Chemical Constituents from the Fruits of Amomum kravanh and Their Role in Activating Alcohol Dehydrogenase
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Elucidation
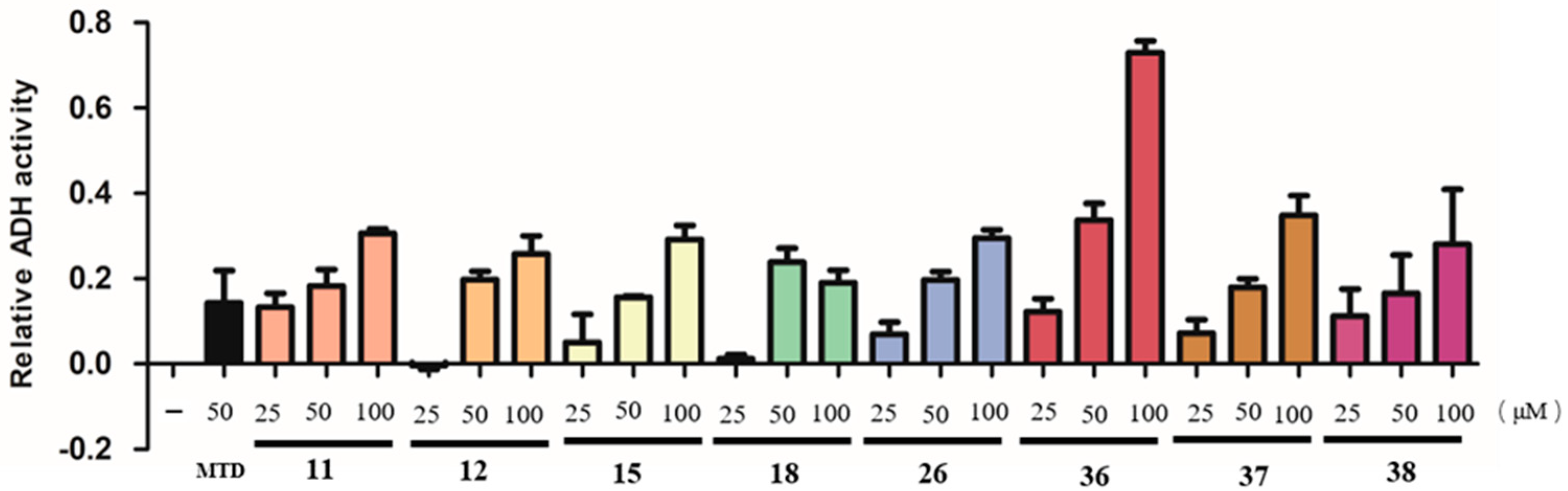
2.2. In Vitro Alcohol Dehydrogenase (ADH) Promoting Activity of Isolated Compounds
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Spectral and Physical Data of Compounds 1–10
3.5. Acetylation Reaction of Compound 10
3.6. Assay for ADH-Promoting Activity In Vitro
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Aslam, A.; Kwo, P.Y. Epidemiology and disease burden of alcohol associated liver disease. J. Clin. Exp. Hepatol. 2023, 13, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.Y.; Kim, S.K.; Yoon, S.J.; Lee, S.B.; Jeong, J.J.; Gupta, H.; Sharma, S.P.; Oh, K.K.; Won, S.M.; Kwon, G.H.; et al. Microbiome-based metabolic therapeutic approaches in alcoholic liver disease. Int. J. Mol. Sci. 2022, 23, 8749. [Google Scholar] [CrossRef]
- Szabo, G. Gut-liver axis in alcoholic liver disease. Gastroenterology 2015, 148, 30–36. [Google Scholar] [CrossRef]
- Rehm, J. The relationship between different dimensions of alcohol use and the burden of disease-an update. Addiction 2017, 112, 968–1001. [Google Scholar] [CrossRef]
- Stockwell, T. Alcohol’s contribution to cancer is underestimated for exactly the same reason that its contribution to cardioprotection is overestimated. Addiction 2017, 112, 230–232. [Google Scholar] [CrossRef]
- Evangelou, E.; Suzuki, H.; Bai, W.; Pazoki, R.; Gao, H.; Matthews, P.M.; Elliott, P. Alcohol consumption in the general population is associated with structural changes in multiple organ systems. Elife 2021, 10, e65325. [Google Scholar] [CrossRef] [PubMed]
