NMR-Based Metabolomic Study on Phaseolus vulgaris Flour Fermented by Lactic Acid Bacteria and Yeasts
Abstract
1. Introduction
2. Results and Discussion
2.1. Phaseolus vulgaris Flour Fermentation
2.2. NMR Spectra Analysis
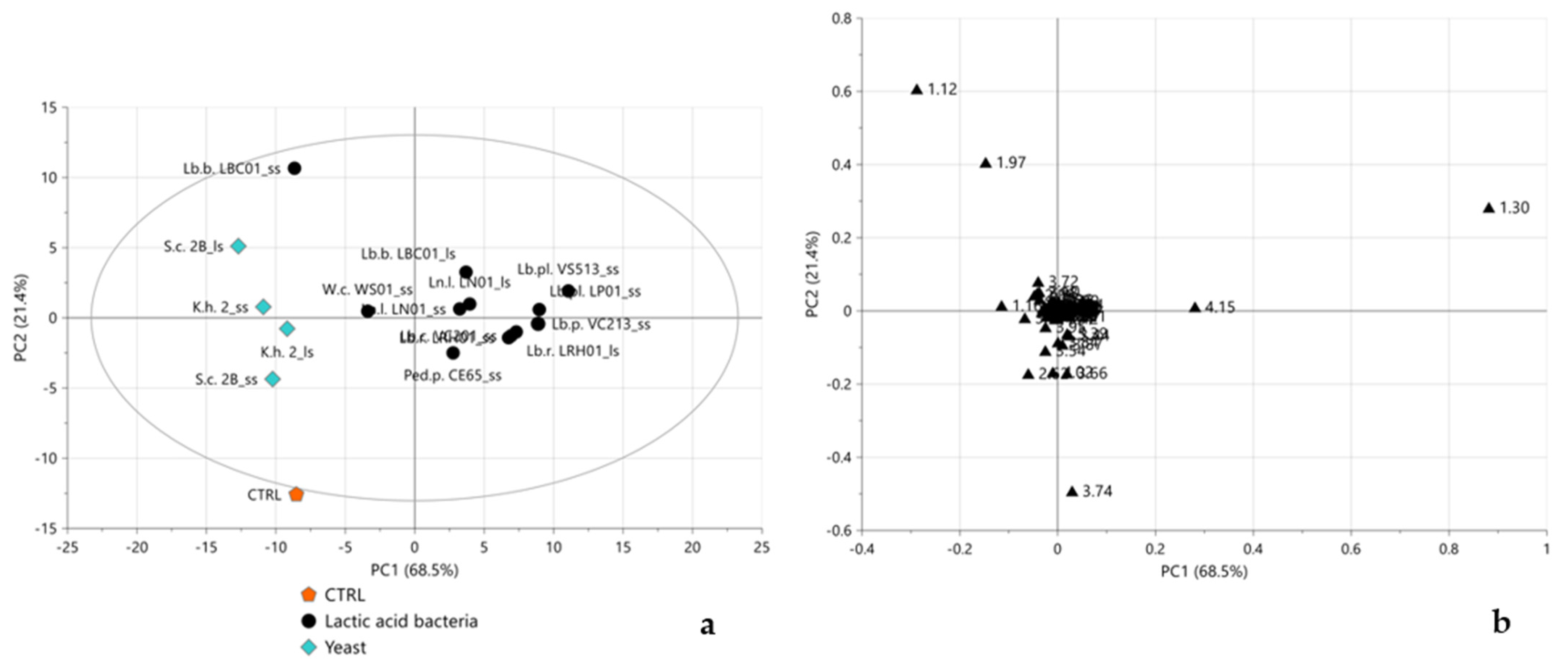
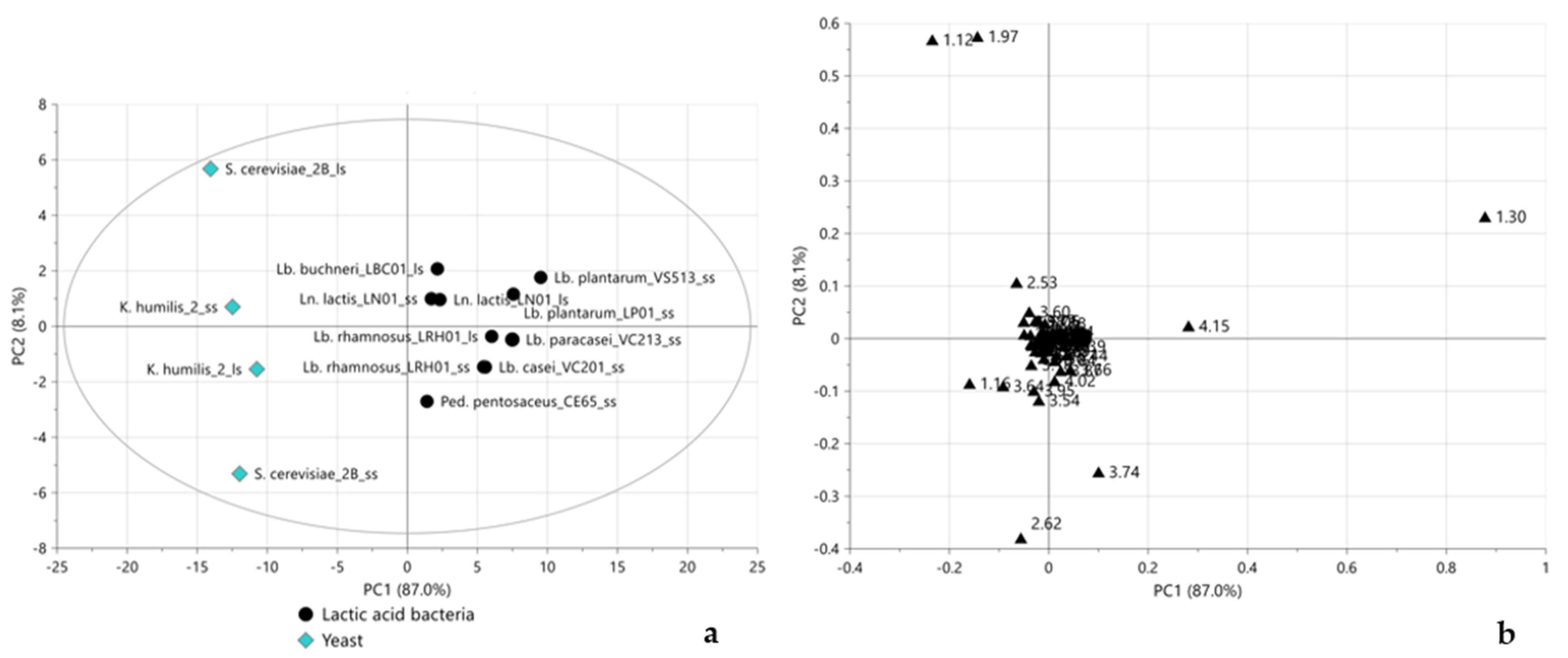
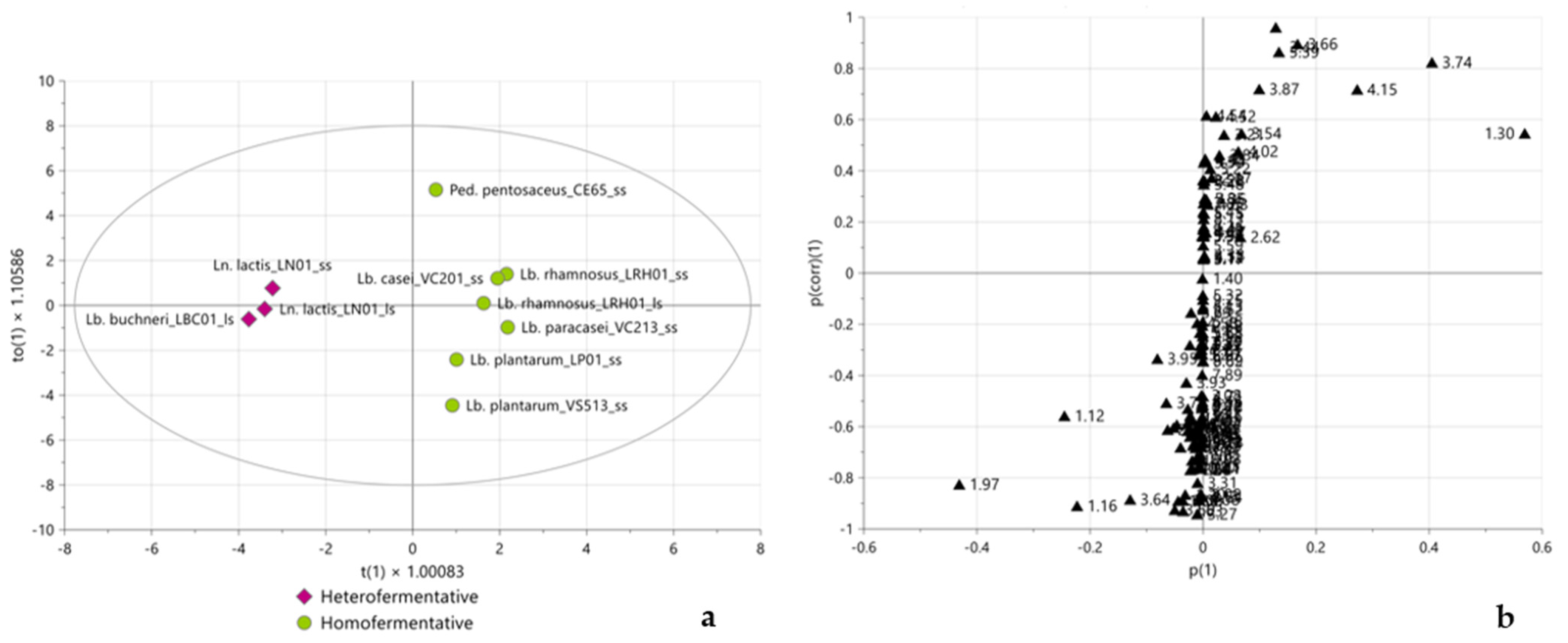
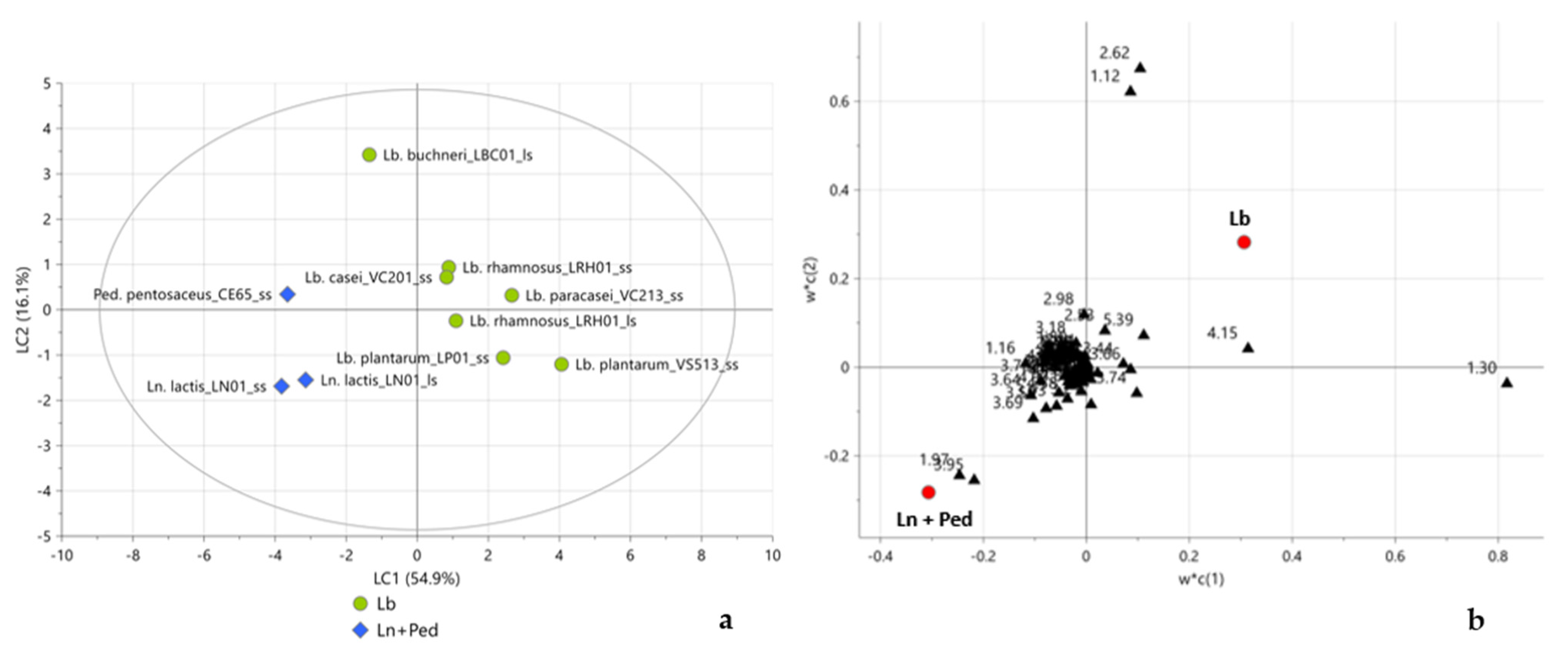
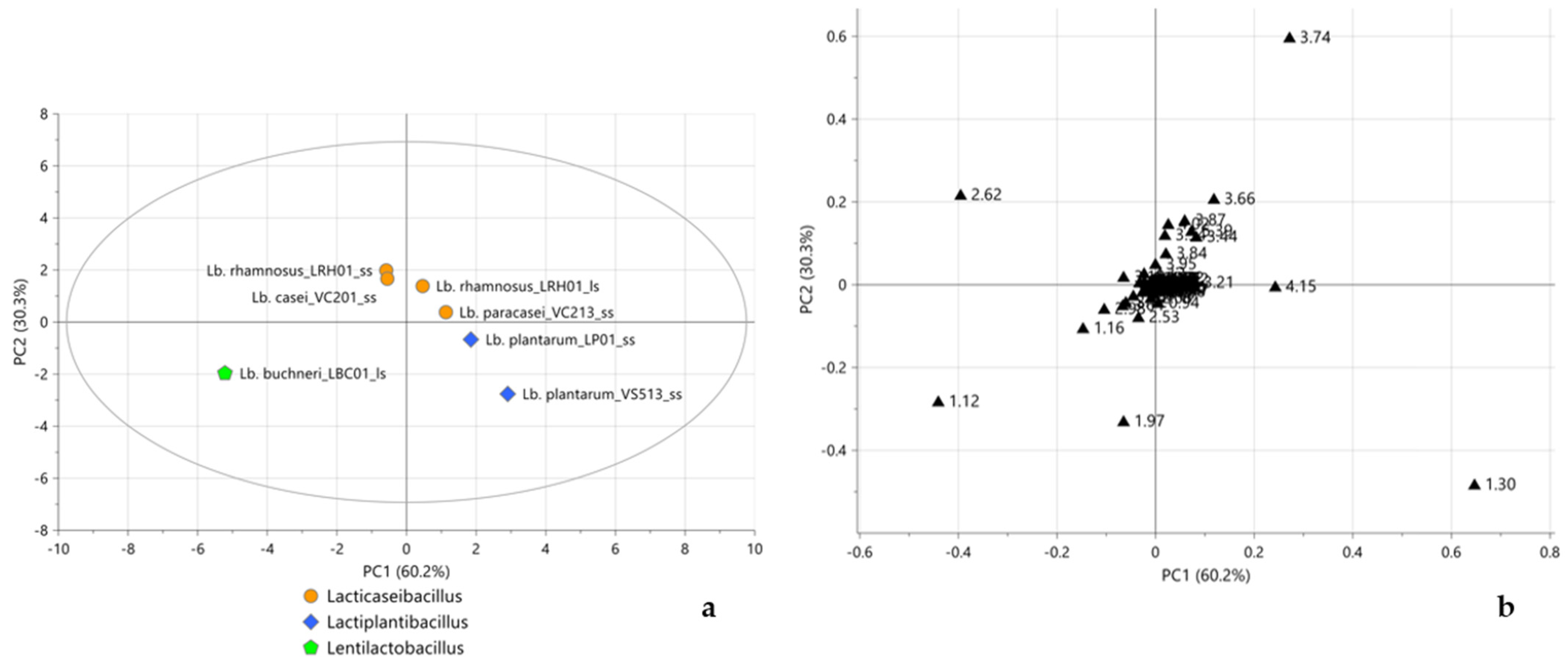
2.3. Multivariate Statistical Analysis
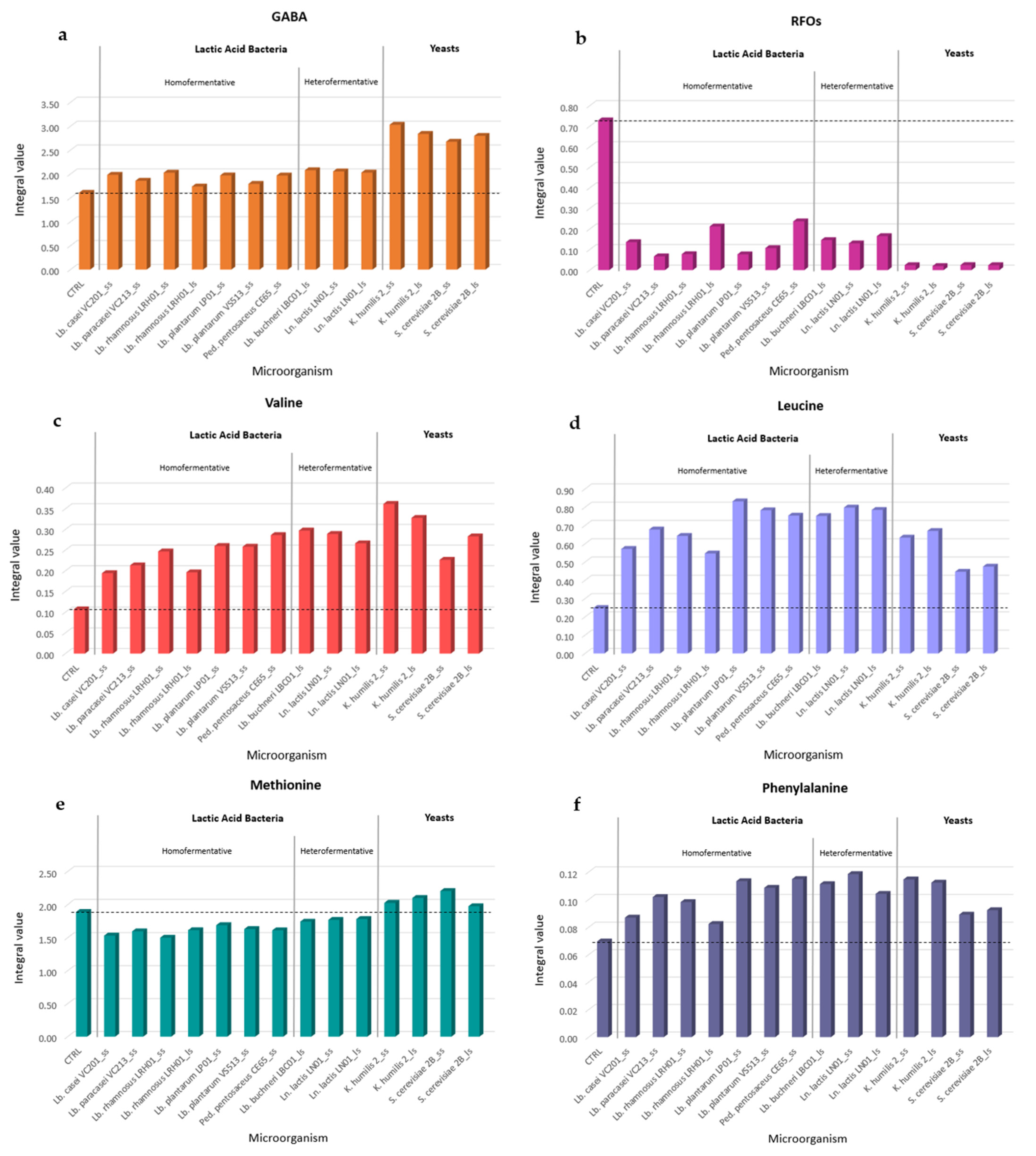
2.4. Essential Amino Acids, GABA, and RFO Analysis
3. Materials and Methods
3.1. Microbial Strains
3.2. Fermentation Protocols
3.2.1. Small-Scale Fermentation Process
3.2.2. Large-Scale Fermentation Process
3.3. NMR Analysis
3.3.1. Sample Preparation
3.3.2. NMR Data Acquisition and Processing
3.4. Statistical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef]
