. Molecules. 2023; 28(13):5045. https://doi.org/10.3390/molecules28135045
Chicago/Turabian StyleGao, Peng, Kun Chang, Shuo Yuan, Yanhang Wang, Kewu Zeng, Yong Jiang, Pengfei Tu, Yingyuan Lu, and Xiaoyu Guo. 2023. "Exploring the Mechanism of Hepatotoxicity Induced by Dictamnus dasycarpus Based on Network Pharmacology, Molecular Docking and Experimental Pharmacology" Molecules 28, no. 13: 5045. https://doi.org/10.3390/molecules28135045
APA StyleGao, P., Chang, K., Yuan, S., Wang, Y., Zeng, K., Jiang, Y., Tu, P., Lu, Y., & Guo, X. (2023). Exploring the Mechanism of Hepatotoxicity Induced by Dictamnus dasycarpus Based on Network Pharmacology, Molecular Docking and Experimental Pharmacology. Molecules, 28(13), 5045. https://doi.org/10.3390/molecules28135045