- Ayares, G.; Idalsoaga, F.; Díaz, L.A.; Arnold, J.; Arab, J.P. Current medical treatment for alcohol-associated liver disease. J. Clin. Exp. Hepatol. 2022, 12, 1333–1348. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.C.; Hu, X.Y.; Wang, Y.Y.; Luo, J.X.; Lin, W.; Jia, L.Y.; Gong, X.Y. Effects of aqueous extracts from Panax ginseng and hippophae rhamnoides on acute alcohol intoxication: An experimental study using mouse model. J. Ethnopharmacol. 2016, 192, 67–73. [Google Scholar] [CrossRef]
- Liang, J.; Olsen, R.W. Alcohol use disorders and current pharmacological therapies: The role of GABAA receptors. Acta Pharmacol. Sin. 2014, 35, 981–993. [Google Scholar] [CrossRef]
- Yin, H.; Luo, J.G.; Kong, L.Y. Tetracyclic diterpenoids with isomerized isospongian skeleton and labdane diterpenoids from the fruits of Amomum kravanh. J. Nat. Prod. 2013, 76, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.S.; Cao, X.X.; Zhang, H. Chemical constituents from the fruits of Amomum kravanh. Biochem. Syst. Ecol. 2020, 92, 92. [Google Scholar] [CrossRef]
- Zhang, J.S.; Cao, X.X.; Yu, J.H.; Yu, Z.P.; Zhang, H. Diarylheptanoids with NO production inhibitory activity from Amomum kravanh. Bioorg. Med. Chem. Lett. 2020, 30, 127026. [Google Scholar] [CrossRef]
- Mathew, J.; Shiburaj, S.; George, V. Antimicrobial activity of Amomum cannicarpum. Fitoterapia 2003, 74, 476–478. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, R.; Zou, G. Cytotoxic, apoptotic and antioxidant activity of the essential oil of Amomum tsao-ko. Bioresour. Technol. 2010, 101, 4205–4211. [Google Scholar] [CrossRef]