- Rai, A.K.; Jeyaram, K. Role of Yeasts in Food Fermentation. In Yeast Diversity in Human Welfare; Satyanarayana, T., Kunze, G., Eds.; Springer: Singapore, 2017. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating fermented: Health benefits of LAB-fermented foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S. The Role of Yeasts in Fermentation Processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Kandler, O. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 1983, 49, 209–224. [Google Scholar] [CrossRef]
- Orla-Jensen, S. The Lactic Acid Bacteria; Andr Fred Høst and Son: Copenhagen, Denmark, 1919. [Google Scholar]
- Axelsson, L. Lactic acid bacteria: Classification and physiology. In Lactic Acid Bacteria: Microbiology and Functional Aspects, 2nd ed.; Revised and Expanded; Salminen, S., von Wright, A., Eds.; Marcel Dekker: New York, NY, USA, 1998; pp. 1–72. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Mozzi, F.; Ortiz, M.E.; Bleckwedel, J.; De Vuyst, L.; Pescuma, M. Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res. Int. 2013, 54, 1152–1161. [Google Scholar] [CrossRef]
- Gao, Y.; Hou, L.; Gao, J.; Li, D.; Tian, Z.; Fan, B.; Wang, F.; Li, S. Metabolomics approaches for the comprehensive evaluation of fermented foods: A review. Foods 2021, 10, 2294. [Google Scholar] [CrossRef]
- Qu, Q.; Jin, L. Application of nuclear magnetic resonance in food analysis. Food Sci. Tech. Brazil 2022, 42, e43622. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Eggers, N.; Bertram, H.C. NMR-Based Metabolomics of Food. In NMR-Based Metabolomics; Methods in Molecular Biology; Gowda, G., Raftery, D., Eds.; Humana: New York, NY, USA, 2019; Volume 2037, pp. 335–344. [Google Scholar] [CrossRef]