- Patanasethanot, D.; Nagai, J.; Yumoto, R.; Murakami, T.; Sutthanut, K.; Sripanidkulchai, B.O.; Yenjai, C.; Takano, M. Effects of Kaempferia parviflora extracts and their flavone constituents on P-glycoprotein function. J. Pharm. Sci. 2007, 96, 223–233. [Google Scholar] [CrossRef]
- Sookkongwaree, K.; Geitmann, M.; Roengsumran, S.; Petsom, A.; Danielson, U.H. Inhibition of viral proteases by Zingiberaceae extracts and flavones isolated from Kaempferia parviflora. Pharmazie 2006, 61, 717–721. [Google Scholar]
- JiSuk, L.; Kyoung, A.K.; SeonHui, J.; SungGeum, L.; Hi, J.P.; Nam, J.K.; Sabina, L. Anti-inflammatory, anti-nociceptive, and anti-psychiatric effects by the rhizomes of Alpinia officinarum on complete freund’s adjuvant-induced arthritis in rats. J. Ethnopharmacol. 2009, 126, 258–264. [Google Scholar]
- Borchuluun, S.; Wang, Q.; Xu, Y.; He, X.; Bao, W.; Pa, B. Structure elucidation and NMR assignments of a new sesquiterpene of volatile oil from Artemisia frigida Willd. Nat. Prod. Res. 2021, 35, 2376–2380. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.B.; Herz, W. Bisabolones and other constituents of Mikania shushunensis. Phytochemistry 1988, 27, 3871–3874. [Google Scholar] [CrossRef]
- Takeda, S.; Iimura, Y.; Tanaka, K.; Kurosawa, E.; Suzuki, T. A new naturally occurring racemic compound from the marine red alga laurencia obtusa (hudson) lamouroux. Chem. Lett. 1990, 19, 155–156. [Google Scholar] [CrossRef]
- Cuenca, M.; Catalan, C.; Díaz, J.G.; Herz, W. Monoterpenes and lignans from Mikania saltensis. J. Nat. Prod. 1991, 54, 1162–1164. [Google Scholar] [CrossRef]