- Piras, C.; Marincola, F.C.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Viale, S.; Pisano, M.B. A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 2013, 141, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Trimigno, A.; Bøge Lyndgaard, C.; Atladóttir, G.A.; Aru, V.; Balling Engelsen, S.; Harder Clemmensen, L.K. An NMR metabolomics approach to investigate factors affecting the yoghurt fermentation process and quality. Metabolites 2020, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-O.; Kim, S.-H.; Cho, S.; Lee, J.H.; Kim, Y.-S.; Yun, S.-S.; Choi, H.-K. Classification of fermented soymilk during fermentation by 1H NMR coupled with principal component analysis and elucidation of free-radical scavenging activities. Biosci. Biotechnol. Biochem. 2009, 73, 1184–1188. [Google Scholar] [CrossRef]
- Qadi, W.S.M.; Mediani, A.; Benchoula, K.; Wong, E.H.; Misnan, N.M.; Sani, N.A. Characterization of physicochemical, biological, and chemical changes associated with coconut milk fermentation and correlation revealed by 1H NMR-based metabolomics. Foods 2023, 12, 1971. [Google Scholar] [CrossRef] [PubMed]
- Tomita, S.; Saito, K.; Nakamura, T.; Sekiyama, Y.; Kikuchi, J. Rapid discrimination of strain-dependent fermentation characteristics among Lactobacillus strains by NMR-based metabolomics of fermented vegetable juice. PLoS ONE 2017, 12, e0182229. [Google Scholar] [CrossRef] [PubMed]
- Markkinen, N.; Pariyani, R.; Jokioja, J.; Kortesniemi, M.; Laaksonen, O.; Yang, B. NMR-based metabolomics approach on optimization of malolactic fermentation of sea buckthorn juice with Lactiplantibacillus plantarum. Food Chem. 2022, 366, 130630. [Google Scholar] [CrossRef]
- Muhialdin, B.J.; Kadum, H.; Hussin, A.S.M. Metabolomics profiling of fermented cantaloupe juice and the potential application to extend the shelf life of fresh cantaloupe juice for six months at 8 °C. Food Control 2021, 120, 107555. [Google Scholar] [CrossRef]
- Colosimo, R.; Gabriele, M.; Cifelli, M.; Longo, V.; Domenici, V.; Pucci, L. The effect of sourdough fermentation on Triticum dicoccum from Garfagnana: 1H NMR characterization and analysis of the antioxidant activity. Food Chem. 2020, 305, 125510. [Google Scholar] [CrossRef]
- Sparvoli, F.; Bollini, R.; Cominelli, E. Nutritional value. In Grain Legumes; Handbook of plant breeding series; De Ron, A., Ed.; Springer: New York, NY, USA; Berlin/Heidelberg, Germany; Dordrecht, The Netherlands; London, UK, 2015; pp. 291–326. [Google Scholar] [CrossRef]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of gamma-aminobutyric acid from lactic acid bacteria: A systematic review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef]
- BMRB—Biological Magnetic Resonance Data Bank. Available online: https://bmrb.io (accessed on 16 June 2023).