- Bohlmann, F.; Kramp, W.; Gupta, R.K.; King, R.M.; Robinson, H. Four guaianolides and other constituents from three kaunia species. Phytochemistry 1981, 20, 2375–2378. [Google Scholar] [CrossRef]
- Liu, S.S.; Sheng, W.L.; Li, Y.; Zhang, S.S.; Zhang, M. Chemical constituents from Alismatis Rhizoma and their anti-inflammatory activities in vitro and in vivo. Bio. Chem. 2019, 92, 103226. [Google Scholar] [CrossRef] [PubMed]
- Rédei, D.; Kúsz, N.; Rafai, T.; Bogdanov, A.; Burián, K.; Csorba, A.; Mándi, A.; Kurtán, T.; Vasas, A.; Hohmann, J. 14-Noreudesmanes and a phenylpropane heterodimer from sea buckthorn berry inhibit Herpes simplex type 2 virus replication. Tetrahedron 2019, 75, 1364–1370. [Google Scholar] [CrossRef]
- Nuñez, Y.O.; Salabarria, I.S.; Collado, I.G.; Hernández-Galán, R. Sesquiterpenes from the wood of Juniperus lucayana. Phytochemistry 2007, 68, 2409–2414. [Google Scholar] [CrossRef]
- Alexander-Lindo, R.L.; Morrison, E.Y.; Nair, M.G. Hypoglycaemic effect of stigmast-4-en-3-one and its corresponding alcohol from the bark of Anacardium occidentale (cashew). Phytother. Res. 2004, 18, 403–407. [Google Scholar] [CrossRef]
- Suguru, T.; Chiharu, G.; Yasuhisa, N.; Takahiro, N.; Shozo, F. Synthesis of some C28 steroids with C-24 (28) double bonds. J. Jpn. Oil Chem. Soc. 2009, 49, 1884–1996. [Google Scholar]
- Huo, J.; Tang, H.; Li, L.; Liu, B.S.; Zhang, W. Study on bioactive constituents of the south China sea soft coral Scleronephthya sp. Acad. J. Second Mil. Med. Univ. 2011, 31, 21–24. [Google Scholar] [CrossRef]
- Anne, G.; Jacqueline, S.; Maurice, A. Isolation of bioactive 5α,8α-epidioxy sterols from the marine sponge Luffariella cf. variabilis. Can. J. Chem. 2000, 78, 986–992. [Google Scholar]
- Yaoita, Y.; Amemiya, K.; Ohnuma, H.; Furumura, K.; Masaki, A.; Matsuki, T.; Kikuchi, M. Cheminform abstract: Constituents of mushrooms. Part 3. sterol constituents from five edible mushrooms. Chem. Pharm. Bull. 1998, 46, 944–950. [Google Scholar] [CrossRef]
- Haven, H.; Rullk-Tter, J. The diagenetic fate of taraxer-14-ene and oleanene isomers. Geochim. Cosmochim. Acta 1988, 52, 2543–2548. [Google Scholar] [CrossRef]
- Parmar, V.S.; Vardhan, A.; Nagarajan, G.R.; Jain, R. Dihydroflavonols from Prunus Domestica. Phytochemistry 1992, 31, 2185–2186. [Google Scholar] [CrossRef]
- Afifi, F.Ü.; Al-Khalil, S.; Abdul-Haq, B.K.; Mahasneh, A. Antifungal flavonoids from Varthemia iphionoides. Phyto. Res. 1991, 5, 173–175. [Google Scholar] [CrossRef]
- Yenjai, C.; Prasanphen, K.; Daodee, S.; Wongpanich, V.; Kittakoop, P. Bioactive flavonoids from Kaempferia parviflora. Fitoterapia 2004, 75, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Lo, W.L.; Wang, Y.D.; Chen, C.Y. A novel cytotoxic monoterpenoid from the leaves of Cinnamomum subavenium. Nat. Prod. Res. 2008, 22, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Versini, G.; Rapp, A.; Reniero, F.; Mandery, H. Structural identification and presence of some p-menth-1-enediols in grape products. Vitis 1991, 30, 143–149. [Google Scholar]
- Hoskins, J.A. The occurrence, metabolism and toxicity of cinnamic acid and related compounds. J. Appl. Toxicol. 1984, 4, 283–292. [Google Scholar] [CrossRef]
- Wahidullah, S.; Desouza, L.; Kamat, S.Y. Dipeptides from the red alga Acanthopora spicifera. Phytochemistry 1991, 30, 3323–3325. [Google Scholar] [CrossRef]