- Hou, Y.; Wu, G. Nutritionally essential amino acids. Adv. Nutr. 2018, 9, 849–851. [Google Scholar] [CrossRef]
- Corsetti, A.; Settanni, L. Lactobacilli in sourdough fermentation. Food Res. Int. 2007, 40, 539–558. [Google Scholar] [CrossRef]
- Melini, F.; Melini, V.; Luziatelli, F.; Ficca, A.G.; Ruzzi, M. Health-promoting components in fermented foods: An up-to-date systematic review. Nutrients 2019, 11, 1189. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Galand, S.; Asensio-Grau, A.; Calvo-Lerma, J.; Heredia, A.; Andrés, A. The potential of fermentation on nutritional and technological improvement of cereal and legume flours: A review. Food Res. Int. 2021, 145, 110398. [Google Scholar] [CrossRef] [PubMed]
- Emkani, M.; Oliete, B.; Saurel, R. Effect of lactic acid fermentation on legume protein properties, a review. Fermentation 2022, 8, 244. [Google Scholar] [CrossRef]
- Ji, X.-J.; Huang, H.; Ouyang, P.-K. Microbial 2,3-butanediol production: A state-of-the-art review. Biotechnol. Adv. 2011, 29, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Park, J.M.; Chung, S.C.; Lee, S.Y.; Song, H. Microbial production of 2,3-butanediol for industrial applications. J. Ind. Microbiol. Biotechnol. 2019, 46, 1583–1601. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.C.; Mishra, R.; Mohapatra, S. Microbial citric acid: Production, properties, application, and future perspectives. Food Front. 2021, 2, 62–76. [Google Scholar] [CrossRef]
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int. J. Food Microbiol. 2014, 171, 136–146. [Google Scholar] [CrossRef]
- Curiel, J.A.; Coda, R.; Centomani, I.; Summo, C.; Gobbetti, M.; Rizzello, C.G. Exploitation of the nutritional and functional characteristics of traditional Italian legumes: The potential of sourdough fermentation. Int. J. Food Microbiol. 2015, 196, 51–61. [Google Scholar] [CrossRef]
- Kitum, V.C.; Kinyanjui, P.K.; Mathara, J.M.; Sila, D.N. Effect of Lb. plantarum BFE 5092 fermentation on antinutrient and oligosaccharide composition of whole red haricot bean (Phaseolus vulgaris L.). Int. J. Food Sci. 2020, 2020, 8876394. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Pini, N.; Guerrini, S.; Granchi, L. Exploitation of sourdough lactic acid bacteria to reduce raffinose family oligosaccharides (RFOs) content in breads enriched with chickpea flour. Eur. Food Res. Technol. 2019, 245, 2353–2363. [Google Scholar] [CrossRef]
- Espinosa-Páez, E.; Alanis-Guzmán, M.G.; Hernández-Luna, C.E.; Báez-González, J.G.; Amaya-Guerra, C.A.; Andrés-Grau, A.M. Increasing antioxidant activity and protein digestibility in Phaseolus vulgaris and Avena sativa by fermentation with the Pleurotus ostreatus fungus. Molecules 2017, 22, 2275. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Lee, N.-R.; Lee, C.H. Comprehensive metabolite profiling of four different beans fermented by Aspergillus oryzae. Molecules 2022, 27, 7917. [Google Scholar] [CrossRef]
- Espinosa-Páez, E.; Hernández-Luna, C.E.; Longoria-García, S.; Martínez-Silva, P.A.; Ortiz-Rodríguez, I.; Villarreal-Vera, M.T.; Cantú-Saldaña, C.M. Pleurotus ostreatus: A potential concurrent biotransformation agent/ingredient on development of functional foods (cookies). LWT 2021, 148, 111727. [Google Scholar] [CrossRef]
- Espinosa-Páez, E.; Hernández-Luna, C.E.; Longoria-García, S.; Torres-Alvarez, C.; Velez-Argumedo, C.; González-Martínez, B.E. Improving nutritional and functional quality charateristics in bread by using flours obtained from fermentation of kidney beans and oats with Pleurotus ostreatus. CYTA J. Food 2023, 21, 151–158. [Google Scholar] [CrossRef]
| Sample | Microorganism | Hexose Fermentation | Early Taxonomy Genera | Latest Taxonomy Genera | Scale Small (ss)/Large (ls) | Initial Cell Counting (107 CFU/mL) | Final Cell Counting (109 CFU/g) | pH |