- Tan, S.B.; Guo, T.; Tang, X.F.; Song, T.T.; Wang, Y. Chemical constituents of Zanthoxylum armatum. II. Chem. Nat. Compd. 2018, 54, 1027–1028. [Google Scholar] [CrossRef]
- Ouyang, M.A.; Wein, Y.S.; Su, R.K.; Kuo, Y.H. Rhusemialins A-C, new cyclolignan esters from the roots of Rhus javanica var. roxburghiana. Chem. Pharm. Bull. 2007, 55, 804–807. [Google Scholar] [CrossRef]
- Ai, W.; Wei, X.; Lin, X.; Sheng, L.; Wang, Z.; Tu, Z.; Yang, X.; Zhou, X.; Li, J.; Liu, Y. Guignardins A–F, spirodioxynaphthalenes from the endophytic fungus Guignardia sp. KcF8 as a new class of PTP1B and SIRT1 inhibitors. Tetrahedron 2014, 70, 5806–5814. [Google Scholar] [CrossRef]
- Vollenweider, S.; Weber, H.; Stolz, S.; Chételat, A.; Farmer, E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000, 24, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yu, F.; Wu, Y.; Dai, L.; Feng, Y.; Chen, S.; Wang, G.; Ma, H.; Li, X.; Dai, C. Identification of a novel peptide that activates alcohol dehydrogenase from crucian carp swim bladder and how it protects against acute alcohol-induced liver injury in mice. J. Pharm. Biomed. Anal. 2022, 207, 114426. [Google Scholar] [CrossRef] [PubMed]
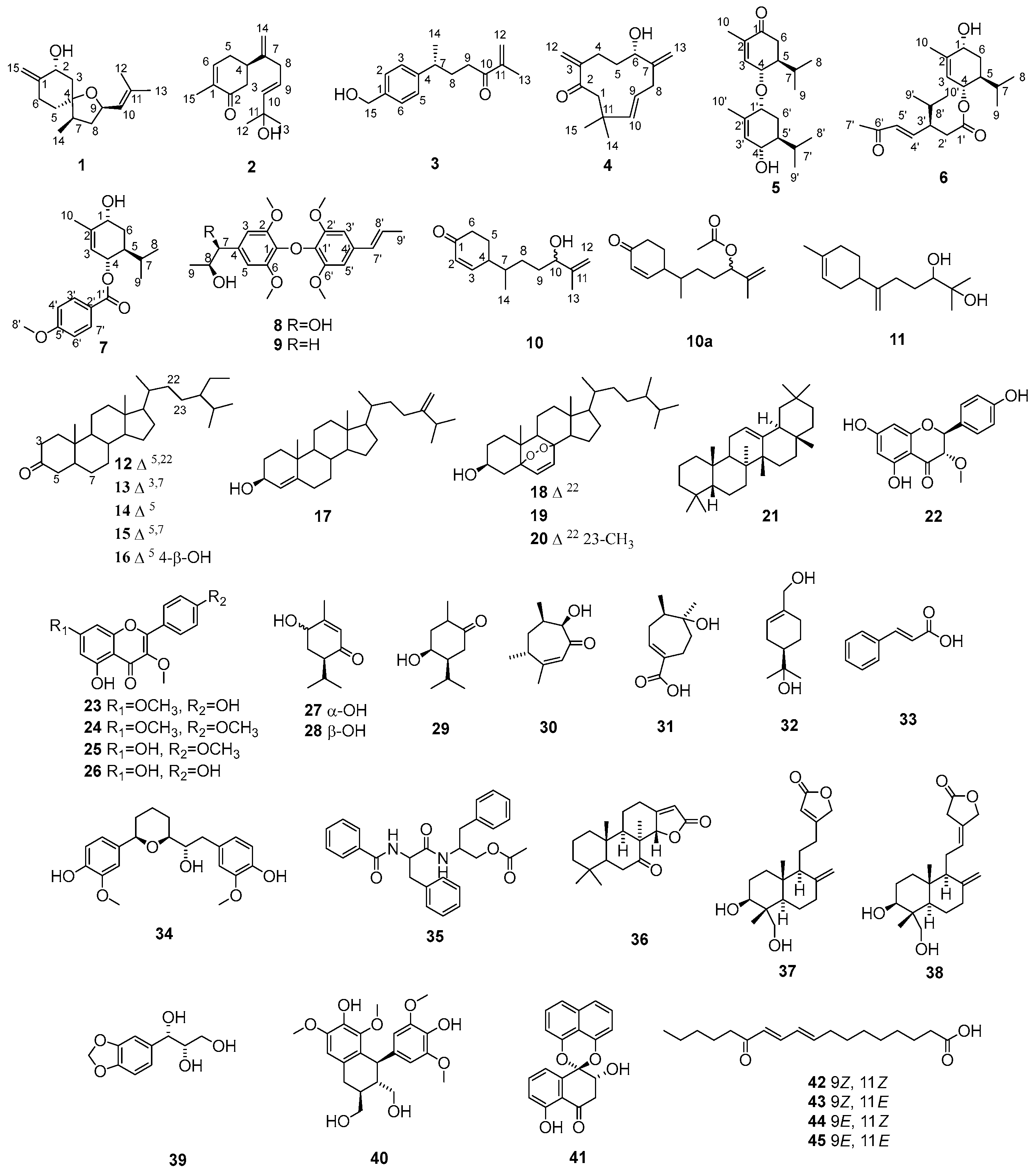
 ) and 1H-1H COSY (
) and 1H-1H COSY ( ) correlations of compounds 1–4.
) correlations of compounds 1–4.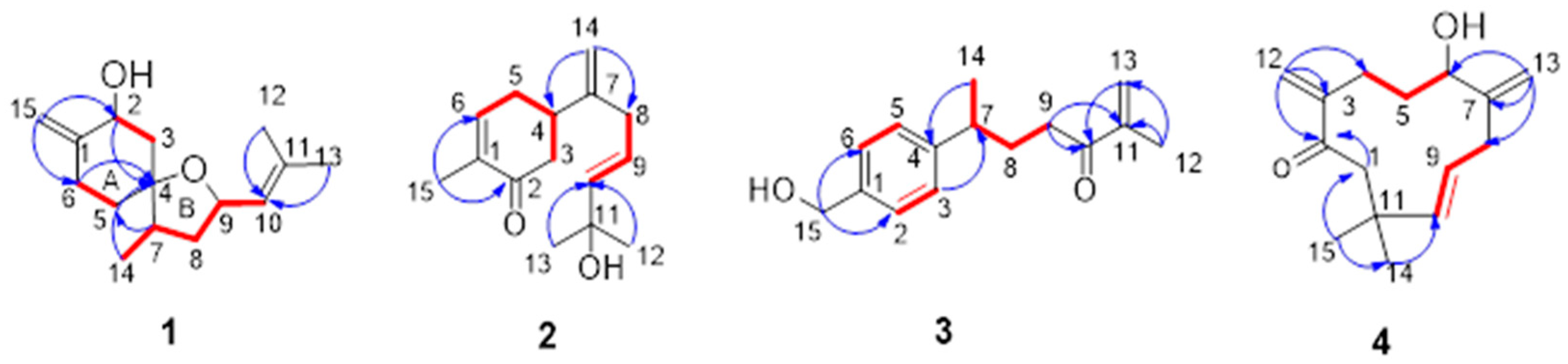
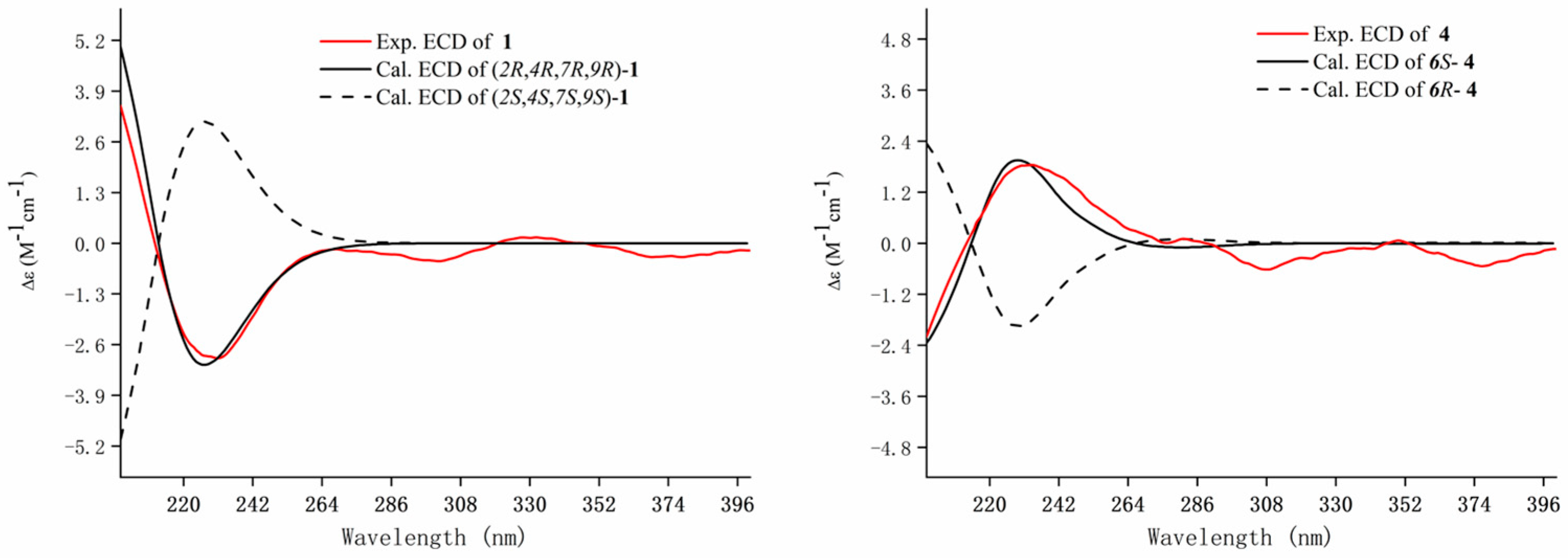
 ) and 1H-1H COSY (
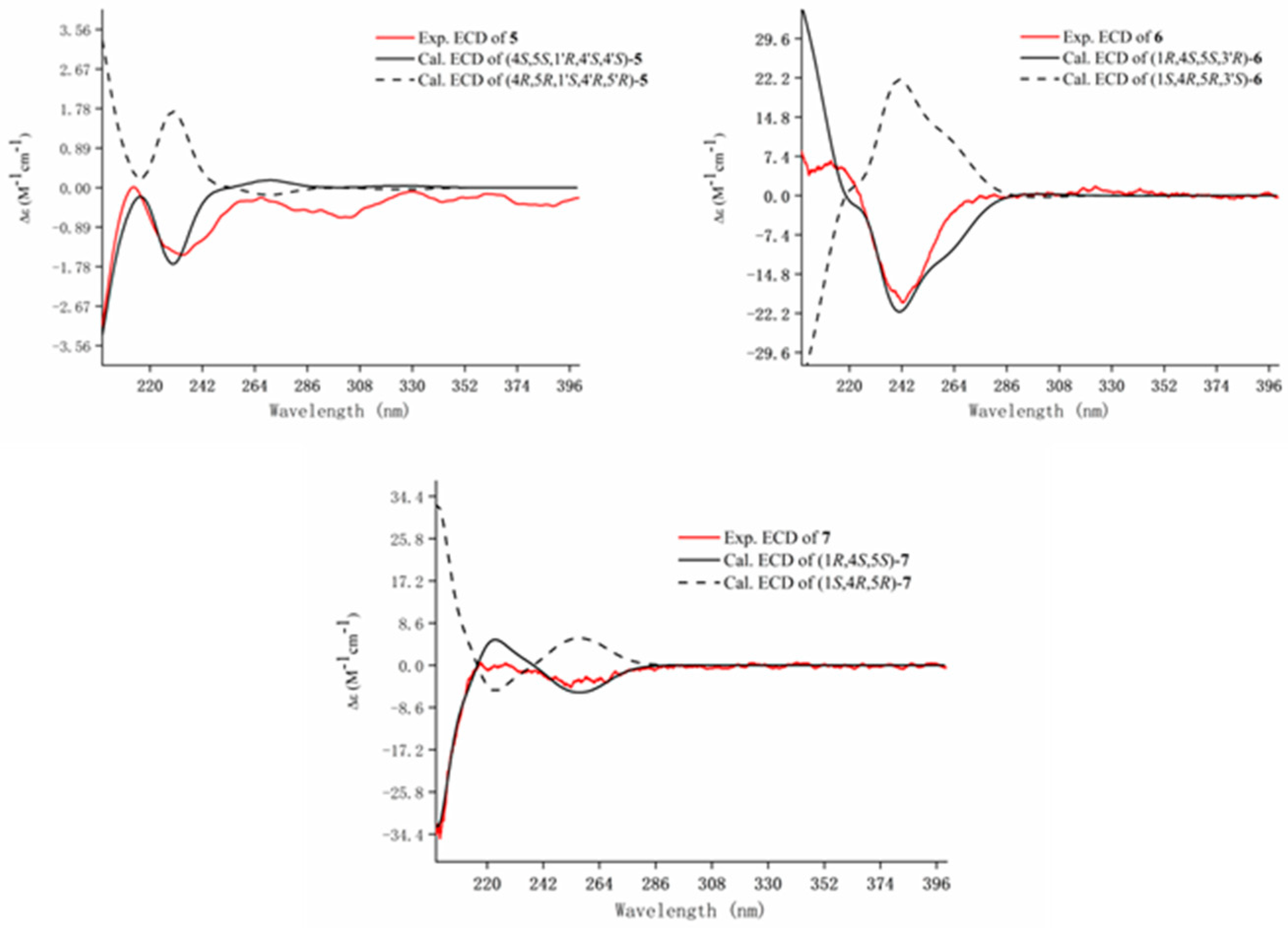
) and 1H-1H COSY ( ) correlations of compounds 5–7.
) correlations of compounds 5–7.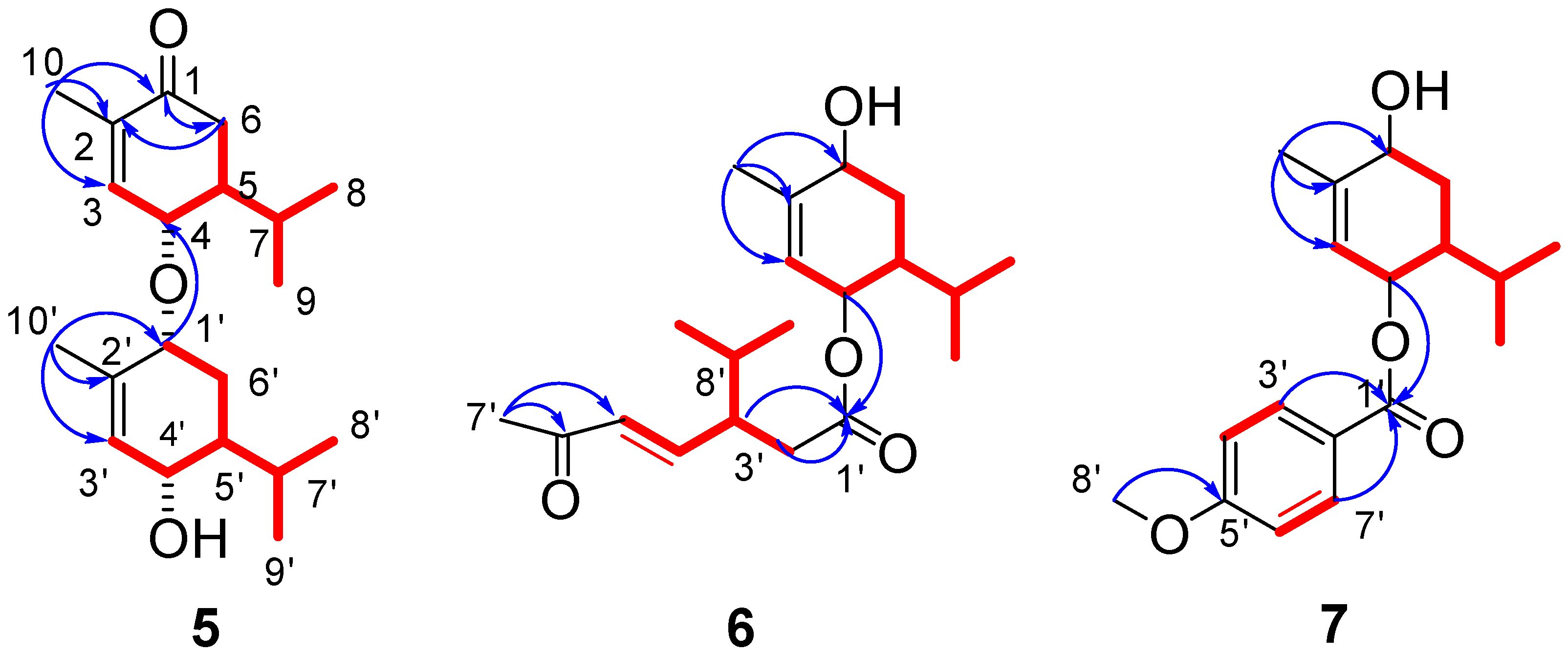
 ) and 1H-1H COSY (
) and 1H-1H COSY ( ) correlations of compounds 8–10.
) correlations of compounds 8–10.
| No. | 1 (CDCl3) | 2 (CDCl3) | 3 (CDCl3) | 4 (CDCl3) | 10 (MeOD) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | |
| 1 | 151.8 | 135.4 | 138.6 | 202.8 | 202.8 | |||||
| 2 | 69.8 | 4.46, dd, 4.9, 11.5 | 199.7 | 127.2 | 7.18, d, 7.9 | 150.4 | 129.9 | 6.00, d, 10.0 | ||
| 3 | 29.5 | 1.97, m | 31.5 | 2.59, dd, 1.44, 3.92 | 127.3 | 7.3, d, 7.9 | 29.3 | 2.84, ddt, 2.8, 5.8, 13.0 | 157.0 | 7.03, brd, 10.0 |
| 1.42, m | 2.36, dd, 13.2, 15.9 | 2.17, td, 2.8, 13.0 | ||||||||
| 4 | 84.1 | 40.8 | 2.7, m | 146.2 | 35.4 | 1.44, m | 42.5 | 2.54, m | ||
| 5 | 40.7 | 1.54, m | 43.3 | 2.47, 2.28 | 127.3 | 7.3, d, 7.9 | 70.8 | 3.92, d, 8.1 | 25.0 | 2.01, m |
| 1.27, m | (25.1) | 1.79, m | ||||||||
| 6 | 46.8 | 2.43, dt, 3.3, 13.6 | 140.3 | 6.74, brd, 4.5 | 127.3 | 7.18, d, 7.9 | 151.8 | 38.2 | 2.49, brt, 16.5 | |
| 2.28, ddd, 2.7, 4.3, 13.3 | 2.41, dt, 4.2, 16.5 | |||||||||
| 7 | 43.6 | 1.99, m | 149.5 | 39.2 | 2.72, m | 41.5 | 2.87, ddd, 1.2, 4.8, 13.1 | 37.7 | 1.71, m | |
| 2.74, brt, 11.6 | (37.8) | |||||||||
| 8 | 33.5 | 2.10, m | 37.3 | 2.79, d, 6.6 | 32.7 | 1.93, m | 124.8 | 5.06, ddd, 4.8, 11.6, 15.4 | 31.0 | 1.42, m |
| 1.34, m | 1.86, m | (31.1) | 1.32, m | |||||||
| 9 | 73.9 | 4.66, ddd, 5.9, 8.7, 10.0 | 124.3 | 5.61, dt, 6.6, 15.4 | 35.5 | 2.85, m | 140.1 | 5.38, dd, 15.4 | 33.9 | 1.63, m |
| 2.50, m | (33.9) | 1.54, m | ||||||||
| 10 | 127.4 | 5.16, brd, 1.5, 8.5 | 144.5 | 5.65, d, 15.4 | 202.0 | 37.3 | 76.8 | 4.02, t, 6.4 | ||
| (77.0) | ||||||||||
| 11 | 134.6 | 70.6 | 144.4 | 47.9 | 3.20, d, 11.0; | 148.7 | ||||
| 2.01, d, 4.5 | (148.8) | |||||||||
| 12 | 18.1 | 1.68, q, 1.2 | 29.8 | 1.33, s | 124.4 | 5.81, s | 124.3 | 5.84, s | 111.4 | 4.94, s |
| 5.69, s | 5.70, s | (111.6) | 4.83, s | |||||||
| 13 | 25.8 | 1.72, q, 1.2 | 29.8 | 1.33, s | 17.6 | 1.83, s | 109.2 | 5.10, s | 17.6 | 1.73, s |
| 4.90, s | (17.7) | |||||||||
| 14 | 14.3 | 0.93, d, 7.0 | 110.6 | 4.86, s | 22.5 | 1.27, s | 23.6 | 1.15, s | 16.5 | 0.94, d, 6.8 |
| (16.5) | ||||||||||
| 15 | 103.2 | 4.78, q, 1.5; | 15.6 | 1.79, s | 65.2 | 4.68, s | 31.6 | 1.09, s | ||
| 4.92, q, 1.5 | ||||||||||
| No. | 5 (CDCl3) | 6 (CDCl3) | 7 (CDCl3) | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1 | 200.0 | 67.5 | 4.00, t, 3 | 67.7 | 4.08, d, 3 | |
| 2 | 135.3 | 139.2 | 138.5 | |||
| 3 | 145.0 | 6.73, s | 124.7 | 5.37, s | 125.6 | 5.57, s |
| 4 | 74.4 | 3.98, d, 9.4 | 71.9 | 5.16, brd, 8.6 | 71.9 | 5.44, brd, 9.0 |
| 5 | 47.7 | 2.03, m | 38.9 | 1.76, m | 39.0 | 1.98, m |
| 6 | 35.7 | 2.45, dd, 3.2, 15.5 | 30.0 | 1.79, 1.56, m | 30.4 | 1.89, 1.65, m |
| 2.11, d, 15.2 | ||||||
| 7 | 25.1 | 2.27, dm, 3.2, 6.9 | 26.6 | 1.72, m | 26.8 | 1.85, m |
| 8 | 16.5 | 0.87, d, 6.9 | 20.6 | 0.92, d, 6.9 | 17.9 | 0.87, d, 6.9 |
| 9 | 20.7 | 0.91, d, 6.9 | 17.3 | 0.79, d, 6.9 | 20.3 | 0.96, d, 6.9 |