|---|---|---|---|---|---|---|---|---|
| Lacticaseibacillus casei VC201 | LAB | Homofermentative | Lactobacillus | Lacticaseibacillus | ss | 5.1 | 5.8 | 4.20 |
| Lacticaseibacillus paracasei VC213 | LAB | Homofermentative | Lactobacillus | Lacticaseibacillus | ss | 12 | 15.5 | 4.17 |
| Lactiplantibacillus plantarum LP01 | LAB | Homofermentative | Lactobacillus | Lactiplantibacillus | ss | 7.8 | 2.8 | 4.18 |
| Lactiplantibacillus plantarum VS513 | LAB | Homofermentative | Lactobacillus | Lactiplantibacillus | ss | 14.5 | 10.4 | 4.21 |
| Lacticaseibacillus rhamnosus LRH01 | LAB | Homofermentative | Lactobacillus | Lacticaseibacillus | ss/ls | 6/7.8 | 7.4/2.6 | 4.29/4.41 |
| Lentilactobacillus buchneri LBC01 | LAB | Heterofermentative | Lactobacillus | Lentilactobacillus | ss/ls | 0.1/1.2 | 0.12/2.7 | 4.83/4.23 |
| Leuconostoc lactis LN01 | LAB | Heterofermentative | Leuconostoc | Leuconostoc | ss/ls | 2.1/1.3 | 4.1/7.6 | 4.22/4.48 |
| Pediococcus pentosaceus CE65 | LAB | Homofermentative | Pediococcus | Pediococcus | ss | 7.2 | 4.6 | 4.55 |
| Weissella confusa WS01 | LAB | Heterofermentative | Weissella | Weissella | ss | 1 | 3.3 | 5.14 |
| Kazachstania humilis 2 | Yeast | - | - | - | ss/ls | 8.6/3.8 | 0.40/0.25 | 6.34/6.04 |
| Saccharomyces cerevisiae 2B | Yeast | - | - | - | ss/ls | 1.7/2.6 | 0.11/0.18 | 6.35/6.18 |
| Peak | Compound | Assignment | ppm |
|---|---|---|---|
| 1 | 2,3-Butanediol | CH3 | 1.14 |
| 2 | Acetate | CH3 | 2.01 |
| 3 | Acetoacetate | CH3 | 2.54 |
| 4 | Acetoin | CH3 | 2.23, 1.38 |
| 5 | Adenine | aromatics | 8.24, 8.21 |
| 6 | Alanine | βCH3 | 1.48 |
| 7 | Asparagine | β/β′CH2 | 2.96, 2.89 |
| 8 | Citrate | CH2 | 2.81, 2.69 |
| 9 | Ethanol | CH2 | 3.65 |
| CH3 | 1.18 | ||
| 10 | Formate | HCOO | 8.43 |
| 11 | GABA | all CH2 | 3.00, 2.38, 1.94 |
| 12 | Galactose | αH1 | 5.27 |
| βH1 | 4.60 | ||
| 13 | Glucose | αH1 | 5.25 |
| βH1 | 4.66 | ||
| 14 | Isoleucine | γCH3 | 1.01 |
| 15 | Lactate | CH | 4.20 |
| CH3 | 1.35 | ||
| 16 | Leucine | βCH2 | 1.70 |
| δ/δ′CH3 | 0.96 | ||
| 17 | Methionine | S-CH3 | 2.13 |
| 18 | Phenylalanine | aromatics | 7.32–7.44 |
| β/β′CH2 | 3.22, 3.25 | ||
| 19 | Pipecolic acid | all CH2 | 3.57, 3.41, 3.00, 2.21, 1.87, 1.63 |
| 20 | Raffinose | H1α Glu | 5.44 |
| 21 | Stachyose | H1α Glu | 5.43 |
| 22 | Sucrose | H1α Glu | 5.41 |
| 23 | Trigonelline | aromatics | 9.12, 8.84, 8.09 |
| N-CH3 | 4.44 | ||
| 24 | Tyramine | aromatics | 7.20, 6.90 |
| 25 | Tyrosine | aromatics | 7.18, 6.87 |
| 26 | Uracil | aromatics | 7.55, 5.81 |
| 27 | Uridine | aromatics | 7.86, 5.92 |
| 28 | Valine | αCH | 3.61 |
| γ, γ′CH3 | 0.99, 1.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatulli, G.; Cagliani, L.R.; Sparvoli, F.; Brasca, M.; Consonni, R. NMR-Based Metabolomic Study on Phaseolus vulgaris Flour Fermented by Lactic Acid Bacteria and Yeasts. Molecules 2023, 28, 4864. https://doi.org/10.3390/molecules28124864
Tatulli G, Cagliani LR, Sparvoli F, Brasca M, Consonni R. NMR-Based Metabolomic Study on Phaseolus vulgaris Flour Fermented by Lactic Acid Bacteria and Yeasts. Molecules. 2023; 28(12):4864. https://doi.org/10.3390/molecules28124864
Chicago/Turabian StyleTatulli, Giuseppina, Laura Ruth Cagliani, Francesca Sparvoli, Milena Brasca, and Roberto Consonni. 2023. "NMR-Based Metabolomic Study on Phaseolus vulgaris Flour Fermented by Lactic Acid Bacteria and Yeasts" Molecules 28, no. 12: 4864. https://doi.org/10.3390/molecules28124864
APA StyleTatulli, G., Cagliani, L. R., Sparvoli, F., Brasca, M., & Consonni, R. (2023). NMR-Based Metabolomic Study on Phaseolus vulgaris Flour Fermented by Lactic Acid Bacteria and Yeasts. Molecules, 28(12), 4864. https://doi.org/10.3390/molecules28124864