| 10 | 20.6 | 1.83, s | 20.3 | 1.79, s | 20.5 | 1.83, s |
| 1′ | 73.1 | 3.77, brs | 172.0 | 166.6 | ||
| 2′ | 134.3 | 37.2 | 2.56, dd, 4.7, 14.8 | 122.8 | ||
| 2.40, dd, 9.5, 14.8 | ||||||
| 3′ | 131.2 | 5.63, s | 45.5 | 2.6, m, 4.7 | 131.6 | 8.01, d, 9.0 |
| 4′ | 69.3 | 3.92, d, 9.14 | 148.0 | 6.65, dd, 8.91, 16.0 | 113.6 | 6.93, d, 9.0 |
| 5′ | 42.7 | 1.61, m | 132.3 | 6.09, d, 16.0 | 163.3 | |
| 6′ | 25.4 | 1.83,1.29, m | 198.1 | 113.6 | 6.93, d, 9.0 | |
| 7′ | 26.3 | 2.15, dm,2.7,6.9 | 27.1 | 2.24, s | 131.6 | 8.01, d, 9.0 |
| 8′ | 16.8 | 0.85, d, 6.9 | 31.5 | 1.76, m | 55.4 | 3.86, s |
| 9′ | 20.9 | 0.98, d, 6.9 | 20.2 | 0.94, d, 6.1 | ||
| 10′ | 15.6 | 1.81, s | 19.3 | 0.89, d, 6.1 | ||
| No. | 8 (MEOD) | 9 (CDCl3) | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 135.5 | 132.9 | ||
| 2 | 149.3 | 146.7 | ||
| 3 | 104.7 | 6.60, s | 106.7 | 6.45, s |
| 4 | 135.5 | 130.1 | ||
| 5 | 104.7 | 6.60, s | 106.7 | 6.45, s |
| 6 | 149.3 | 146.7 | ||
| 7 | 75.8 | 4.79, d, 3.6 | 43.4 | 3.10, dd, 13.4, 5.1 |
| 2.71, dd, 13.4, 8.1 | ||||
| 8 | 83.8 | 4.32, dq, 3.6, 6.3 | 80.3 | 4.35, m |
| 9 | 13.9 | 1.15, d, 6.3 | 20.4 | 1.21, d, 6.2 |
| 1′ | 135.5 | 135.2 | ||
| 2′ | 154.9 | 153.7 | ||
| 3′ | 104.2 | 6.67, s | 103.0 | 6.56, s |
| 4′ | 135.5 | 133.4 | ||
| 5′ | 104.2 | 6.67, s | 103.0 | 6.56, s |
| 6′ | 154.9 | 153.7 | ||
| 7′ | 132.3 | 6.36, d, 15.7 | 130.9 | 6.32, d, 15.7 |
| 8′ | 126.2 | 6.24, dq, 6.5, 15.7 | 125.1 | 6.17, dq, 6.5, 15.7 |
| 9′ | 18.4 | 1.88, d, 6.5 | 18.6 | 1.88, d, 6.5 |
| 2,6-OCH3 | 56.7 | 3.80, s | 56.3 | 3.86, s |
| 2′,6′-OCH3 | 56.5 | 3.85, s | 56.0 | 3.81, s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, H.-M.; Li, H.-Y.; Lin, Z.-R.; Liu, X.-M.; Bai, L.-P.; Zhang, W.; Jiang, Z.-H.; Zhu, G.-Y. Chemical Constituents from the Fruits of Amomum kravanh and Their Role in Activating Alcohol Dehydrogenase. Molecules 2023, 28, 4878. https://doi.org/10.3390/molecules28124878
Xiong H-M, Li H-Y, Lin Z-R, Liu X-M, Bai L-P, Zhang W, Jiang Z-H, Zhu G-Y. Chemical Constituents from the Fruits of Amomum kravanh and Their Role in Activating Alcohol Dehydrogenase. Molecules. 2023; 28(12):4878. https://doi.org/10.3390/molecules28124878
Chicago/Turabian StyleXiong, Hao-Ming, Hui-Ying Li, Zhi-Rong Lin, Xiao-Mei Liu, Li-Ping Bai, Wei Zhang, Zhi-Hong Jiang, and Guo-Yuan Zhu. 2023. "Chemical Constituents from the Fruits of Amomum kravanh and Their Role in Activating Alcohol Dehydrogenase" Molecules 28, no. 12: 4878. https://doi.org/10.3390/molecules28124878
APA StyleXiong, H.-M., Li, H.-Y., Lin, Z.-R., Liu, X.-M., Bai, L.-P., Zhang, W., Jiang, Z.-H., & Zhu, G.-Y. (2023). Chemical Constituents from the Fruits of Amomum kravanh and Their Role in Activating Alcohol Dehydrogenase. Molecules, 28(12), 4878. https://doi.org/10.3390/molecules28124878







